合成生物学 ›› 2022, Vol. 3 ›› Issue (1): 35-52.DOI: 10.12211/2096-8280.2021-097
合成生物学在肠道微生态疗法研发中的应用
高梦学1, 王丽娜1, 黄鹤1,2
- 1.天津大学化工学院,教育部合成生物学前沿科学中心,系统生物工程教育部重点实验室,天津 300072
2.天津大学浙江绍兴研究院,浙江 绍兴 312000
-
收稿日期:2021-10-21修回日期:2021-12-08出版日期:2022-02-28发布日期:2022-03-14 -
通讯作者:黄鹤 -
作者简介:高梦学 (1996—),女,博士研究生。研究方向为产维生素类营养物质活菌制剂的制备及应用。E-mail:2018207213@tju.edu.cn黄鹤 (1971—),女,教授,博士生导师。研究方向为合成生物肠道菌(群)构建及应用、纳米抗体等功能生物大分子的理性设计及高效制备、耐药致病微生物代谢机理与应对策略等。E-mail:huang@tju.edu.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2019YFA09005600)
Advances in synthetic biology assisted intestinal microecological therapy
GAO Mengxue1, WANG Lina1, HUANG He1,2
- 1.Frontiers Science Center for Synthetic Biology,Key Laboratory of Systems Bioengineering (Ministry of Education),School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
2.Shaoxing Research Institute Tianjin University,Shaoxing 312000,Zhejiang,China
-
Received:2021-10-21Revised:2021-12-08Online:2022-02-28Published:2022-03-14 -
Contact:HUANG He
摘要:
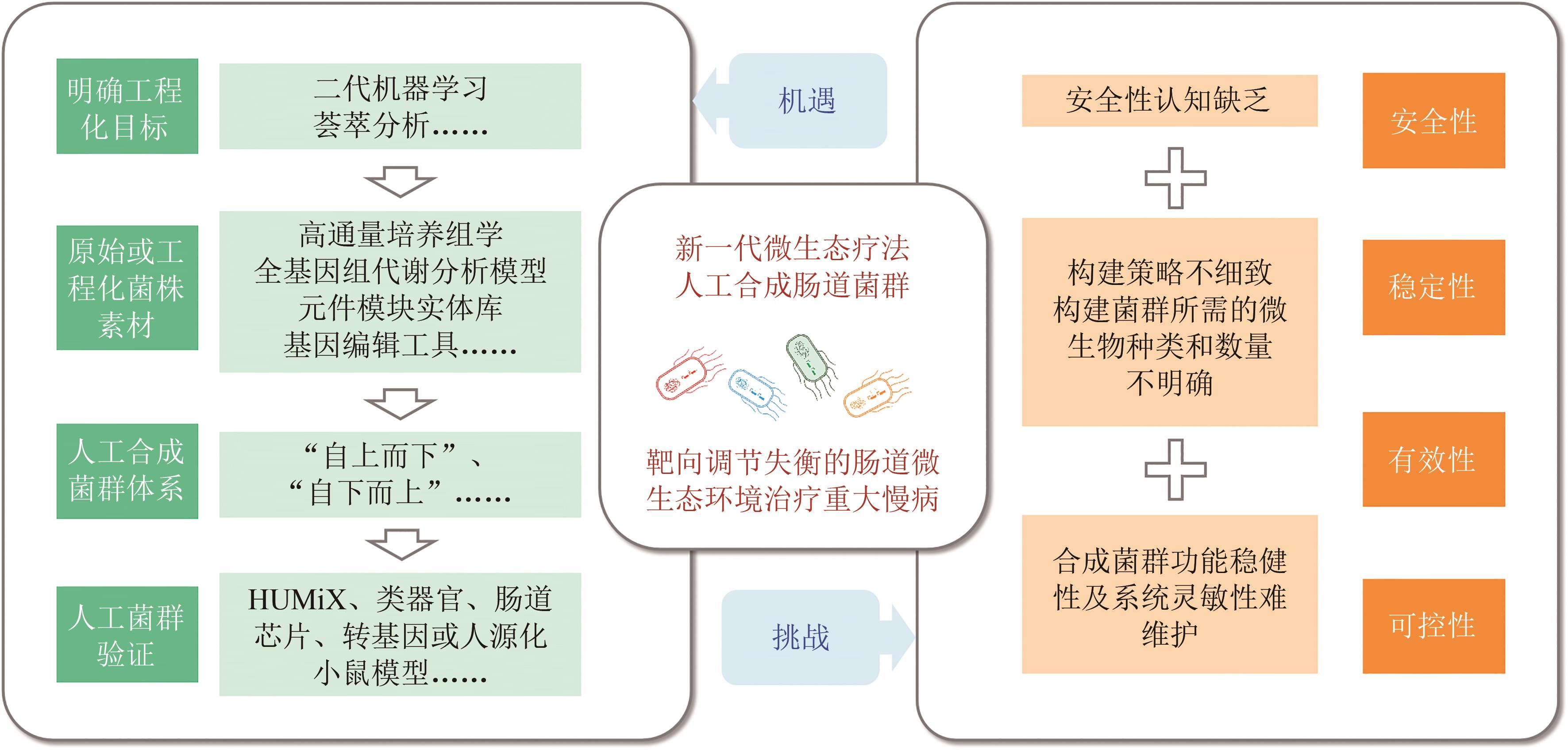
肠道菌群是人体的重要“器官”。工业化世界的发展加速了对肠道菌群研究从“传统结构”向“工业结构”的转变,而纠正肠道微生态失衡已成为解决重大慢性非传染性疾病传播难题的核心策略之一。然而,目前开发的靶向调节肠道菌群结构与功能的传统微生态疗法,如益生元疗法、益生菌疗法和粪菌移植疗法,只有少数被用于临床多发难治性重大慢病的防治,且出现了可控性差、菌群遗传背景不清晰等安全问题。合成生物学技术手段的迭代发展推动了新型微生态药物的研发,成为对重大慢病进行精准识别和精准施策的关键。本文首先以消化系统疾病、代谢性疾病和精神疾病等重大慢性疾病的干预和治疗为切入点,回顾了基于合成生物学方法设计构建工程益生菌的研究进展,并对以工程益生菌为核心的微生态疗法在上述重大慢病中的应用进行了综述。同时,考虑到单一工程益生菌的负荷和抗干扰能力等问题,本文还提出了利用工程益生菌构建人工合成肠道菌群开发新一代微生态疗法的设想,分析了这一过程所面临的机遇与挑战,旨在为工程益生菌和人工合成肠道群落基础研究和临床应用的双向转化提供借鉴,从而推动新型微生态药物的研发。
中图分类号:
引用本文
高梦学, 王丽娜, 黄鹤. 合成生物学在肠道微生态疗法研发中的应用[J]. 合成生物学, 2022, 3(1): 35-52.
GAO Mengxue, WANG Lina, HUANG He. Advances in synthetic biology assisted intestinal microecological therapy[J]. Synthetic Biology Journal, 2022, 3(1): 35-52.
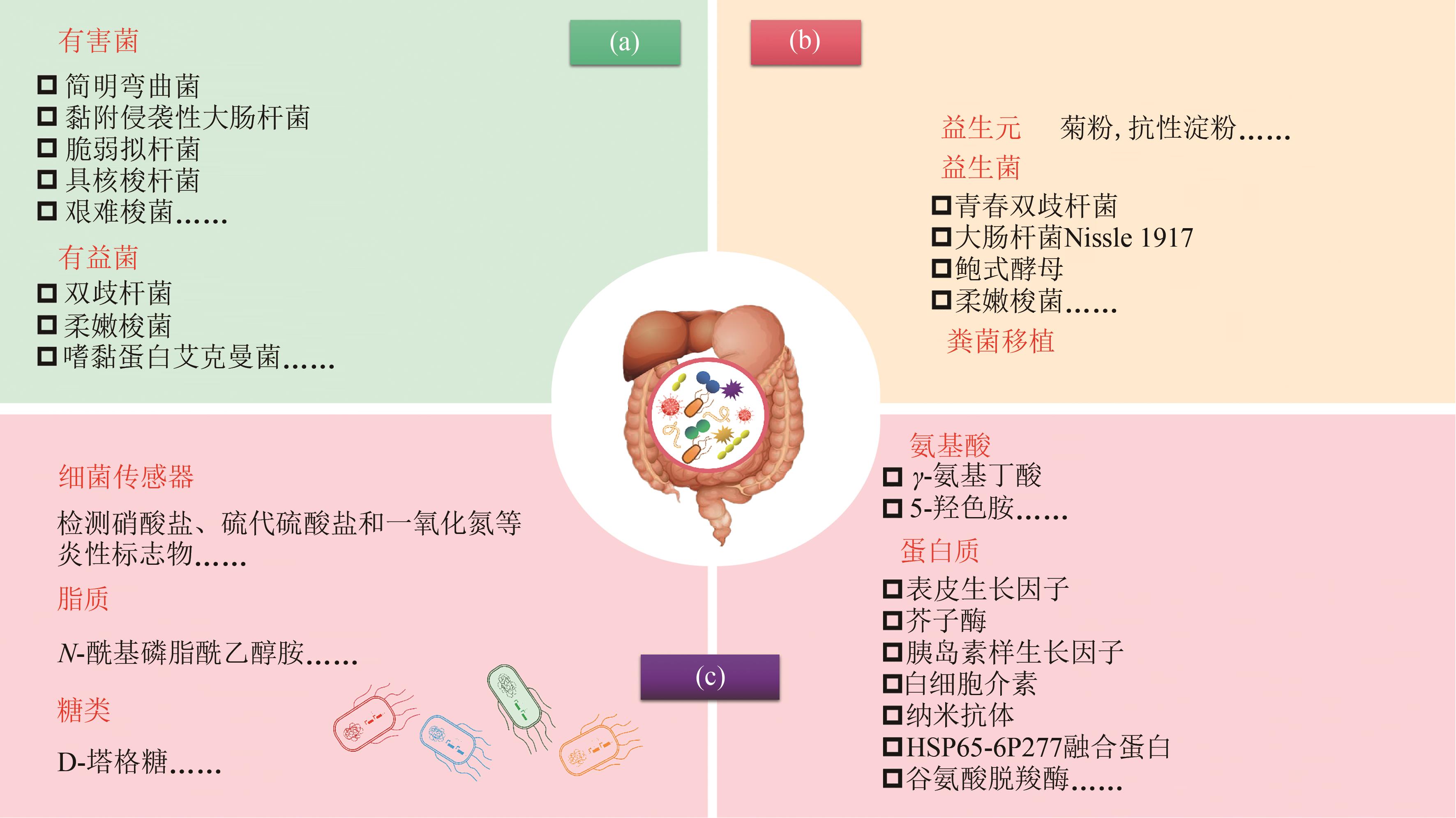
图1 肠道微生态疗法(a)肠道菌群失衡;(b)常见微生态疗法;(c)工程益生菌疗法
Fig. 1 Gut microbiome therapeutics(a) Gut microbiota dysbiosis; (b) Traditional modalities to manipulate microbiome; (c) Engineered probiotics
| 底盘细胞 | 治疗策略 | 遗传操作 | 疾病 类型 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 Nissle 1917 | 利用ThsSR、TtrSR双组分调控系统分别构建细菌传感器,以响应硫代硫酸盐等炎性标志物 | pKD226/pKD227/pKD233.7-3/pKD239-1g-2/pKD238-1a/pKD184/pKD182/pKD237-3a-2/pKD236-4b等 | IBD (诊断) | [ |
| 大肠杆菌 Nissle 1917 | 利用NarXL、TtrSR双组分调控系统构建细菌传感器,用于同时检测硫代硫酸盐和硝酸盐等炎性标志物 | pSEVA141-J23114-NPG/pSEVA131-J23106-NPG/pSEVA131-AND/pSEVA131-J23113-TPG/pSEVA131-J23113-N(N509D)PG等 | IBD (诊断) | [ |
| 大肠杆菌 Nissle 1917 | 利用TtrSR双组分调控系统构建细菌传感器,与具有信号读取、信息传输功能的微电子设备整合,发明了一种可植入胃肠道并直接检测硫代硫酸盐等炎性标志物的微型设备 | pZE2-hrtR-RBS3-chuA/pKD236-4b/pKD237-3a-3-Lux/pZE1-LuxR-Plux-luxCDABE等 | IBD (诊断) | [ |
| 乳酸乳球菌MG1363 | 由食品级益生菌原位合成治疗剂mIL-10 | pT1MIL10 | IBD (治疗) | [ |
| 乳酸乳球菌MG1363 | 在结肠黏膜处局部递送抗mTNF纳米抗体 | pMT1/pMT1–MT1 | IBD (治疗) | [ |
| 酵母菌BS004 | 基于CRISPR-Cas9编辑技术将传感(P2Y2)和响应(RROP1)元件共同引入酵母菌株基因组中,根据肠道内促炎代谢物三磷酸腺苷(eATP)水平启动三磷酸腺苷双磷酸酶的表达和分泌并产生抑制炎症的腺苷 | mfa2::HIS3-pTDH3 RROP1/mfa2::HIS3-pFUS1 RROP1等 | IBD (诊疗) | [ |
| 大肠杆菌 Nissle 1917 | 在患有结肠炎的小鼠肠腔中可持续生酮体3-羟基丁酸(3-HB) | 敲除AdhE和ldhA基因,在malEK、rhtCB或yicS/nepI位点上整合3HB生物合成途径涉及的酶基因(乙酰辅酶A乙酰基转移酶、3HB-CoA脱氢酶和硫酯酶) | IBD (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 受近红外光和pH同时调控的光诊疗纳米系统 | pDawn-mil-10 | IBD (诊疗) | [ |
| 大肠杆菌 Nissle 1917 | 肠内原位分泌三叶因子在较短时间内修复肠黏膜 | 在csgBACDEFG位点上引入CmR基因,分别导入pBbB8k-CsgA-TFF1、pBbB8k-CsgA-TFF2、pBbB8k-CsgA-TFF3、pBbB8kN22-TFF3质粒 | IBD (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 肠内原位分泌表皮生长因子在较短时间内修复肠黏膜 | 在OmpC基因处插入基因串EGF-LARD3-prtDEF | IBD (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 与CRC细胞特异性结合,将十字花科蔬菜的天然成分转化为具有抗癌活性的萝卜硫烷 | peAlrt-J23105-YebF-I1-J23108-INP-HlpA | CRC (预防) | [ |
| 戊糖片球菌SL4 | 分泌抗癌蛋白P8,抑制癌细胞的增殖 | pCBT24-2-PK-p8-PK-p8 | CRC (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 利用葡萄糖生产5-氨基乙酰丙酸(5-ALA),结合光动力疗法治疗CRC | pCL1920-gltX、pCL1920-hemA、pCL1920-hemL | CRC (治疗) | [ |
表1 靶向消化系统疾病的微生态疗法
Tab. 1 Intestinal microecological treatment for digestive system diseases
| 底盘细胞 | 治疗策略 | 遗传操作 | 疾病 类型 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 Nissle 1917 | 利用ThsSR、TtrSR双组分调控系统分别构建细菌传感器,以响应硫代硫酸盐等炎性标志物 | pKD226/pKD227/pKD233.7-3/pKD239-1g-2/pKD238-1a/pKD184/pKD182/pKD237-3a-2/pKD236-4b等 | IBD (诊断) | [ |
| 大肠杆菌 Nissle 1917 | 利用NarXL、TtrSR双组分调控系统构建细菌传感器,用于同时检测硫代硫酸盐和硝酸盐等炎性标志物 | pSEVA141-J23114-NPG/pSEVA131-J23106-NPG/pSEVA131-AND/pSEVA131-J23113-TPG/pSEVA131-J23113-N(N509D)PG等 | IBD (诊断) | [ |
| 大肠杆菌 Nissle 1917 | 利用TtrSR双组分调控系统构建细菌传感器,与具有信号读取、信息传输功能的微电子设备整合,发明了一种可植入胃肠道并直接检测硫代硫酸盐等炎性标志物的微型设备 | pZE2-hrtR-RBS3-chuA/pKD236-4b/pKD237-3a-3-Lux/pZE1-LuxR-Plux-luxCDABE等 | IBD (诊断) | [ |
| 乳酸乳球菌MG1363 | 由食品级益生菌原位合成治疗剂mIL-10 | pT1MIL10 | IBD (治疗) | [ |
| 乳酸乳球菌MG1363 | 在结肠黏膜处局部递送抗mTNF纳米抗体 | pMT1/pMT1–MT1 | IBD (治疗) | [ |
| 酵母菌BS004 | 基于CRISPR-Cas9编辑技术将传感(P2Y2)和响应(RROP1)元件共同引入酵母菌株基因组中,根据肠道内促炎代谢物三磷酸腺苷(eATP)水平启动三磷酸腺苷双磷酸酶的表达和分泌并产生抑制炎症的腺苷 | mfa2::HIS3-pTDH3 RROP1/mfa2::HIS3-pFUS1 RROP1等 | IBD (诊疗) | [ |
| 大肠杆菌 Nissle 1917 | 在患有结肠炎的小鼠肠腔中可持续生酮体3-羟基丁酸(3-HB) | 敲除AdhE和ldhA基因,在malEK、rhtCB或yicS/nepI位点上整合3HB生物合成途径涉及的酶基因(乙酰辅酶A乙酰基转移酶、3HB-CoA脱氢酶和硫酯酶) | IBD (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 受近红外光和pH同时调控的光诊疗纳米系统 | pDawn-mil-10 | IBD (诊疗) | [ |
| 大肠杆菌 Nissle 1917 | 肠内原位分泌三叶因子在较短时间内修复肠黏膜 | 在csgBACDEFG位点上引入CmR基因,分别导入pBbB8k-CsgA-TFF1、pBbB8k-CsgA-TFF2、pBbB8k-CsgA-TFF3、pBbB8kN22-TFF3质粒 | IBD (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 肠内原位分泌表皮生长因子在较短时间内修复肠黏膜 | 在OmpC基因处插入基因串EGF-LARD3-prtDEF | IBD (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 与CRC细胞特异性结合,将十字花科蔬菜的天然成分转化为具有抗癌活性的萝卜硫烷 | peAlrt-J23105-YebF-I1-J23108-INP-HlpA | CRC (预防) | [ |
| 戊糖片球菌SL4 | 分泌抗癌蛋白P8,抑制癌细胞的增殖 | pCBT24-2-PK-p8-PK-p8 | CRC (治疗) | [ |
| 大肠杆菌 Nissle 1917 | 利用葡萄糖生产5-氨基乙酰丙酸(5-ALA),结合光动力疗法治疗CRC | pCL1920-gltX、pCL1920-hemA、pCL1920-hemL | CRC (治疗) | [ |
| 底盘细胞 | 治疗策略 | 遗传操作 | 疾病类型 | 参考文献 |
|---|---|---|---|---|
| 乳酸乳球菌 MG1363 | 构建分泌GLP-1的基因工程菌改善肥胖,通过促进肥胖小鼠的脂肪酸氧化、增加肠道微生物多样性改善肥胖状况 | pMG36e-GLP-1 | 肥胖 | [ |
| 大肠杆菌 Nissle 1917 | 对GLP-1的序列进行改进,形成新的GLP-1GM序列,并利用益生菌进行表达分泌 | pGEX-GLP-1 GM | 肥胖 | [ |
| 长双歧杆菌NCC2705 | 建立了一种新型的双歧杆菌口服给药系统,用于降低超重小鼠的食物摄入量、体重和血脂水平 | pBBADs-OXM | 肥胖 | [ |
| 乳酸乳球菌 NZ9000 | 通过基因工程改造乳酸乳球菌,以生产羧化或低羧化的骨钙素,以用于糖尿病和肥胖症的治疗研究 | pNZ8148#2:SEC-OC | 糖尿病和肥胖 | [ |
| 乳酸乳球菌 NZ3900 | 使用乳酸乳球菌表达成纤维细胞生长因子21,改善肥胖小鼠的疾病状态 | PNZ8149-Human FGF21 | 肥胖 | [ |
| 大肠杆菌 Nissle 1917 | 口服表达NAPE的工程化大肠杆菌Nissle 1917可降低高脂肪饮食诱导的小鼠的肥胖水平 | pNAPE | 肥胖 | [ |
| 枯草芽孢杆菌 SCK6 | 通过构建高产丁酸的工程菌,调节与肥胖相关的菌群及代谢物水平,发展减轻肥胖的治疗策略 | 敲除skfA、sdpC、acdA和ackA等基因,敲入丁酰CoA:乙酸CoA转移酶编码基因 | 肥胖 | [ |
乳酸乳球菌MG1363 乳酸乳球菌sAGX0037 | 开发一种基于乳酸乳球菌的致耐受性细菌递送技术,用于控制肠道中自身抗原GAD65370-575和抗炎细胞因子白细胞介素10的分泌 | pGAD65370-575 | 1型糖尿病 | [ |
乳酸乳球菌MG1363 乳酸乳球菌sAGX0037 | 通过分泌完整的胰岛素原自身抗原和免疫调节细胞因子IL-10,开发一种使用黏膜递送恢复耐受性的治疗策略 | pThyAhPINS | 1型糖尿病 | [ |
乳酸乳球菌pCYT:SNase 乳酸乳球菌 NZ9000 | 递送葡萄球菌核酸酶有效预防1型糖尿病,改善炎症症状,并有助于调节免疫平衡 | pCYT:SNase | 1型糖尿病 (预防) | [ |
| 乳酸乳球菌 NZ9000 | 组成型或通过乳链菌肽诱导表达HSP65-6P277,可预防高血糖、改善葡萄糖耐量和减少胰岛炎 | pCYT:HSP65-6P277 pHJ:HSP65-6P277 | 1型糖尿病 (预防) | [ |
| 乳酸乳球菌 | 开发胰高血糖素样肽-1的活体口服给药系统,用于治疗2型糖尿病 | pUBGLP-1 | 2型糖尿病 | [ |
| 加氏乳杆菌 ATCC 33323 | 分泌胰高血糖素样肽-1将肠细胞重编程为葡萄糖反应性胰岛素分泌细胞来改善糖尿病大鼠的高血糖水平 | pFD-GLPL/pBluescript-GLPL/pORI28-LGLPL | 2型糖尿病 | [ |
| 乳酸乳球菌 NZ9000 | 乳酸乳球菌分泌胰高血糖素样蛋白-1受体激动剂,增强葡萄糖依赖性胰岛素分泌、激活PI3-K/AKT信号通路 | pNZ8048-rExd4 | 2型糖尿病 | [ |
| 乳酸乳球菌 NZ9000 | 分泌L-阿拉伯糖异构酶的乳酸乳球菌菌株具有在体内产生D-塔格糖的能力,并赋予小鼠抗高血糖作用 | pSEC:LEISS:araA | 糖尿病 | [ |
表2 靶向代谢性疾病的微生态疗法
Tab. 2 Intestinal microecological treatment for metabolic diseases
| 底盘细胞 | 治疗策略 | 遗传操作 | 疾病类型 | 参考文献 |
|---|---|---|---|---|
| 乳酸乳球菌 MG1363 | 构建分泌GLP-1的基因工程菌改善肥胖,通过促进肥胖小鼠的脂肪酸氧化、增加肠道微生物多样性改善肥胖状况 | pMG36e-GLP-1 | 肥胖 | [ |
| 大肠杆菌 Nissle 1917 | 对GLP-1的序列进行改进,形成新的GLP-1GM序列,并利用益生菌进行表达分泌 | pGEX-GLP-1 GM | 肥胖 | [ |
| 长双歧杆菌NCC2705 | 建立了一种新型的双歧杆菌口服给药系统,用于降低超重小鼠的食物摄入量、体重和血脂水平 | pBBADs-OXM | 肥胖 | [ |
| 乳酸乳球菌 NZ9000 | 通过基因工程改造乳酸乳球菌,以生产羧化或低羧化的骨钙素,以用于糖尿病和肥胖症的治疗研究 | pNZ8148#2:SEC-OC | 糖尿病和肥胖 | [ |
| 乳酸乳球菌 NZ3900 | 使用乳酸乳球菌表达成纤维细胞生长因子21,改善肥胖小鼠的疾病状态 | PNZ8149-Human FGF21 | 肥胖 | [ |
| 大肠杆菌 Nissle 1917 | 口服表达NAPE的工程化大肠杆菌Nissle 1917可降低高脂肪饮食诱导的小鼠的肥胖水平 | pNAPE | 肥胖 | [ |
| 枯草芽孢杆菌 SCK6 | 通过构建高产丁酸的工程菌,调节与肥胖相关的菌群及代谢物水平,发展减轻肥胖的治疗策略 | 敲除skfA、sdpC、acdA和ackA等基因,敲入丁酰CoA:乙酸CoA转移酶编码基因 | 肥胖 | [ |
乳酸乳球菌MG1363 乳酸乳球菌sAGX0037 | 开发一种基于乳酸乳球菌的致耐受性细菌递送技术,用于控制肠道中自身抗原GAD65370-575和抗炎细胞因子白细胞介素10的分泌 | pGAD65370-575 | 1型糖尿病 | [ |
乳酸乳球菌MG1363 乳酸乳球菌sAGX0037 | 通过分泌完整的胰岛素原自身抗原和免疫调节细胞因子IL-10,开发一种使用黏膜递送恢复耐受性的治疗策略 | pThyAhPINS | 1型糖尿病 | [ |
乳酸乳球菌pCYT:SNase 乳酸乳球菌 NZ9000 | 递送葡萄球菌核酸酶有效预防1型糖尿病,改善炎症症状,并有助于调节免疫平衡 | pCYT:SNase | 1型糖尿病 (预防) | [ |
| 乳酸乳球菌 NZ9000 | 组成型或通过乳链菌肽诱导表达HSP65-6P277,可预防高血糖、改善葡萄糖耐量和减少胰岛炎 | pCYT:HSP65-6P277 pHJ:HSP65-6P277 | 1型糖尿病 (预防) | [ |
| 乳酸乳球菌 | 开发胰高血糖素样肽-1的活体口服给药系统,用于治疗2型糖尿病 | pUBGLP-1 | 2型糖尿病 | [ |
| 加氏乳杆菌 ATCC 33323 | 分泌胰高血糖素样肽-1将肠细胞重编程为葡萄糖反应性胰岛素分泌细胞来改善糖尿病大鼠的高血糖水平 | pFD-GLPL/pBluescript-GLPL/pORI28-LGLPL | 2型糖尿病 | [ |
| 乳酸乳球菌 NZ9000 | 乳酸乳球菌分泌胰高血糖素样蛋白-1受体激动剂,增强葡萄糖依赖性胰岛素分泌、激活PI3-K/AKT信号通路 | pNZ8048-rExd4 | 2型糖尿病 | [ |
| 乳酸乳球菌 NZ9000 | 分泌L-阿拉伯糖异构酶的乳酸乳球菌菌株具有在体内产生D-塔格糖的能力,并赋予小鼠抗高血糖作用 | pSEC:LEISS:araA | 糖尿病 | [ |
| 底盘细胞 | 治疗策略 | 遗传操作 | 疾病 类型 | 参考文献 |
|---|---|---|---|---|
| 短乳杆菌CGMCC1306 | 构建高产γ-氨基丁酸的细胞工厂,有利于开发工程化活菌制剂用于精神疾病的治疗 | pMG36e-gadA/pMG36e-gadB/pMG36e-gadC/pMG36e-gadCA/pMG36e-gadCB | 抑郁症 | [ |
| 乳酸乳球菌乳脂 亚种MG1363 | 递送胰高血糖素样蛋白-1恢复LPS引起的小鼠空间学习和记忆障碍,治疗阿尔兹海默症 | pMG36e-GLP- 1 | 阿尔兹海默症 | [ |
| 枯草芽孢杆菌60015/YF256 | 操纵肌醇代谢构建工程菌,以允许从肌醇到鲨肌醇的生物转化,作为细胞工厂提供鲨肌醇,是治疗阿尔兹海默症的潜在的活菌生物制剂 | iolE41 iolI::spc/iolR::cat/iolW::pMutin2(erm::tet)/iolX::pMutin4(erm) | 阿尔兹海默症 | [ |
| 乳酸乳球菌乳脂 亚种MG1363 | 递送p62蛋白减少氧化和炎症标志物、调节肠道菌群结构,对阿尔兹海默症产生积极影响 | pExu:p62 | 阿尔兹海默症 | [ |
表3 靶向精神疾病的微生态疗法
Tab. 3 Intestinal microecological treatment for mental illness
| 底盘细胞 | 治疗策略 | 遗传操作 | 疾病 类型 | 参考文献 |
|---|---|---|---|---|
| 短乳杆菌CGMCC1306 | 构建高产γ-氨基丁酸的细胞工厂,有利于开发工程化活菌制剂用于精神疾病的治疗 | pMG36e-gadA/pMG36e-gadB/pMG36e-gadC/pMG36e-gadCA/pMG36e-gadCB | 抑郁症 | [ |
| 乳酸乳球菌乳脂 亚种MG1363 | 递送胰高血糖素样蛋白-1恢复LPS引起的小鼠空间学习和记忆障碍,治疗阿尔兹海默症 | pMG36e-GLP- 1 | 阿尔兹海默症 | [ |
| 枯草芽孢杆菌60015/YF256 | 操纵肌醇代谢构建工程菌,以允许从肌醇到鲨肌醇的生物转化,作为细胞工厂提供鲨肌醇,是治疗阿尔兹海默症的潜在的活菌生物制剂 | iolE41 iolI::spc/iolR::cat/iolW::pMutin2(erm::tet)/iolX::pMutin4(erm) | 阿尔兹海默症 | [ |
| 乳酸乳球菌乳脂 亚种MG1363 | 递送p62蛋白减少氧化和炎症标志物、调节肠道菌群结构,对阿尔兹海默症产生积极影响 | pExu:p62 | 阿尔兹海默症 | [ |
| 1 | BRODY H. The gut microbiome[J]. Nature, 2020, 577(7792): S5. |
| 2 | CRESCI G A, BAWDEN E. Gut microbiome: what we do and don′t know[J]. Nutrition in Clinical Practice, 2015, 30(6): 734-746. |
| 3 | VÁZQUEZ-CASTELLANOS J F, BICLOT A, VRANCKEN G, et al. Design of synthetic microbial consortia for gut microbiota modulation[J]. Current Opinion in Pharmacology, 2019, 49: 52-59. |
| 4 | DEWEERDT S. Microbiome: a complicated relationship status[J]. Nature, 2014, 508(7496): S61-S63. |
| 5 | ZHAO L P. The gut microbiota and obesity: from correlation to causality[J]. Nature Reviews Microbiology, 2013, 11(9): 639-647. |
| 6 | GURUNG M, LI Z P, YOU H, et al. Role of gut microbiota in type 2 diabetes pathophysiology[J]. EBioMedicine, 2020, 51: 102590. |
| 7 | LIU R T, ROWAN-NASH A D, SHEEHAN A E, et al. Reductions in anti-inflammatory gut bacteria are associated with depression in a sample of young adults[J]. Brain, Behavior, and Immunity, 2020, 88: 308-324. |
| 8 | VOGT N M, KERBY R L, DILL-MCFARLAND K A, et al. Gut microbiome alterations in Alzheimer's disease[J]. Scientific Reports, 2017, 7: 13537. |
| 9 | ZHAO Y H, CONG L, JABER V, et al. Microbiome-derived lipopolysaccharide enriched in the perinuclear region of Alzheimer's disease brain[J]. Frontiers in Immunology, 2017, 8: 1064. |
| 10 | SONNENBURG J L, SONNENBURG E D. Vulnerability of the industrialized microbiota[J]. Science, 2019, 366(6464): eaaw9255. |
| 11 | GIBSON G R, HUTKINS R, SANDERS M E, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics[J]. Nature Reviews Gastroenterology & Hepatology, 2017, 14(8): 491-502. |
| 12 | CLAUS S P. Inulin prebiotic: is it all about bifidobacteria? [J]. Gut, 2017, 66(11): 1883-1884. |
| 13 | WILSON B, ROSSI M, KANNO T, et al. β-Galactooligosaccharide in conjunction with low FODMAP diet improves irritable bowel syndrome symptoms but reduces fecal bifidobacteria[J]. American Journal of Gastroenterology, 2020, 115(6): 906-915. |
| 14 | LILLY D M, STILLWELL R H. Probiotics: growth-promoting factors produced by microorganisms[J]. Science, 1965, 147(3659): 747-748. |
| 15 | SANDERS M E, MERENSTEIN D J, REID G, et al. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic[J]. Nature Reviews Gastroenterology & Hepatology, 2019, 16(10): 605-616. |
| 16 | KRUIS W, FRIC P, POKROTNIEKS J, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine[J]. Gut, 2004, 53(11): 1617-1623. |
| 17 | MARTÍN R, MIQUEL S, BENEVIDES L, et al. Functional characterization of novel faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic[J]. Frontiers in Microbiology, 2017, 8: 1226. |
| 18 | HENN M R, O'BRIEN E J, DIAO L Y, et al. A phase 1b safety study of SER-287, a spore-based microbiome therapeutic, for active mild to moderate ulcerative colitis[J]. Gastroenterology, 2021, 160(1): 115-127. |
| 19 | CURRÒ D, IANIRO G, PECERE S, et al. Probiotics, fibre and herbal medicinal products for functional and inflammatory bowel disorders[J]. British Journal of Pharmacology, 2017, 174(11): 1426-1449. |
| 20 | CHEN P, XU H Y, TANG H, et al. Modulation of gut mucosal microbiota as a mechanism of probiotics-based adjunctive therapy for ulcerative colitis[J]. Microbial Biotechnology, 2020, 13(6): 2032-2043. |
| 21 | SHIN J H, NAM M H, LEE H, et al. Amelioration of obesity-related characteristics by a probiotic formulation in a high-fat diet-induced obese rat model[J]. European Journal of Nutrition, 2018, 57(6): 2081-2090. |
| 22 | KARBASCHIAN Z, MOKHTARI Z, PAZOUKI A, et al. Probiotic supplementation in morbid obese patients undergoing one anastomosis gastric bypass-mini gastric bypass (OAGB-MGB) surgery: a randomized, double-blind, placebo-controlled, clinical trial[J]. Obesity Surgery, 2018, 28(9): 2874-2885. |
| 23 | SABICO S, AL-MASHHARAWI A, AL-DAGHRI N M, et al. Effects of a multi-strain probiotic supplement for 12 weeks in circulating endotoxin levels and cardiometabolic profiles of medication naïve T2DM patients: a randomized clinical trial[J]. Journal of Translational Medicine, 2017, 15(1): 249. |
| 24 | PERRAUDEAU F, MCMURDIE P, BULLARD J, et al. Improvements to postprandial glucose control in subjects with type 2 diabetes: a multicenter, double blind, randomized placebo-controlled trial of a novel probiotic formulation[J]. BMJ Open Diabetes Research & Care, 2020, 8(1): e001319. |
| 25 | MALDONADO-GÓMEZ M X, MARTÍNEZ I, BOTTACINI F, et al. Stable engraftment of bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome[J]. Cell Host & Microbe, 2016, 20(4): 515-526. |
| 26 | MARX W, SCHOLEY A, FIRTH J, et al. Prebiotics, probiotics, fermented foods and cognitive outcomes: a meta-analysis of randomized controlled trials[J]. Neuroscience & Biobehavioral Reviews, 2020, 118: 472-484. |
| 27 | WILSON B, ROSSI M, DIMIDI E, et al. Prebiotics in irritable bowel syndrome and other functional bowel disorders in adults: a systematic review and meta-analysis of randomized controlled trials[J]. American Journal of Clinical Nutrition, 2019, 109(4): 1098-1111. |
| 28 | COSTELOE K, HARDY P, JUSZCZAK E, et al. Bifidobacterium breve BBG-001 in very preterm infants: a randomised controlled phase 3 trial[J]. Lancet, 2016, 387(10019): 649-660. |
| 29 | FREEDMAN S B, FINKELSTEIN Y, PANG X L, et al. Pathogen-specific effects of probiotics in children with acute gastroenteritis seeking emergency care: a randomized trial[J]. Clinical Infectious Diseases, 2021, . |
| 30 | JOHNSTONE J, MEADE M, LAUZIER F, et al. Effect of probiotics on incident ventilator-associated pneumonia in critically ill patients: a randomized clinical trial[J]. JAMA, 2021, 326(11): 1024-1033. |
| 31 | ZHANG F M, CUI B T, HE X X, et al. Microbiota transplantation: concept, methodology and strategy for its modernization[J]. Protein & Cell, 2018, 9(5): 462-473. |
| 32 | MULLISH B H, QURAISHI M N, SEGAL J P, et al. The use of faecal microbiota transplant as treatment for recurrent or refractory Clostridium difficile infection and other potential indications: joint British Society of Gastroenterology (BSG) and Healthcare Infection Society (HIS) guidelines[J]. Gut, 2018, 67(11): 1920-1941. |
| 33 | DEFILIPP Z, BLOOM P P, TORRES SOTO M, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant[J]. New England Journal of Medicine, 2019, 381(21): 2043-2050. |
| 34 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 35 | RIGLAR D T, SILVER P A. Engineering bacteria for diagnostic and therapeutic applications[J]. Nature Reviews Microbiology, 2018, 16(4): 214-225. |
| 36 | MCCARTY N S, LEDESMA-AMARO R. Synthetic biology tools to engineer microbial communities for biotechnology[J]. Trends in Biotechnology, 2019, 37(2): 181-197. |
| 37 | PEDROLLI D B, RIBEIRO N V, SQUIZATO P N, et al. Engineering microbial living therapeutics: the synthetic biology toolbox[J]. Trends in Biotechnology, 2019, 37(1): 100-115. |
| 38 | GOH Y J, BARRANGOU R. Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli[J]. Current Opinion in Biotechnology, 2019, 56: 163-171. |
| 39 | PETERS J M, KOO B M, PATINO R, et al. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi[J]. Nature Microbiology, 2019, 4(2): 244-250. |
| 40 | GAO C, XU P, YE C, et al. Genetic circuit-assisted smart microbial engineering[J]. Trends in Microbiology, 2019, 27(12): 1011-1024. |
| 41 | WINDSOR J W, KAPLAN G G. Evolving epidemiology of IBD[J]. Current Gastroenterology Reports, 2019, 21(8): 40. |
| 42 | KEUM N, GIOVANNUCCI E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies[J]. Nature Reviews Gastroenterology & Hepatology, 2019, 16(12): 713-732. |
| 43 | SOMMER F, RÜHLEMANN M C, BANG C, et al. Microbiomarkers in inflammatory bowel diseases: caveats come with caviar[J]. Gut, 2017, 66(10): 1734-1738. |
| 44 | TILG H, ADOLPH T E, GERNER R R, et al. The intestinal microbiota in colorectal cancer[J]. Cancer Cell, 2018, 33(6): 954-964. |
| 45 | HODSON R. Inflammatory bowel disease[J]. Nature, 2016, 540(7634): S97. |
| 46 | DORDEVIĆ D, JANČÍKOVÁ S, VÍTĚZOVÁ M, et al. Hydrogen sulfide toxicity in the gut environment: meta-analysis of sulfate-reducing and lactic acid bacteria in inflammatory processes[J]. Journal of Advanced Research, 2021, 27: 55-69. |
| 47 | DAEFFLER K N M, GALLEY J D, SHETH R U, et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation[J]. Molecular Systems Biology, 2017, 13(4): 923. |
| 48 | WOO S G, MOON S J, KIM S K, et al. A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing[J]. Biosensors and Bioelectronics, 2020, 168: 112523. |
| 49 | MIMEE M, NADEAU P, HAYWARD A, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health[J]. Science, 2018, 360(6391): 915-918. |
| 50 | STEIDLER L, HANS W, SCHOTTE L, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10[J]. Science, 2000, 289(5483): 1352-1355. |
| 51 | VANDENBROUCKE K, DE HAARD H, BEIRNAERT E, et al. Orally administered L. lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis[J]. Mucosal Immunology, 2010, 3(1): 49-56. |
| 52 | SCOTT B M, GUTIÉRREZ-VÁZQUEZ C, SANMARCO L M, et al. Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease[J]. Nature Medicine, 2021, 27(7): 1212-1222. |
| 53 | MAYS Z J, NAIR N U. Synthetic biology in probiotic lactic acid bacteria: at the frontier of living therapeutics[J]. Current Opinion in Biotechnology, 2018, 53: 224-231. |
| 54 | DOU J, BENNETT M R. Synthetic biology and the gut microbiome[J]. Biotechnology Journal, 2018, 13(5): e1700159. |
| 55 | YAN X, LIU X Y, ZHANG D, et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis[J]. Cellular & Molecular Immunology, 2021, 18(10): 2344-2357. |
| 56 | CUI M H, PANG G J, ZHANG T, et al. Optotheranostic nanosystem with phone visual diagnosis and optogenetic microbial therapy for ulcerative colitis at-home care[J]. ACS Nano, 2021, 15(4): 7040-7052. |
| 57 | PRAVESCHOTINUNT P, DURAJ-THATTE A M, GELFAT I, et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut[J]. Nature Communications, 2019, 10: 5580. |
| 58 | YAN Z F, YIN M, CHEN J N, et al. Assembly and substrate recognition of curli biogenesis system[J]. Nature Communications, 2020, 11: 241. |
| 59 | YU M, KIM J, AHN J H, et al. Nononcogenic restoration of the intestinal barrier by E. coli-delivered human EGF[J]. JCI Insight, 2019, 4(16): e125166. |
| 60 | NADEEM M S, KUMAR V, AL-ABBASI F A, et al. Risk of colorectal cancer in inflammatory bowel diseases[J]. Seminars in Cancer Biology, 2020, 64: 51-60. |
| 61 | DAI Z W, COKER O O, NAKATSU G, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers[J]. Microbiome, 2018, 6(1): 70. |
| 62 | HO C L, TAN H Q, CHUA K J, et al. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention[J]. Nature Biomedical Engineering, 2018, 2(1): 27-37. |
| 63 | AN B C, RYU Y, YOON Y S, et al. Colorectal cancer therapy using a pediococcus pentosaceus SL4 drug delivery system secreting lactic acid bacteria-derived protein p8[J]. Molecules and Cells, 2019, 42(11): 755-762. |
| 64 | CHEN J H, LI X H, LIU Y M, et al. Engineering a probiotic strain of Escherichia coli to induce the regression of colorectal cancer through production of 5-aminolevulinic acid[J]. Microbial Biotechnology, 2021, 14(5): 2130-2139. |
| 65 | CANI P D. Microbiota and metabolites in metabolic diseases[J]. Nature Reviews Endocrinology, 2019, 15(2): 69-70. |
| 66 | HE Y, WU W, WU S, et al. Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis[J]. Microbiome, 2018, 6(1): 172. |
| 67 | BROUSSARD J L, DEVKOTA S. The changing microbial landscape of Western society: diet, dwellings and discordance[J]. Molecular Metabolism, 2016, 5(9): 737-742. |
| 68 | CABALLERO B. Humans against obesity: who will win?[J]. Advances in Nutrition, 2019, 10(): S4-S9. |
| 69 | FEI N, BRUNEAU A, ZHANG X J, et al. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease[J]. mBio, 2020, 11(1): e03263-e03219. |
| 70 | ZHAO L P, ZHAO N S. Demonstration of causality: back to cultures[J]. Nature Reviews Gastroenterology & Hepatology, 2021, 18(2): 97-98. |
| 71 | LE CHATELIER E, NIELSEN T, QIN J J, et al. Richness of human gut microbiome correlates with metabolic markers[J]. Nature, 2013, 500(7464): 541-546. |
| 72 | WADDEN T A, VOLGER S, TSAI A G, et al. Managing obesity in primary care practice: an overview with perspective from the POWER-UP study[J]. International Journal of Obesity, 2013, 37(1): S3-S11. |
| 73 | THEREAUX J, LESUFFLEUR T, CZERNICHOW S, et al. Long-term adverse events after sleeve gastrectomy or gastric bypass: A 7-year nationwide, observational, population-based, cohort study[J]. The Lancet Diabetes & Endocrinology, 2019, 7(10): 786-795. |
| 74 | HIRA T, PINYO J, HARA H. What is GLP-1 really doing in obesity?[J]. Trends in Endocrinology & Metabolism, 2020, 31(2): 71-80. |
| 75 | WANG L F, CHEN T T, WANG H, et al. Engineered bacteria of MG1363-pMG36e-GLP-1 attenuated obesity-induced by high fat diet in mice[J]. Frontiers in Cellular and Infection Microbiology, 2021, 11: 595575. |
| 76 | MA J, LI C Y, WANG J R, et al. Genetically engineered Escherichia coli nissle 1917 secreting GLP-1 analog exhibits potential antiobesity effect in high-fat diet-induced obesity mice[J]. Obesity, 2020, 28(2): 315-322. |
| 77 | LONG R T, ZENG W S, CHEN L Y, et al. Bifidobacterium as an oral delivery carrier of oxyntomodulin for obesity therapy: inhibitory effects on food intake and body weight in overweight mice[J]. International Journal of Obesity, 2010, 34(4): 712-719. |
| 78 | NAMAI F, SHIGEMORI S, SUDO K, et al. Recombinant mouse osteocalcin secreted by Lactococcus lactis promotes glucagon-like peptide-1 induction in STC-1 cells[J]. Current Microbiology, 2018, 75(1): 92-98. |
| 79 | CAO W Y, DONG M, HU Z Y, et al. Recombinant Lactococcus lactis NZ3900 expressing bioactive human FGF21 reduced body weight of Db/Db mice through the activity of brown adipose tissue[J]. Beneficial Microbes, 2020, 11(1): 67-78. |
| 80 | CHEN Z Y, GUO L L, ZHANG Y Q, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity[J]. Journal of Clinical Investigation, 2014, 124(8): 3391-3406. |
| 81 | DOSOKY N S, GUO L L, CHEN Z Y, et al. Dietary fatty acids control the species of N-acyl-phosphatidylethanolamines synthesized by therapeutically modified bacteria in the intestinal tract[J]. ACS Infectious Diseases, 2018, 4(1): 3-13. |
| 82 | BAI L, GAO M X, CHENG X M, et al. Engineered butyrate-producing bacteria prevents high fat diet-induced obesity in mice[J]. Microbial Cell Factories, 2020, 19(1): 94. |
| 83 | GUTHRIE R A, GUTHRIE D W. Pathophysiology of diabetes mellitus[J]. Critical Care Nursing Quarterly, 2004, 27(2): 113-125. |
| 84 | PAUN A, DANSKA J S. Modulation of type 1 and type 2 diabetes risk by the intestinal microbiome[J]. Pediatric Diabetes, 2016, 17(7): 469-477. |
| 85 | NEEDELL J C, ZIPRIS D. The role of the intestinal microbiome in type 1 diabetes pathogenesis[J]. Current Diabetes Reports, 2016, 16(10): 89. |
| 86 | TAN S Y, WONG J L M, SIM Y J, et al. Type 1 and 2 diabetes mellitus: a review on current treatment approach and gene therapy as potential intervention[J]. Diabetes & Metabolic Syndrome: Clinical Research & Reviews, 2019, 13(1): 364-372. |
| 87 | ROBERT S, GYSEMANS C, TAKIISHI T, et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice[J]. Diabetes, 2014, 63(8): 2876-2887. |
| 88 | TAKIISHI T, KORF H, BELLE T L VAN, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice[J]. Journal of Clinical Investigation, 2012, 122(5): 1717-1725. |
| 89 | LANG J C, WANG X K, LIU K F, et al. Oral delivery of staphylococcal nuclease by Lactococcus lactis prevents type 1 diabetes mellitus in NOD mice[J]. Applied Microbiology and Biotechnology, 2017, 101(20): 7653-7662. |
| 90 | MA Y J, LIU J J, HOU J, et al. Oral administration of recombinant Lactococcus lactis expressing HSP65 and tandemly repeated P277 reduces the incidence of type I diabetes in non-obese diabetic mice[J]. PLoS One, 2014, 9(8): e105701. |
| 91 | AGARWAL P, KHATRI P, BILLACK B, et al. Oral delivery of glucagon like peptide-1 by a recombinant Lactococcus lactis [J]. Pharmaceutical Research, 2014, 31(12): 3404-3414. |
| 92 | DUAN F F, LIU J H, MARCH J C. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes[J]. Diabetes, 2015, 64(5): 1794-1803. |
| 93 | ZENG Z, YU R, ZUO F L, et al. Recombinant Lactococcus lactis expressing bioactive exendin-4 to promote insulin secretion and beta-cell proliferation in vitro [J]. Applied Microbiology and Biotechnology, 2017, 101(19): 7177-7186. |
| 94 | RHIMI M, BERMUDEZ-HUMARAN L G, HUANG Y, et al. The secreted L-arabinose isomerase displays anti-hyperglycemic effects in mice[J]. Microbial Cell Factories, 2015, 14: 204. |
| 95 | GMITROWICZ A, KUCHARSKA A. Developmental disorders in the fourth edition of the American classification: diagnostic and statistical manual of mental disorders (DSM IV — optional book)[J]. Psychiatria Polska, 1994, 28(5): 509-521. |
| 96 | SMITH K. Mental health: a world of depression[J]. Nature, 2014, 515(7526): 181. |
| 97 | CHOPRA K, KUMAR B, KUHAD A. Pathobiological targets of depression[J]. Expert Opinion on Therapeutic Targets, 2011, 15(4): 379-400. |
| 98 | SIMPSON C A, DIAZ-ARTECHE C, ELIBY D, et al. The gut microbiota in anxiety and depression-a systematic review[J]. Clinical Psychology Review, 2021, 83: 101943. |
| 99 | ZHENG P, ZENG B, ZHOU C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism[J]. Molecular Psychiatry, 2016, 21(6): 786-796. |
| 100 | STRANDWITZ P, KIM K H, TEREKHOVA D, et al. GABA-modulating bacteria of the human gut microbiota[J]. Nature Microbiology, 2019, 4(3): 396-403. |
| 101 | MEDINA-RODRIGUEZ E M, MADORMA D, O'CONNOR G, et al. Identification of a signaling mechanism by which the microbiome regulates Th17 cell-mediated depressive-like behaviors in mice[J]. The American Journal of Psychiatry, 2020, 177(10): 974-990. |
| 102 | DINAN T G, STANTON C, CRYAN J F. Psychobiotics: a novel class of psychotropic[J]. Biological Psychiatry, 2013, 74(10): 720-726. |
| 103 | BAMBURY A, SANDHU K, CRYAN J F, et al. Finding the needle in the haystack: systematic identification of psychobiotics[J]. British Journal of Pharmacology, 2018, 175(24): 4430-4438. |
| 104 | XU D, FANG M J, WANG H J, et al. Enhanced production of 5-hydroxytryptophan through the regulation of L-tryptophan biosynthetic pathway[J]. Applied Microbiology and Biotechnology, 2020, 104(6): 2481-2488. |
| 105 | LÜ C J, ZHAO W R, HU S, et al. Physiology-oriented engineering strategy to improve gamma-aminobutyrate production in Lactobacillus brevis [J]. Journal of Agricultural and Food Chemistry, 2017, 65(4): 858-866. |
| 106 | DUNG PHAM VAN, SOMASUNDARAM S, LEE S H, et al. Efficient production of gamma-aminobutyric acid using Escherichia coli by co-localization of glutamate synthase, glutamate decarboxylase, and GABA transporter[J]. Journal of Industrial Microbiology and Biotechnology, 2016, 43(1): 79-86. |
| 107 | DEN H, DONG X H, CHEN M L, et al. Efficacy of probiotics on cognition, and biomarkers of inflammation and oxidative stress in adults with Alzheimer's disease or mild cognitive impairment - a meta-analysis of randomized controlled trials[J]. Aging, 2020, 12(4): 4010-4039. |
| 108 | ZHAO Y H, JABER V, LUKIW W J. Secretory products of the human GI tract microbiome and their potential impact on Alzheimer's disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus[J]. Frontiers in Cellular and Infection Microbiology, 2017, 7: 318. |
| 109 | FANG X, ZHOU X T, MIAO Y Q, et al. Therapeutic effect of GLP-1 engineered strain on mice model of Alzheimer's disease and Parkinson's disease[J]. AMB Express, 2020, 10(1): 80. |
| 110 | CHEN T T, TIAN P Y, HUANG Z X, et al. Engineered commensal bacteria prevent systemic inflammation-induced memory impairment and amyloidogenesis via producing GLP-1[J]. Applied Microbiology and Biotechnology, 2018, 102(17): 7565-7575. |
| 111 | YAMAOKA M, OSAWA S, MORINAGA T, et al. A cell factory of Bacillus subtilis engineered for the simple bioconversion of myo-inositol to scyllo-inositol, a potential therapeutic agent for Alzheimer's disease[J]. Microbial Cell Factories, 2011, 10: 69. |
| 112 | CECARINI V, BONFILI L, GOGOI O, et al. Neuroprotective effects of p62(SQSTM1)-engineered lactic acid bacteria in Alzheimer's disease: a pre-clinical study[J]. Aging, 2020, 12(16): 15995-16020. |
| 113 | KIM S, KERNS S J, ZIESACK M, et al. Quorum sensing can be repurposed to promote information transfer between bacteria in the mammalian gut[J]. ACS Synthetic Biology, 2018, 7(9): 2270-2281. |
| 114 | CAMACHO D M, COLLINS K M, POWERS R K, et al. Next-generation machine learning for biological networks[J]. Cell, 2018, 173(7): 1581-1592. |
| 115 | JIAO N, BAKER S S, NUGENT C A, et al. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: a meta-analysis[J]. Physiological Genomics, 2018, 50(4): 244-254. |
| 116 | GUO X F, LI S H, ZHANG J C, et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas[J]. BMC Genomics, 2017, 18(1): 800. |
| 117 | KAPSE N G, ENGINEER A S, GOWDAMAN V, et al. Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243[J]. Genomics, 2019, 111(4): 921-929. |
| 118 | MEDLOCK G L, CAREY M A, MCDUFFIE D G, et al. Inferring metabolic mechanisms of interaction within a defined gut microbiota[J]. Cell Systems, 2018, 7(3): 245-257. |
| 119 | URIBE R V, HELM E VAN DER, MISIAKOU M A, et al. Discovery and characterization of Cas9 inhibitors disseminated across seven bacterial phyla[J]. Cell Host & Microbe, 2019, 25(2): 233-241. |
| 120 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12): 725-741. |
| 121 | SHAH P, FRITZ J V, GLAAB E, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface[J]. Nature Communications, 2016, 7: 11535. |
| 122 | TSAKMAKI A, FONSECA PEDRO P, BEWICK G A. 3D intestinal organoids in metabolic research: virtual reality in a dish[J]. Current Opinion in Pharmacology, 2017, 37: 51-58. |
| 123 | INGBER D E. Reverse engineering human pathophysiology with organs-on-chips[J]. Cell, 2016, 164(6): 1105-1109. |
| 124 | UZBAY T. Germ-free animal experiments in the gut microbiota studies[J]. Current Opinion in Pharmacology, 2019, 49: 6-10. |
| 125 | KOVATCHEVA-DATCHARY P, SHOAIE S, LEE S, et al. Simplified intestinal microbiota to study microbe-diet-host interactions in a mouse model[J]. Cell Reports, 2019, 26(13): 3772-3783. |
| 126 | REARDON S. Genetically modified bacteria enlisted in fight against disease[J]. Nature, 2018, 558(7711): 497-498. |
| 127 | KURTZ C B, MILLET Y A, PUURUNEN M K, et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans[J]. Science Translational Medicine, 2019, 11(475): eaau7975. |
| 128 | ISABELLA V M, HA B N, CASTILLO M J, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria[J]. Nature Biotechnology, 2018, 36(9): 857-864. |
| 129 | BERMÚDEZ-HUMARÁN L G, LANGELLA P. Live bacterial biotherapeutics in the clinic[J]. Nature Biotechnology, 2018, 36(9): 816-818. |
| 130 | WEGMANN U, CARVALHO A L, STOCKS M, et al. Use of genetically modified bacteria for drug delivery in humans: Revisiting the safety aspect[J]. Scientific Reports, 2017, 7: 2294. |
| 131 | ALAYANDE K A, AIYEGORO O A, NENGWEKHULU T M, et al. Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1 [J]. PLoS One, 2020, 15(7): e0235873. |
| 132 | CARMO F L R DO, RABAH H, DE OLIVEIRA CARVALHO R D, et al. Extractable bacterial surface proteins in probiotic-host interaction[J]. Frontiers in Microbiology, 2018, 9: 645. |
| 133 | SONG W Z, WEMHEUER B, ZHANG S, et al. MetaCHIP: community-level horizontal gene transfer identification through the combination of best-match and phylogenetic approaches[J]. Microbiome, 2019, 7(1): 36. |
| 134 | TANOUE T, MORITA S, PLICHTA D R, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity[J]. Nature, 2019, 565(7741): 600-605. |
| 135 | WU S B, LIU J H, LIU C J, et al. Quorum sensing for population-level control of bacteria and potential therapeutic applications[J]. Cellular and Molecular Life Sciences, 2020, 77(7): 1319-1343. |
| 136 | ZIESACK M, GIBSON T, OLIVER J K W, et al. Engineered inter species amino acid cross-feeding increases population evenness in a synthetic bacterial consortium[J]. mSystems, 2019, 4(4): e00352-e00319. |
| 137 | DURAJ-THATTE A M, COURCHESNE N M D, PRAVESCHOTINUNT P, et al. Genetically programmable self-regenerating bacterial hydrogels[J]. Advanced Materials, 2019, 31(40): 1970289. |
| 138 | CHIEN T, HARIMOTO T, KEPECS B, et al. Enhancing the tropism of bacteria via genetically programmed biosensors[J]. Nature Biomedical Engineering, 2021: 1-11. |
| 139 | MAYS Z J S, CHAPPELL T C, NAIR N U. Quantifying and engineering mucus adhesion of probiotics[J]. ACS Synthetic Biology, 2020, 9(2): 356-367. |
| 140 | CHAPPELL T C, NAIR N U. Engineered lactobacilli display anti-biofilm and growth suppressing activities against Pseudomonas aeruginosa[J]. NPJ Biofilms and Microbiomes, 2020, 6: 48. |
| 141 | PUURUNEN M K, VOCKLEY J, SEARLE S L, et al. Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study[J]. Nature Metabolism, 2021, 3(8): 1125-1132. |
| 142 | ADOLFSEN K J, CALLIHAN I, MONAHAN C E, et al. Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering[J]. Nature Communications, 2021, 12: 6215. |
| 143 | WANG L L W, JANES M E, KUMBHOJKAR N, et al. Cell therapies in the clinic[J]. Bioengineering & Translational Medicine, 2021, 6(2): e10214. |
| [1] | 林思思, 潘超, 张一帆, 刘尽尧. 基于表面涂层益生菌的肿瘤抗原口服递送系统[J]. 合成生物学, 2022, 3(4): 810-820. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||