微生物组生物合成基因簇发掘方法及应用前景
|
|
赖奇龙, 姚帅, 查毓国, 白虹, 宁康
|
Microbiome-based biosynthetic gene cluster data mining techniques and application potentials
|
|
LAI Qilong, YAO Shuai, ZHA Yuguo, BAI Hong, NING Kang
|
|
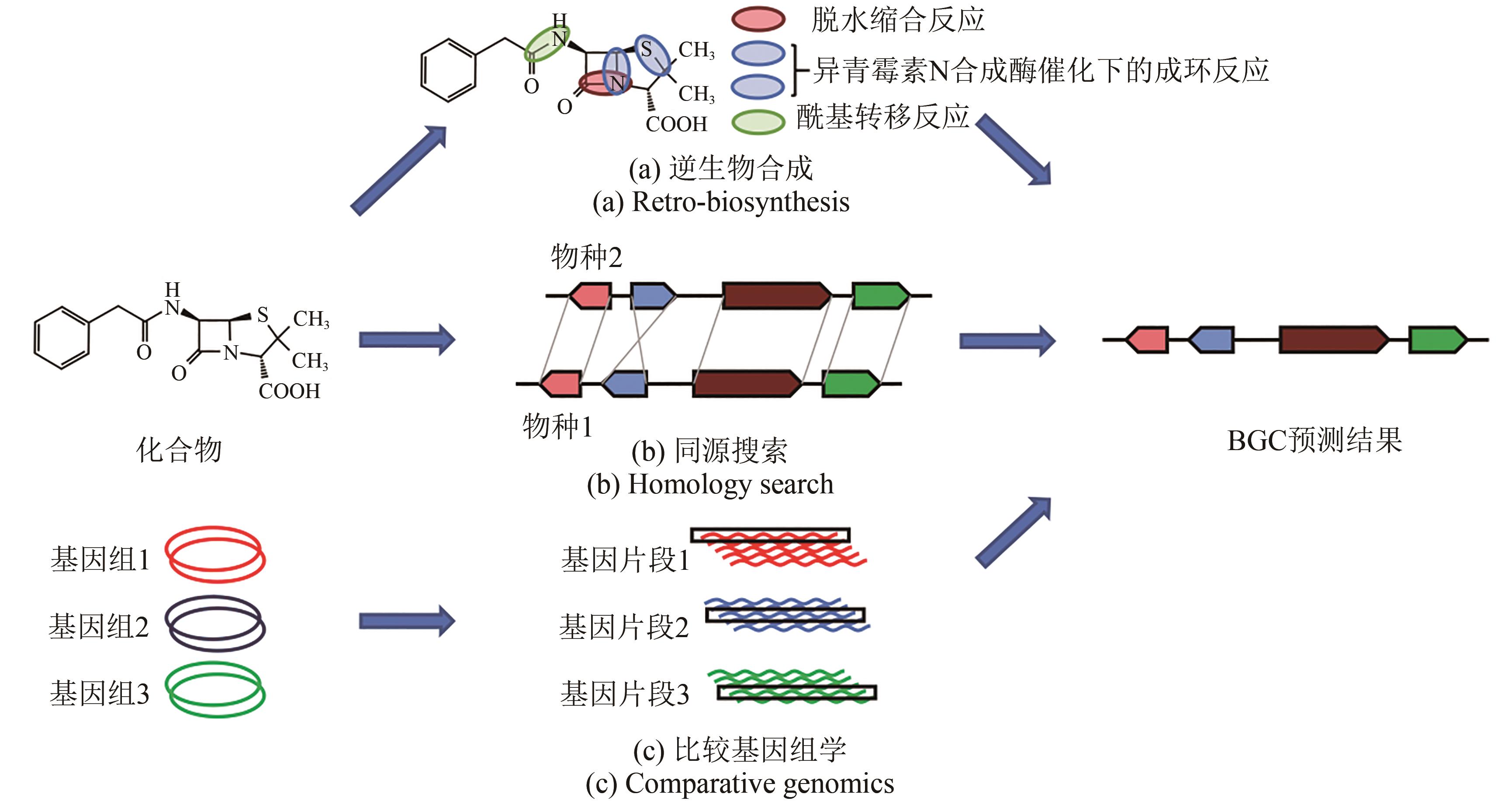
图4 建立BGC和次生代谢产物关联性的分析方法[58]
(a)逆生物合成:从已知化合物开始,预测生产该化合物所需的活性酶(主干酶和裁剪酶),并从这些预测中找到与基因组中需求匹配的假定簇。本图中选用的案例为青霉素G[59]。(b)同源搜索:从物种1产生的已知化合物和物种2产生的相同或相似的化合物开始,使用来自物种2的已知基因集群在物种1的基因组中搜索相似的基因集群,从而确定感兴趣的基因集群。(c)比较基因组学:从一组生物开始,其中一些生物产生目标化合物,而另一些生物则不产生,有可能在生产中识别同源基因簇,并在非生产中没有同源基因的基础上进行筛选,从而识别候选基因簇
|
Fig. 4 Analytical methods for establishing correlation between BGC and the production of secondary metabolites[58]
(a) Retro-biosynthesis: starting with a known compound but no related gene clusters identified, it is possible for predicting enzyme(s) to catalyze the synthesis of such a compound (backbone and tailoring enzymes), and with these predictions putative gene clusters matching the requirements can be found in the genome. The selected case in this figure is penicillin G[59]. (b) Homology searching: starting with a known compound produced by organism 1 and the same or similar compound produced by organism 2 with gene cluster identified, it is possible to use the known gene cluster from organism 2 to search for a similar gene cluster in the genome of organism 1, and thereby identify the gene cluster of interest. (c) Comparative genomics: starting with a group of organisms, some of which produce compounds of interest and some of which do not, it is possible to identify homologous gene clusters in the species that produce them and to screen on the basis of the absence of homologous genes in the species that does not produce them, thereby identifying candidate gene clusters.
|
|
|
|
|
