合成生物学 ›› 2024, Vol. 5 ›› Issue (6): 1461-1484.DOI: 10.12211/2096-8280.2024-052
体外多酶分子机器产氢应用中的氢酶研究
李怡霏1,2,3,4, 陈艾1,2,3,4, 孙俊松1,2, 张以恒2,3,4,5
- 1.中国科学院上海高等研究院低碳生物转化团队,上海 201210
2.中国科学院大学,北京 100049
3.中国科学院天津工业生物技术研究所低碳合成工程生物学(全国)重点实验室,天津 300308
4.中国科学院天津工业生物技术研究所体外合成生物学中心,天津 300308
5.合成生物学海河实验室,天津 300308
-
收稿日期:2024-07-09修回日期:2024-09-25出版日期:2024-12-31发布日期:2025-01-10 -
通讯作者:孙俊松,张以恒 -
作者简介:李怡霏 (2000—),女,硕士研究生。研究方向为氢酶参与的体外多酶分子机器构建。 E-mail:liyf@sari.ac.cn孙俊松 (1974—),男,博士,研究员。研究方向为氢酶表达、微生物代谢改造及生物合成。 E-mail:sunjs@sari.ac.cn张以恒 (1971—),男,博士,研究员,中国科学院天津工业生物技术研究所低碳合成工程生物学(全国)重点实验室主任,曾任美国弗吉尼亚理工大学终身正教授。研究方向为体外合成生物学、新质生物制造、生物炼制和淀粉储能。 E-mail:zhang_xw@tib.cas.cn -
基金资助:国家重点研发计划“合成生物学”重点专项“糖水氢电系统——体外多酶高效产氢及氢电装置的基础及工程研究”(2022YFA0912000)
Studies on hydrogenases for hydrogen production using in vitro synthetic enzymatic biosystems
LI Yifei1,2,3,4, CHEN Ai1,2,3,4, SUN Junsong1,2, ZHANG Yi-Heng P. Job2,3,4,5
- 1.Low Carbon Biotransformation Group,Shanghai Advanced Research Institute,Chinese Academy of Sciences,Shanghai 201210,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
3.Key Laboratory of Engineering Biology for Low-Carbon Manufacturing,Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
4.In Vitro Synthetic Biology Center,Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
5.Haihe Laboratory of Synthetic Biology,Tianjin 300308,China
-
Received:2024-07-09Revised:2024-09-25Online:2024-12-31Published:2025-01-10 -
Contact:SUN Junsong, ZHANG Yi-Heng P. Job
摘要:
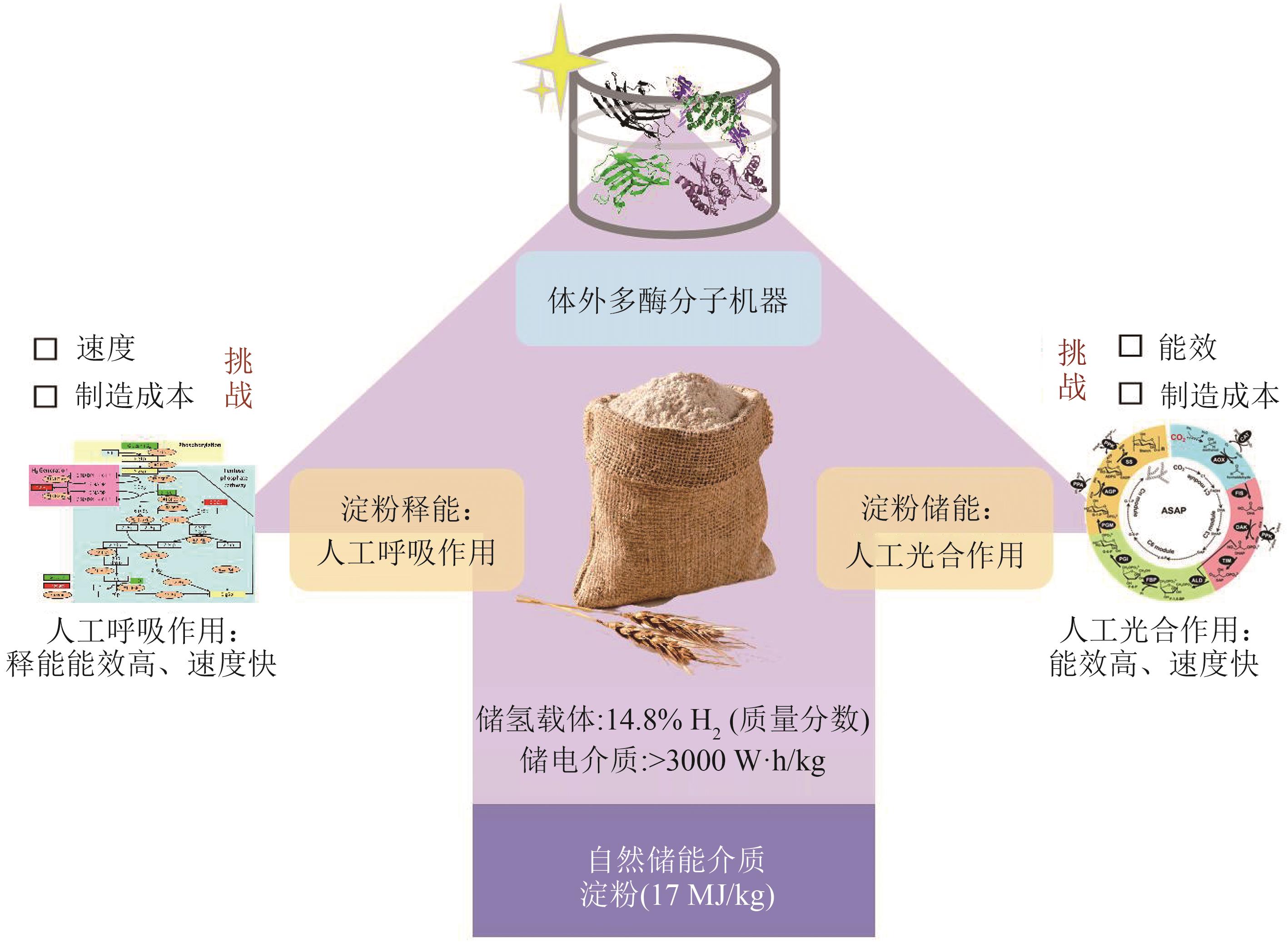
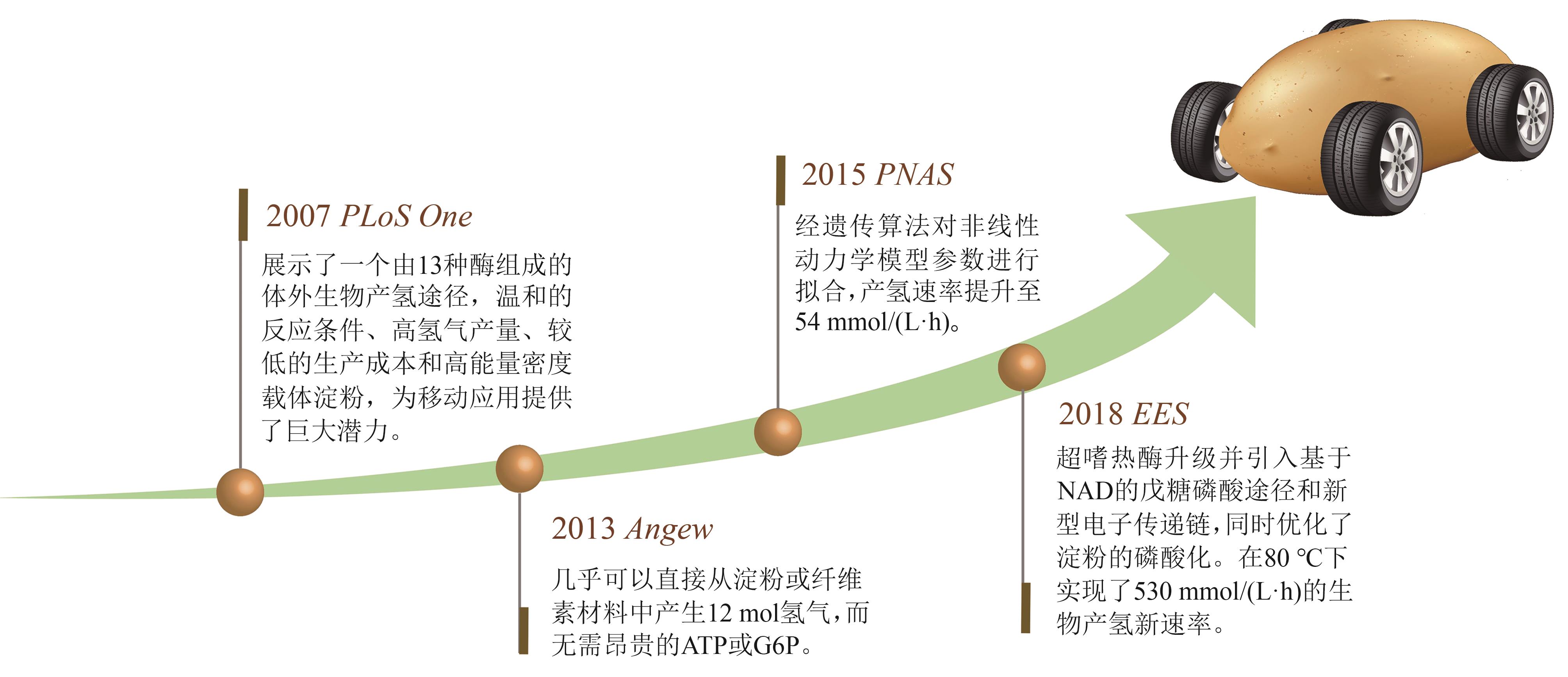
氢酶是生物制氢和氢能利用的最关键酶,它是一类广泛分布的对氧敏感的多亚基金属复合酶。体外多酶分子机器是体外生物转化技术中的高效酶生物催化系统,利用该分子机器生产氢气是一种新型高效的绿氢生产技术,它突破微生物产氢的Thauer极限,将葡萄糖产氢的转化率提高至接近化学理论值(1 mol葡萄糖裂解水生产12 mol氢气),代表着生物产氢的未来方向。氢酶的制备及催化性能是限制多酶分子机器产氢技术广泛应用的主要瓶颈;氧气不仅抑制氢酶的活性,也是氢酶转录翻译及翻译后加工的重要影响因素。体外多酶分子机器对氢酶的耐氧性能、热稳定性及高周转性能等参数提出高要求。本文结合氢酶在多酶分子机器制氢应用中的技术障碍,针对迫切的基础科学问题,分别从氢酶分类、结构功能、重组表达技术进展、(仿生)辅酶的适配等方面对其进行了相关的总结,并初步对氧的抑制机制、微生物重组表达氢酶以及产氢人工电子传递链的优化等难点问题的研究进行了跟踪,期待能够为氢酶在体外合成生物学的应用提供参考。
中图分类号:
引用本文
李怡霏, 陈艾, 孙俊松, 张以恒. 体外多酶分子机器产氢应用中的氢酶研究[J]. 合成生物学, 2024, 5(6): 1461-1484.
LI Yifei, CHEN Ai, SUN Junsong, ZHANG Yi-Heng P. Job. Studies on hydrogenases for hydrogen production using in vitro synthetic enzymatic biosystems[J]. Synthetic Biology Journal, 2024, 5(6): 1461-1484.
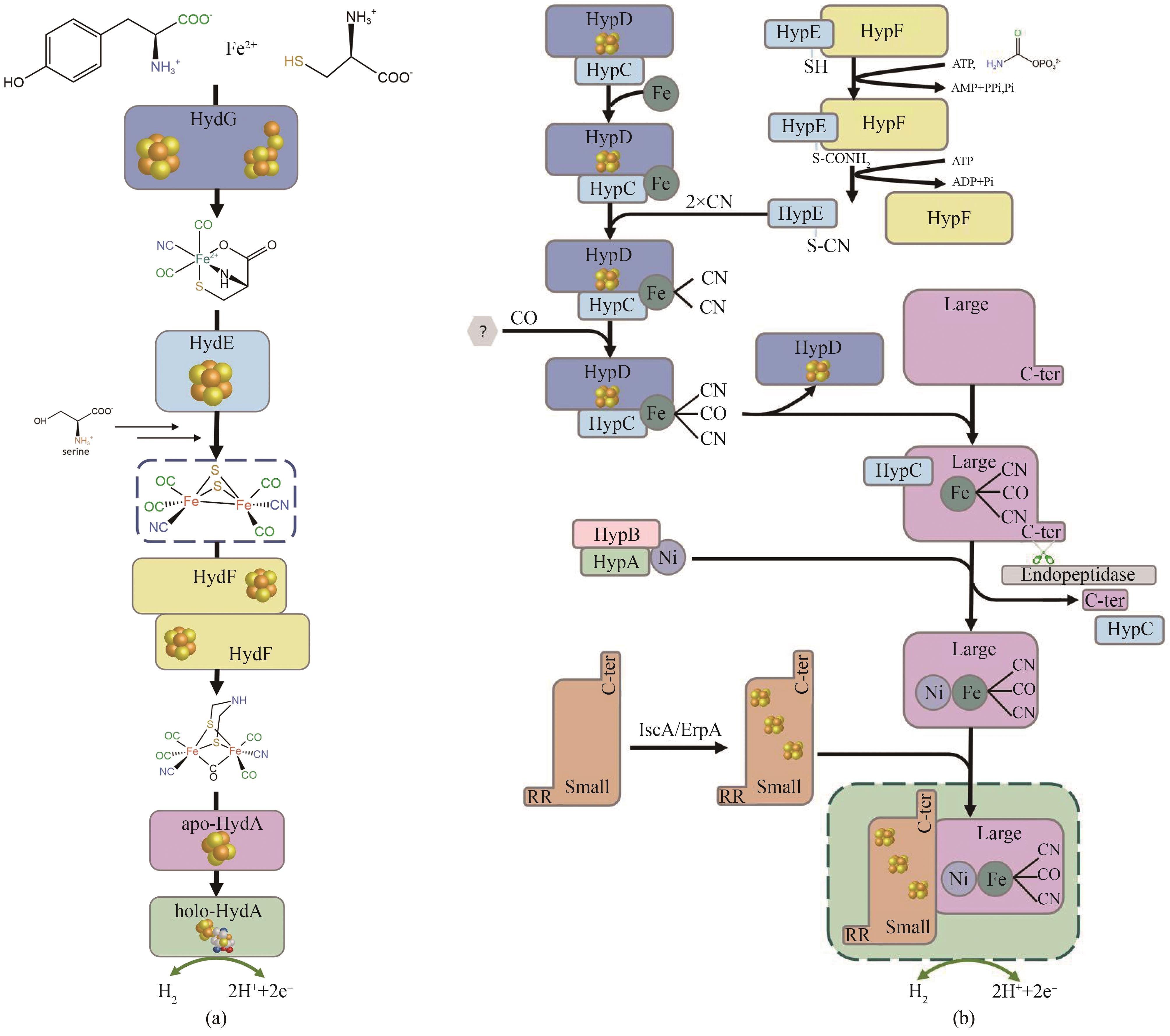
图2 [FeFe]氢酶成熟过程示意(a)和[NiFe]氢酶成熟过程示意(b)
Fig. 2 Schematic of the maturation process of FeFe-hydrogenase (a) and schematic of the maturation process of NiFe-hydrogenase (b)
氢酶 Hydrogenase | 表达宿主 Expression host | 成熟蛋白 Maturation protein | 全细胞活性 Whole cell activity | 纯化酶活性 Purified enzyme activity | 参考文献 References |
|---|---|---|---|---|---|
| CacHydA | C. acetobutylicum | Host | NR | 162 ② | [ |
| CacHydA | E. coli | C. acetobutylicum | 96 ① | 75 ② | [ |
| CacHydA | E. coli ∆iscR | C. acetobutylicum | 1.3 ①,* | 96 ② | [ |
| CacHydA | S. elongatus | C. reinhardtii | NR | 0.05 ② | [ |
| CacHydB | E. coli | C. acetobutylicum | NR | 8.6 ② | [ |
| CbuHydA | E. coli | Host | 500 ① | NR | [ |
| CpaHydA | C. pasteurianum | Host | 1681 ① | 1236 ② | [ |
| CpaHydI | E. coli | S. oneidensis | NR | 1087 ② | [ |
| CpaHydI | Synechococcus sp. | Host | NR | 4.6 ② | [ |
| CreHydA1 | C. reinhardtii | Host | 13.8 ②,* | 741 ② | [ |
| CreHydA1 | C. acetobutylicum | Host | NR | 625~760 ② | [ |
| CreHydA1 | E. coli | C. reinhardtii | NR | 0.4 ② | [ |
| CreHydA1 | E. coli | C. acetobutylicum | 61 ① | 150 ② | [ |
| CreHydA1-Fd | E. coli | C. acetobutylicum | NR | 1000 ② | [ |
| CreHydA1 | E. coli | S. oneidensis | NR | 641 ② | [ |
| CreHydA1 | S. oneidensis | Host | NA | 740 ② | [ |
| CreHydA1 | Synechocystis sp. | Host | NR | 0.1 ② | [ |
| CreHydA2 | E. coli | C. acetobutylicum | 108 ① | 116 ② | [ |
| CsuHydA | E. coli | S. oneidensis | NA | 6.5 ② | [ |
| EhaHyd | E. coli | Host | NR | 70 ② | [ |
| EhiHyd | E. coli | Host | NR | 0.04 ② | [ |
| PgrHyd | E. coli | Host | NR | 2131 ② | [ |
| SobHydA1 | C. acetobutylicum | Host | NR | 633 ② | [ |
| SonHydA | Anabaena sp. | S. oneidensis | NR | 0.06 ② | [ |
表1 重组[FeFe]氢酶活性
Table 1 Recombinant [FeFe] hydrogenase activity
氢酶 Hydrogenase | 表达宿主 Expression host | 成熟蛋白 Maturation protein | 全细胞活性 Whole cell activity | 纯化酶活性 Purified enzyme activity | 参考文献 References |
|---|---|---|---|---|---|
| CacHydA | C. acetobutylicum | Host | NR | 162 ② | [ |
| CacHydA | E. coli | C. acetobutylicum | 96 ① | 75 ② | [ |
| CacHydA | E. coli ∆iscR | C. acetobutylicum | 1.3 ①,* | 96 ② | [ |
| CacHydA | S. elongatus | C. reinhardtii | NR | 0.05 ② | [ |
| CacHydB | E. coli | C. acetobutylicum | NR | 8.6 ② | [ |
| CbuHydA | E. coli | Host | 500 ① | NR | [ |
| CpaHydA | C. pasteurianum | Host | 1681 ① | 1236 ② | [ |
| CpaHydI | E. coli | S. oneidensis | NR | 1087 ② | [ |
| CpaHydI | Synechococcus sp. | Host | NR | 4.6 ② | [ |
| CreHydA1 | C. reinhardtii | Host | 13.8 ②,* | 741 ② | [ |
| CreHydA1 | C. acetobutylicum | Host | NR | 625~760 ② | [ |
| CreHydA1 | E. coli | C. reinhardtii | NR | 0.4 ② | [ |
| CreHydA1 | E. coli | C. acetobutylicum | 61 ① | 150 ② | [ |
| CreHydA1-Fd | E. coli | C. acetobutylicum | NR | 1000 ② | [ |
| CreHydA1 | E. coli | S. oneidensis | NR | 641 ② | [ |
| CreHydA1 | S. oneidensis | Host | NA | 740 ② | [ |
| CreHydA1 | Synechocystis sp. | Host | NR | 0.1 ② | [ |
| CreHydA2 | E. coli | C. acetobutylicum | 108 ① | 116 ② | [ |
| CsuHydA | E. coli | S. oneidensis | NA | 6.5 ② | [ |
| EhaHyd | E. coli | Host | NR | 70 ② | [ |
| EhiHyd | E. coli | Host | NR | 0.04 ② | [ |
| PgrHyd | E. coli | Host | NR | 2131 ② | [ |
| SobHydA1 | C. acetobutylicum | Host | NR | 633 ② | [ |
| SonHydA | Anabaena sp. | S. oneidensis | NR | 0.06 ② | [ |
氢酶 Hydrogenase | 表达宿主 Expression host | 成熟蛋白 Maturation protein | 全细胞活性 Whole cell activity | 纯化酶活性 Purified enzyme activity | 参考文献 References |
|---|---|---|---|---|---|
| AmaHynSL | A. macleodi ∆HynSL | Host | 0.03 ①,* | 0.1 ③ | [ |
| AmaHynSL | E. coli | A. macleodii | 3×10-3~70×10-3 ①,* | NR | [ |
| AmaHyaAB | T. roseopersicina | Host, A. macleodii | 5×10-3 ① | NR | [ |
| AflHydSL | E. coli | Host | NA | 77 ① | [ |
| DgiHynAB | D. gigas ∆HynAB | Host | 1.9 ①,* | 91 ① | [ |
| DgiHynAB | D. fructosovorans ∆HynAB | Host | 0.2 ② | NR | [ |
| EcoHyd1 | E. coli ∆Hyd1 | Host | 4×10-2~7×10-2 ①,* | 1×10-2~3×10-2 ① | [ |
| HmaMBH | E. coli | E. coli | 0.07 ①,* | 0.03 ① | [ |
| NpuHupSL | E. coli | E. coli | 208 ① | NR | [ |
| PfuSH | E. coli | P. furiosus | 2.9 ① | 100 ① | [ |
| PfuSHI | T. kodakarensis | Host | 23.6 ④ | 880 ④ | [ |
| ReuMBH | R. Eutropha H16 | Host | 1.0 ③,* | 170 ③ | [ |
| ReuMBH | P. stutzeri | R. eutropha | 17~19 ③,* | NR | [ |
| ReuRH | E. coli | R. eutropha | NR | 0.8 ② | [ |
| ReuRH | E. coli | R. eutropha | 1.2 ②,* | 230 ② | [ |
| RopSH | R. eutropha ∆SH ∆MBH | Host, R. opacus | 5.9 ①,* | NR | [ |
| SynSH | E. coli | Synechocystis sp. | 0.04 ①,* | NR | [ |
表2 重组[NiFe]氢酶活性
Table 2 The activities of the reported recombinant [NiFe] hydrogenases
氢酶 Hydrogenase | 表达宿主 Expression host | 成熟蛋白 Maturation protein | 全细胞活性 Whole cell activity | 纯化酶活性 Purified enzyme activity | 参考文献 References |
|---|---|---|---|---|---|
| AmaHynSL | A. macleodi ∆HynSL | Host | 0.03 ①,* | 0.1 ③ | [ |
| AmaHynSL | E. coli | A. macleodii | 3×10-3~70×10-3 ①,* | NR | [ |
| AmaHyaAB | T. roseopersicina | Host, A. macleodii | 5×10-3 ① | NR | [ |
| AflHydSL | E. coli | Host | NA | 77 ① | [ |
| DgiHynAB | D. gigas ∆HynAB | Host | 1.9 ①,* | 91 ① | [ |
| DgiHynAB | D. fructosovorans ∆HynAB | Host | 0.2 ② | NR | [ |
| EcoHyd1 | E. coli ∆Hyd1 | Host | 4×10-2~7×10-2 ①,* | 1×10-2~3×10-2 ① | [ |
| HmaMBH | E. coli | E. coli | 0.07 ①,* | 0.03 ① | [ |
| NpuHupSL | E. coli | E. coli | 208 ① | NR | [ |
| PfuSH | E. coli | P. furiosus | 2.9 ① | 100 ① | [ |
| PfuSHI | T. kodakarensis | Host | 23.6 ④ | 880 ④ | [ |
| ReuMBH | R. Eutropha H16 | Host | 1.0 ③,* | 170 ③ | [ |
| ReuMBH | P. stutzeri | R. eutropha | 17~19 ③,* | NR | [ |
| ReuRH | E. coli | R. eutropha | NR | 0.8 ② | [ |
| ReuRH | E. coli | R. eutropha | 1.2 ②,* | 230 ② | [ |
| RopSH | R. eutropha ∆SH ∆MBH | Host, R. opacus | 5.9 ①,* | NR | [ |
| SynSH | E. coli | Synechocystis sp. | 0.04 ①,* | NR | [ |
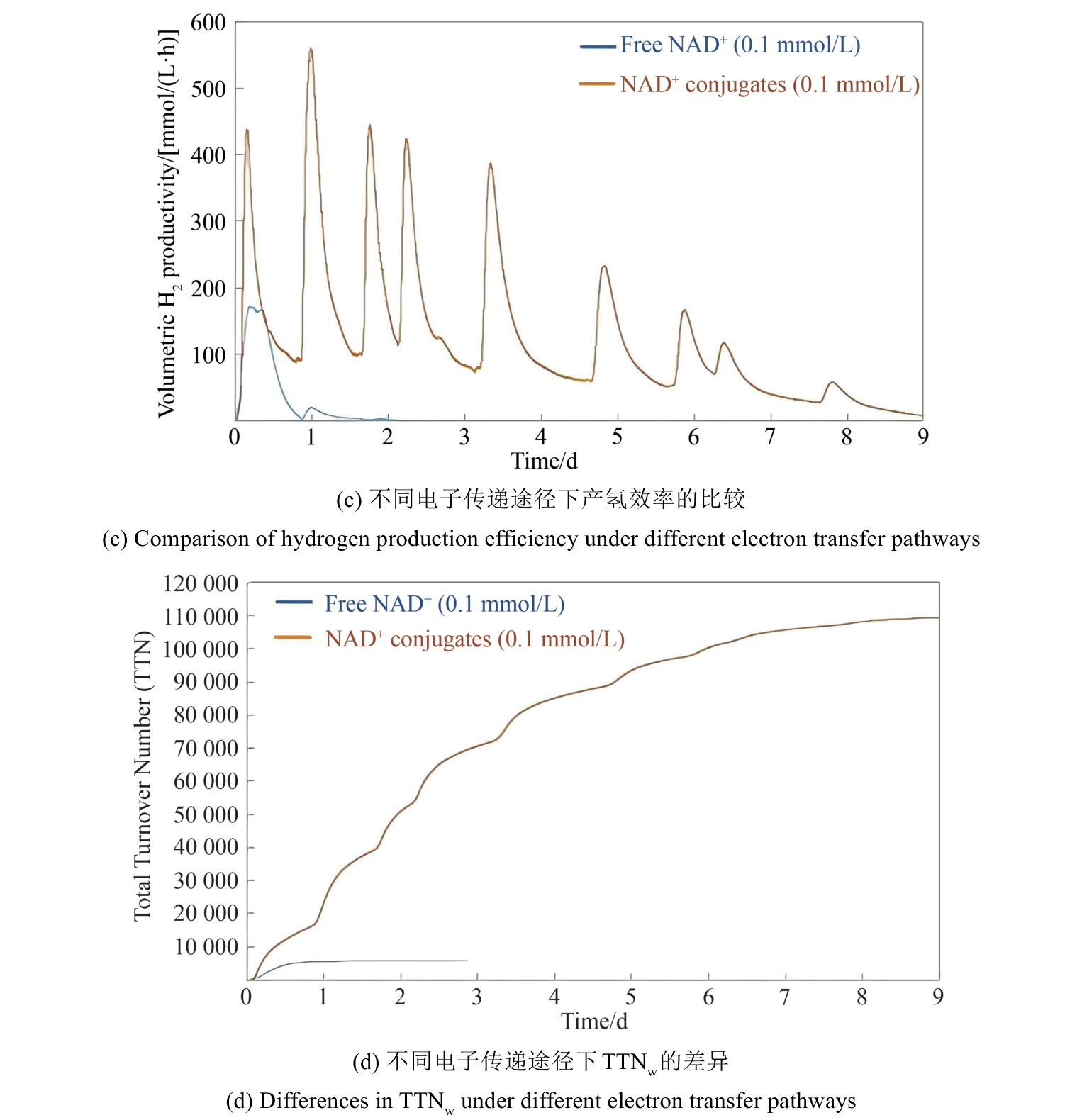
图6 人工电子辅酶对产氢电子传递的效率有提升作用(构建不同的人工电子传递通路对产物合成的影响)G6PDH—葡萄糖6-磷酸脱氢酶;6PGDH—6-磷酸葡萄糖酸脱氢酶;DI—黄递酶;SHⅠ—可溶氢酶Ⅰ;BCV—偶联的苄基紫精
Fig. 6 The application of artificial electron coenzyme to improve the efficiency of electron transport in ivSEB system for hydrogen production(The impact of different artificial electron transfer pathways on the synthesis) G6PDH—Glucose 6-phosphate dehydrogenase; 6PGDH—6-phosphogluconate dehydrogenase; DI—diaphorase; SHⅠ—soluble hydrogenaseⅠ; BCV—the coupled benzyl viologen
| 1 | ARMAROLI N, BALZANI V. The hydrogen issue[J]. ChemSusChem, 2011, 4(1): 21-36. |
| 2 | NIKOLAIDIS P, POULLIKKAS A. A comparative overview of hydrogen production processes[J]. Renewable and Sustainable Energy Reviews, 2017, 67: 597-611. |
| 3 | MIDILLI A, KUCUK H, TOPAL M E, et al. A comprehensive review on hydrogen production from coal gasification: challenges and opportunities[J]. International Journal of Hydrogen Energy, 2021, 46(50): 25385-25412. |
| 4 | HAQ B, SALAHU MUHAMMED N, LIU J S, et al. Enhanced natural gas production using CO2 injection: application to sustainable hydrogen production[J]. Fuel, 2023, 347: 128474. |
| 5 | YANG K, GU Z H, LONG Y H, et al. Hydrogen production via chemical looping reforming of coke oven gas[J]. Green Energy & Environment, 2021, 6(5): 678-692. |
| 6 | ZHANG L N, LI R, ZANG H Y, et al. Advanced hydrogen evolution electrocatalysts promising sustainable hydrogen and chlor-alkali co-production[J]. Energy & Environmental Science, 2021, 14(12): 6191-6210. |
| 7 | NIBLETT D, DELPISHEH M, RAMAKRISHNAN S, et al. Review of next generation hydrogen production from offshore wind using water electrolysis[J]. Journal of Power Sources, 2024, 592: 233904. |
| 8 | WANG S, LU A L, ZHONG C J. Hydrogen production from water electrolysis: role of catalysts[J]. Nano Convergence, 2021, 8(1): 4. |
| 9 | BALAT H, KIRTAY E. Hydrogen from biomass - present scenario and future prospects[J]. International Journal of Hydrogen Energy, 2010, 35(14): 7416-7426. |
| 10 | KIM S H, KUMAR G, CHEN W H, et al. Renewable hydrogen production from biomass and wastes (ReBioH2-2020)[J]. Bioresource Technology, 2021, 331: 125024. |
| 11 | REDDING K E, APPEL J, BOEHM M, et al. Advances and challenges in photosynthetic hydrogen production[J]. Trends in Biotechnology, 2022, 40(11): 1393. |
| 12 | SRIRANGAN K, PYNE M E, CHOU C P. Biochemical and genetic engineering strategies to enhance hydrogen production in photosynthetic algae and cyanobacteria[J]. Bioresource Technology, 2011, 102(18): 8589-8604. |
| 13 | 张以恒. 中国哲学思想“道法术器”对生物制造的启示[J]. 合成生物学, 2023, 5(6):1231-1241. |
| ZHANG Y-H P J. The enlightenment of the Chinese philosophy “Tao-Fa-Shu-Qi” to industrial biomanufacturing [J]. Synthetic Biology Journal, 2023, 5(6):1231-1241. | |
| 14 | SAMBUSITI C, BELLUCCI M, ZABANIOTOU A, et al. Algae as promising feedstocks for fermentative biohydrogen production according to a biorefinery approach: a comprehensive review[J]. Renewable and Sustainable Energy Reviews, 2015, 44: 20-36. |
| 15 | ZHANG T, JIANG D P, ZHANG H, et al. Comparative study on bio-hydrogen production from corn stover: photo-fermentation, dark-fermentation and dark-photo co-fermentation[J]. International Journal of Hydrogen Energy, 2020, 45(6): 3807-3814. |
| 16 | THAUER R K, JUNGERMANN K, DECKER K. Energy conservation in chemotrophic anaerobic bacteria[J]. Bacteriological Reviews, 1977, 41(1): 100-180. |
| 17 | AKAÇIN İ, ERSOY Ş, KESKIN T, et al. Optimizing biohydrogen production yields by employing locally isolated thermophilic bacteria from hot springs[J]. International Journal of Hydrogen Energy, 2024, 52: 502-510. |
| 18 | XING D F, REN N Q, WANG A J, et al. Continuous hydrogen production of auto-aggregative Ethanoligenens harbinense YUAN-3 under non-sterile condition[J]. International Journal of Hydrogen Energy, 2008, 33(5): 1489-1495. |
| 19 | SINGH R, WHITE D, DEMIREL Y, et al. Uncoupling fermentative synthesis of molecular hydrogen from biomass formation in Thermotoga maritima [J]. Applied and Environmental Microbiology, 2018, 84(17): e00998-18. |
| 20 | YE X H, WANG Y R, HOPKINS R C, et al. Spontaneous high-yield production of hydrogen from cellulosic materials and water catalyzed by enzyme cocktails[J]. ChemSusChem, 2009, 2(2): 149-152. |
| 21 | ZHANG Y H, EVANS B R, MIELENZ J R, et al. High-yield hydrogen production from starch and water by a synthetic enzymatic pathway[J]. PLoS One, 2007, 2(5): e456. |
| 22 | ZHANG Y H P. Using extremophile enzymes to generate hydrogen for electricity[J]. Microbe Magazine, 2009, 4(12): 560-565. |
| 23 | Call for views: synthetic biology [OL]. The Royal Society, 2023,6: 2. . |
| 24 | CORREDOR J, HARANKAHAGE D, GLOAGUEN F, et al. Influence of QD photosensitizers in the photocatalytic production of hydrogen with biomimetic[FeFe]-hydrogenase. Comparative performance of CdSe and CdTe[J]. Chemosphere, 2021, 278: 130485. |
| 25 | ZHANG L Y, MORELLO G, CARR S B, et al. Aerobic photocatalytic H2 production by a [NiFe] hydrogenase engineered to place a silver nanocluster in the electron relay[J]. Journal of the American Chemical Society, 2020, 142(29): 12699-12707. |
| 26 | EDWARDES MOORE E, ANDREI V, ZACARIAS S, et al. Integration of a hydrogenase in a lead halide perovskite photoelectrode for tandem solar water splitting[J]. ACS Energy Letters, 2020, 5(1): 232-237. |
| 27 | SAKAI T, MERSCH D, REISNER E. Photocatalytic hydrogen evolution with a hydrogenase in a mediator-free system under high levels of oxygen[J]. Angewandte Chemie International Edition, 2013, 52(47): 12313-12316. |
| 28 | JI H S, WAN L, GAO Y X, et al. Hydrogenase as the basis for green hydrogen production and utilization[J]. Journal of Energy Chemistry, 2023, 85: 348-362. |
| 29 | KIM E J, KIM J E, ZHANG Y H P J. Ultra-rapid rates of water splitting for biohydrogen gas production through in vitro artificial enzymatic pathways[J]. Energy & Environmental Science, 2018, 11(8): 2064-2072. |
| 30 | LUBITZ W, OGATA H, RÜDIGER O, et al. Hydrogenases [J]. Chemical Reviews, 2014, 114(8): 4081-148. |
| 31 | STEPHENSON M, STICKLAND L H. Hydrogenase: a bacterial enzyme activating molecular hydrogen: the properties of the enzyme[J]. Biochemical Journal, 1931, 25(1): 205-214. |
| 32 | FRIEDRICH B, FRITSCH J, LENZ O. Oxygen-tolerant hydrogenases in hydrogen-based technologies[J]. Current Opinion in Biotechnology, 2011, 22(3): 358-364. |
| 33 | ARRIAZA-GALLARDO F J, ZHENG Y C, GEHL M, et al. [Fe]-hydrogenase, cofactor biosynthesis and engineering[J]. ChemBioChem, 2023, 24(20): e202300330. |
| 34 | PICHÉ-CHOQUETTE S, CONSTANT P. Molecular hydrogen, a neglected key driver of soil biogeochemical processes[J]. Applied and Environmental Microbiology, 2019, 85(6): e02418-18. |
| 35 | BEATON S E, EVANS R M, FINNEY A J, et al. The structure of hydrogenase-2 from Escherichia coli: implications for H2-driven proton pumping[J]. Biochemical Journal, 2018, 475(7): 1353-1370. |
| 36 | ROHAC R, MARTIN L, LIU L, et al. Crystal structure of the [FeFe]-hydrogenase maturase HydE bound to complex-B[J]. Journal of the American Chemical Society, 2021, 143(22): 8499-8508. |
| 37 | MAEDA T, SANCHEZ-TORRES V, WOOD T K. Hydrogen production by recombinant Escherichia coli strains[J]. Microbial Biotechnology, 2012, 5(2): 214-225. |
| 38 | ANWAR M, LOU S L, CHEN L, et al. Recent advancement and strategy on bio-hydrogen production from photosynthetic microalgae[J]. Bioresource Technology, 2019, 292: 121972. |
| 39 | VALLE A, CANTERO D, BOLÍVAR J. Metabolic engineering for the optimization of hydrogen production in Escherichia coli: a review[J]. Biotechnology Advances, 2019, 37(5): 616-633. |
| 40 | XUAN J S, HE L L, WEN W, et al. Hydrogenase and nitrogenase: key catalysts in biohydrogen production[J]. Molecules, 2023, 28(3): 1392. |
| 41 | WANG Y M, SONG Y H, MA C L, et al. A heterologously-expressed thermostable Pyrococcus furiosus cytoplasmic [NiFe]-hydrogenaseⅠused as the catalyst of H2/air biofuel cells[J]. International Journal of Hydrogen Energy, 2021, 46(4): 3035-3044. |
| 42 | JENNEY F E JR, ADAMS M W W. Hydrogenases of the model hyperthermophiles[J]. Annals of the New York Academy of Sciences, 2008, 1125: 252-266. |
| 43 | SUN J S, HOPKINS R C, JENNEY F E, et al. Heterologous expression and maturation of an NADP-dependent [NiFe]- hydrogenase: a key enzyme in biofuel production[J]. PLoS One, 2010, 5(5): e10526. |
| 44 | CHANDRAYAN S K, MCTERNAN P M, HOPKINS R C, et al. Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase[J]. Journal of Biological Chemistry, 2012, 287(5): 3257-3264. |
| 45 | SONG Y H, LIU M X, XIE L P, et al. A recombinant 12-His tagged Pyrococcus furiosus soluble [NiFe]-hydrogenaseⅠoverexpressed in Thermococcus kodakarensis KOD1 facilitates hydrogen-powered in vitro NADH regeneration[J]. Biotechnology Journal, 2019, 14(4): e1800301. |
| 46 | MEYER J. [FeFe] hydrogenases and their evolution: a genomic perspective[J]. Cellular and Molecular Life Sciences, 2007, 64(9): 1063-1084. |
| 47 | GREENING C, CABOTAJE P R, VALENTIN ALVARADO L E, et al. Minimal and hybrid hydrogenases are active from Archaea[J]. Cell, 2024, 187(13): 3357-3372.e19. |
| 48 | KIM E J, WU C H, ADAMS M W, et al. Exceptionally high rates of biological hydrogen production by biomimetic in vitro synthetic enzymatic pathways[J]. Chemistry, 2016, 22(45): 16047-16051. |
| 49 | KOO J, SHIIGI S, ROHOVIE M, et al. Characterization of [FeFe] hydrogenase O2 sensitivity using a new, physiological approach[J]. Journal of Biological Chemistry, 2016, 291(41): 21563-21570. |
| 50 | SHIMA S, THAUER R K. A third type of hydrogenase catalyzing H2 activation[J]. Chemical Record, 2007, 7(1): 37-46. |
| 51 | LEONE L, SGUEGLIA G, LA GATTA S, et al. Enzymatic and bioinspired systems for hydrogen production[J]. International Journal of Molecular Sciences, 2023, 24(10): 8605. |
| 52 | WEI W, SUN P Q, LI Z, et al. A surface-display biohybrid approach to light-driven hydrogen production in air[J]. Science Advances, 2018, 4(2): eaap9253. |
| 53 | SZCZESNY J, BIRRELL J A, CONZUELO F, et al. Redox-polymer-based high-current-density gas-diffusion H2-oxidation bioanode using [FeFe] hydrogenase from Desulfovibrio desulfuricans in a membrane-free biofuel cell[J]. Angewandte Chemie International Edition, 2020, 59(38): 16506-16510. |
| 54 | AL-SHAMERI A, SIEBERT D L, SUTIONO S, et al. Hydrogenase-based oxidative biocatalysis without oxygen[J]. Nature Communications, 2023, 14: 2693. |
| 55 | LI F, LUTZ P B, PEPELYAYEVA Y, et al. Redox active motifs in selenoproteins[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(19): 6976-6981. |
| 56 | CHUNG C Z, KRAHN N. The selenocysteine toolbox: a guide to studying the 21st amino acid[J]. Archives of Biochemistry and Biophysics, 2022, 730: 109421. |
| 57 | HONDAL R J, MARINO S M, GLADYSHEV V N. Selenocysteine in thiol/disulfide-like exchange reactions[J]. Antioxidants & Redox Signaling, 2013, 18(13): 1675-1689. |
| 58 | MARQUES M C, TAPIA C, GUTIÉRREZ-SANZ O, et al. The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis[J]. Nature Chemical Biology, 2017, 13(5): 544-550. |
| 59 | BROOKE E J, EVANS R M, ISLAM S T A, et al. Importance of the active site “canopy” residues in an O2-tolerant [NiFe]-hydrogenase[J]. Biochemistry, 2017, 56(1): 132-142. |
| 60 | MCGUIRE A D, GENET H, LYU Z, et al. Assessing historical and projected carbon balance of Alaska: a synthesis of results and policy/management implications[J]. Ecological Applications, 2018, 28(6): 1396-1412. |
| 61 | PATEL A, MULDER D W, SÖLL D, et al. Harnessing selenocysteine to enhance microbial cell factories for hydrogen production[J]. Frontiers in Catalysis, 2022, 2: 1089176. |
| 62 | ABOU HAMDAN A, BURLAT B, GUTIÉRREZ-SANZ O, et al. O2-independent formation of the inactive states of NiFe hydrogenase[J]. Nature Chemical Biology, 2013, 9(1): 15-17. |
| 63 | FRITSCH J, LENZ O, FRIEDRICH B. Structure, function and biosynthesis of O₂-tolerant hydrogenases[J]. Nature Reviews Microbiology, 2013, 11(2): 106-114. |
| 64 | PANDELIA M E, NITSCHKE W, INFOSSI P, et al. Characterization of a unique [FeS] cluster in the electron transfer chain of the oxygen tolerant [NiFe] hydrogenase from Aquifex aeolicus [J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(15): 6097-6102. |
| 65 | WULFF P, THOMAS C, SARGENT F, et al. How the oxygen tolerance of a [NiFe]-hydrogenase depends on quaternary structure[J]. Journal of Biological Inorganic Chemistry, 2016, 21(1): 121-134. |
| 66 | KUBAS A, ORAIN C, DE SANCHO D, et al. Mechanism of O2 diffusion and reduction in FeFe hydrogenases[J]. Nature Chemistry, 2017, 9(1): 88-95. |
| 67 | STRIPP S T, GOLDET G, BRANDMAYR C, et al. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(41): 17331-17336. |
| 68 | VINCENT K A, PARKIN A, LENZ O, et al. Electrochemical definitions of O2 sensitivity and oxidative inactivation in hydrogenases[J]. Journal of the American Chemical Society, 2005, 127(51): 18179-18189. |
| 69 | ORAIN C, SAUJET L, GAUQUELIN C, et al. Electrochemical measurements of the kinetics of inhibition of two FeFe hydrogenases by O2 demonstrate that the reaction is partly reversible[J]. Journal of the American Chemical Society, 2015, 137(39): 12580-12587. |
| 70 | BINGHAM A S, SMITH P R, SWARTZ J R. Evolution of an [FeFe] hydrogenase with decreased oxygen sensitivity[J]. International Journal of Hydrogen Energy, 2012, 37(3): 2965-2976. |
| 71 | FONTECILLA-CAMPS J C, VOLBEDA A, CAVAZZA C, et al. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases[J]. Chemical Reviews, 2007, 107(10): 4273-4303. |
| 72 | PAPINI C, SOMMER C, PECQUEUR L, et al. Bioinspired artificial [FeFe]-hydrogenase with a synthetic H-cluster[J]. ACS Catalysis, 2019, 9(5): 4495-4501. |
| 73 | LIEBGOTT P P, LEROUX F, BURLAT B, et al. Relating diffusion along the substrate tunnel and oxygen sensitivity in hydrogenase[J]. Nature Chemical Biology, 2010, 6(1): 63-70. |
| 74 | BAFFERT C, DEMUEZ M, COURNAC L, et al. Hydrogen-activating enzymes: activity does not correlate with oxygen sensitivity[J]. Angewandte Chemie International Edition, 2008, 47(11): 2052-2054. |
| 75 | LAMPRET O, DUAN J F, HOFMANN E, et al. The roles of long-range proton-coupled electron transfer in the directionality and efficiency of [FeFe]-hydrogenases[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(34): 20520-20529. |
| 76 | RUTZ A, DAS C K, FASANO A, et al. Increasing the O2 resistance of the [FeFe]-hydrogenase CbA5H through enhanced protein flexibility[J]. ACS Catalysis, 2023, 13(2): 856-865. |
| 77 | LU Y, KOO J. O2 sensitivity and H2 production activity of hydrogenases - a review[J]. Biotechnology and Bioengineering, 2019, 116(11): 3124-3135. |
| 78 | CRACKNELL J A, WAIT A F, LENZ O, et al. A kinetic and thermodynamic understanding of O2 tolerance in [NiFe]- hydrogenases[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(49): 20681-20686. |
| 79 | GORIS T, WAIT A F, SAGGU M, et al. A unique iron-sulfur cluster is crucial for oxygen tolerance of a [NiFe]-hydrogenase[J]. Nature Chemical Biology, 2011, 7(5): 310-318. |
| 80 | WULFF P, DAY C C, SARGENT F, et al. How oxygen reacts with oxygen-tolerant respiratory [NiFe]-hydrogenases[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(18): 6606-6611. |
| 81 | FRIELINGSDORF S, FRITSCH J, SCHMIDT A, et al. Reversible [4Fe-3S]cluster morphing in an O2-tolerant [NiFe] hydrogenase[J]. Nature Chemical Biology, 2014, 10(5): 378-385. |
| 82 | FRITSCH J, SCHEERER P, FRIELINGSDORF S, et al. The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre[J]. Nature, 2011, 479(7372): 249-252. |
| 83 | SIEGBAHN P E M, TYE J W, HALL M B. Computational studies of [NiFe] and [FeFe] hydrogenases[J]. Chemical Reviews, 2007, 107(10): 4414-4435. |
| 84 | SÖDERHJELM P, RYDE U. Combined computational and crystallographic study of the oxidised states of [NiFe] hydrogenase[J]. Journal of Molecular Structure: THEOCHEM, 2006, 770(1/2/3): 199-219. |
| 85 | STEIN M, LUBITZ W. Quantum chemical calculations of [NiFe] hydrogenase[J]. Current Opinion in Chemical Biology, 2002, 6(2): 243-249. |
| 86 | FLANAGAN L A, CHIDWICK H S, WALTON J, et al. Conserved histidine adjacent to the proximal cluster tunes the anaerobic reductive activation of Escherichia coli membrane-bound [NiFe] hydrogenase-1[J]. ChemElectroChem, 2018, 5(6): 855-860. |
| 87 | VOLBEDA A, MOUESCA J M, DARNAULT C, et al. X-ray structural, functional and computational studies of the O2-sensitive E. coli hydrogenase-1 C19G variant reveal an unusual [4Fe-4S] cluster[J]. Chemical Communications, 2018, 54(52): 7175-7178. |
| 88 | RODRÍGUEZ-MACIÁ P, REIJERSE E J, VAN GASTEL M, et al. Sulfide protects [FeFe] hydrogenases from O2 [J]. Journal of the American Chemical Society, 2018, 140(30): 9346-9350. |
| 89 | XIONG W, ZHAO X H, ZHU G X, et al. Silicification-induced cell aggregation for the sustainable production of H2 under aerobic conditions[J]. Angewandte Chemie International Edition, 2015, 54(41): 11961-11965. |
| 90 | PLUMERÉ N, RÜDIGER O, OUGHLI A A, et al. A redox hydrogel protects hydrogenase from high-potential deactivation and oxygen damage[J]. Nature Chemistry, 2014, 6(9): 822-827. |
| 91 | EILENBERG H, WEINER I, BEN-ZVI O, et al. The dual effect of a ferredoxin-hydrogenase fusion protein in vivo: successful divergence of the photosynthetic electron flux towards hydrogen production and elevated oxygen tolerance[J]. Biotechnology for Biofuels, 2016, 9(1): 182. |
| 92 | LI T P, JIANG Q Y, HUANG J F, et al. Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production[J]. Nature Communications, 2020, 11(1): 5448. |
| 93 | GHIRARDI M L. Implementation of photobiological H2 production: the O2 sensitivity of hydrogenases[J]. Photosynthesis Research, 2015, 125(3): 383-393. |
| 94 | WECKER M S A, GHIRARDI M L. High-throughput biosensor discriminates between different algal H2-photoproducing strains[J]. Biotechnology and Bioengineering, 2014, 111(7): 1332-1340. |
| 95 | KOO J, SCHNABEL T, LIONG S, et al. High-throughput screening of catalytic H2 production[J]. Angewandte Chemie International Edition, 2017, 56(4): 1012-1016. |
| 96 | ATTA M, MEYER J. Characterization of the gene encoding the [Fe]-hydrogenase from Megasphaera elsdenii [J]. Biochimica et Biophysica Acta, 2000, 1476(2): 368-371. |
| 97 | VOORDOUW G, HAGEN W R, KRÜSE-WOLTERS K M, et al. Purification and characterization of Desulfovibrio vulgaris (Hildenborough) hydrogenase expressed in Escherichia coli [J]. European Journal of Biochemistry, 1987, 162(1): 31-36. |
| 98 | ASADA Y, KOIKE Y, SCHNACKENBERG J, et al. Heterologous expression of clostridial hydrogenase in the Cyanobacterium synechococcus PCC7942[J]. Biochimica et Biophysica Acta, 2000, 1490(3): 269-278. |
| 99 | GORWA M F, CROUX C, SOUCAILLE P. Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824[J]. Journal of Bacteriology, 1996, 178(9): 2668-2675. |
| 100 | DEMUEZ M, COURNAC L, GUERRINI O, et al. Complete activity profile of Clostridium acetobutylicum [FeFe]-hydrogenase and kinetic parameters for endogenous redox partners[J]. FEMS Microbiology Letters, 2007, 275(1): 113-121. |
| 101 | KING P W, POSEWITZ M C, GHIRARDI M L, et al. Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system[J]. Journal of Bacteriology, 2006, 188(6): 2163-2172. |
| 102 | AKHTAR M K, JONES P R. Deletion of iscR stimulates recombinant clostridial Fe-Fe hydrogenase activity and H2-accumulation in Escherichia coli BL21(DE3)[J]. Applied Microbiology and Biotechnology, 2008, 78(5): 853-862. |
| 103 | DUCAT D C, SACHDEVA G, SILVER P A. Rewiring hydrogenase-dependent redox circuits in cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(10): 3941-3946. |
| 104 | SUBUDHI S, LAL B. Fermentative hydrogen production in recombinant Escherichia coli harboring a [FeFe]-hydrogenase gene isolated from Clostridium butyricum [J]. International Journal of Hydrogen Energy, 2011, 36(21): 14024-14030. |
| 105 | ADAMS M W W, MORTENSON L E. The purification of hydrogenase Ⅱ (uptake hydrogenase) from the anaerobic N2-fixing bacterium Clostridium pasteurianum [J]. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 1984, 766(1): 51-61. |
| 106 | KUCHENREUTHER J M, GRADY-SMITH C S, BINGHAM A S, et al. High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli [J]. PLoS One, 2010, 5(11): e15491. |
| 107 | KAMP C, SILAKOV A, WINKLER M, et al. Isolation and first EPR characterization of the [FeFe]-hydrogenases from green algae[J]. Biochimica et Biophysica Acta, 2008, 1777(5): 410-416. |
| 108 | GIRBAL L, VON ABENDROTH G, WINKLER M, et al. Homologous and heterologous overexpression in Clostridium acetobutylicum and characterization of purified clostridial and algal Fe-only hydrogenases with high specific activities[J]. Applied and Environmental Microbiology, 2005, 71(5): 2777-2781. |
| 109 | VON ABENDROTH G, STRIPP S, SILAKOV A, et al. Optimized over-expression of [FeFe] hydrogenases with high specific activity in Clostridium acetobutylicum [J]. International Journal of Hydrogen Energy, 2008, 33(21): 6076-6081. |
| 110 | POSEWITZ M C, KING P W, SMOLINSKI S L, et al. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase[J]. Journal of Biological Chemistry, 2004, 279(24): 25711-25720. |
| 111 | YACOBY I, TEGLER L T, POCHEKAILOV S, et al. Optimized expression and purification for high-activity preparations of algal [FeFe]-hydrogenase[J]. PLoS One, 2012, 7(4): e35886. |
| 112 | SYBIRNA K, ANTOINE T, LINDBERG P, et al. Shewanella oneidensis: a new and efficient system for expression and maturation of heterologous [Fe-Fe] hydrogenase from Chlamydomonas reinhardtii [J]. BMC Biotechnology, 2008, 8: 73. |
| 113 | BERTO P, D’ADAMO S, BERGANTINO E, et al. The Cyanobacterium synechocystis sp. PCC 6803 is able to express an active [FeFe]-hydrogenase without additional maturation proteins[J]. Biochemical and Biophysical Research Communications, 2011, 405(4): 678-683. |
| 114 | KELLY C L, PINSKE C, MURPHY B J, et al. Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli [J]. Biotechnology Reports, 2015, 8: 94-104. |
| 115 | ZHAO X, XING D F, ZHANG L, et al. Characterization and overexpression of a [FeFe]-hydrogenase gene of a novel hydrogen-producing bacterium Ethanoligenens harbinense [J]. International Journal of Hydrogen Energy, 2010, 35(18): 9598-9602. |
| 116 | NIXON J E, FIELD J, MCARTHUR A G, et al. Iron-dependent hydrogenases of Entamoeba histolytica and Giardia lamblia: activity of the recombinant entamoebic enzyme and evidence for lateral gene transfer[J]. The Biological Bulletin, 2003, 204(1): 1-9. |
| 117 | INOUE J I, SAITA K, KUDO T, et al. Hydrogen production by termite gut protists: characterization of iron hydrogenases of parabasalian symbionts of the termite Coptotermes formosanus [J]. Eukaryotic Cell, 2007, 6(10): 1925-1932. |
| 118 | GÄRTNER K, LECHNO-YOSSEF S, CORNISH A J, et al. Expression of Shewanella oneidensis MR-1[FeFe]- hydrogenase genes in Anabaena sp. strain PCC 7120[J]. Applied and Environmental Microbiology, 2012, 78(24): 8579-8586. |
| 119 | VARGAS W A, WEYMAN P D, TONG Y K, et al. [NiFe] hydrogenase from Alteromonas macleodii with unusual stability in the presence of oxygen and high temperature[J]. Applied and Environmental Microbiology, 2011, 77(6): 1990-1998. |
| 120 | WEYMAN P D, VARGAS W A, CHUANG R Y, et al. Heterologous expression of Alteromonas macleodii and Thiocapsa roseopersicina [NiFe] hydrogenases in Escherichia coli [J]. Microbiology, 2011, 157(Pt 5): 1363-1374. |
| 121 | YONEMOTO I T, MATTERI C W, NGUYEN T A, et al. Dual organism design cycle reveals small subunit substitutions that improve [NiFe] hydrogenase hydrogen evolution[J]. Journal of Biological Engineering, 2013, 7(1): 17. |
| 122 | MARÓTI G, TONG Y K, YOOSEPH S, et al. Discovery of [NiFe] hydrogenase genes in metagenomic DNA cloning and heterologous expression in Thiocapsa roseopersicina [J]. Applied and Environmental Microbiology, 2009, 75(18): 5821-5830. |
| 123 | MURA G M, PEDRONI P, PRATESI C, et al. The [Ni-Fe] hydrogenase from the thermophilic bacterium Acetomicrobium flavidum [J]. Microbiology, 1996, 142( Pt 4): 829-836. |
| 124 | HATCHIKIAN E C, BRUSCHI M, LE GALL J. Characterization of the periplasmic hydrogenase from Desulfovibrio gigas [J]. Biochemical and Biophysical Research Communications, 1978, 82(2): 451-461. |
| 125 | ROUSSET M, MAGRO V, FORGET N, et al. Heterologous expression of the Desulfovibrio gigas [NiFe] hydrogenase in Desulfovibrio fructosovorans MR400[J]. Journal of Bacteriology, 1998, 180(18): 4982-4986. |
| 126 | KIM J Y H, JO B H, CHA H J. Production of biohydrogen by recombinant expression of [NiFe]-hydrogenase 1 in Escherichia coli [J]. Microbial Cell Factories, 2010, 9: 54. |
| 127 | KIM J Y H, JO B H, CHA H J. Production of biohydrogen by heterologous expression of oxygen-tolerant Hydrogenovibrio marinus [NiFe]-hydrogenase in Escherichia coli [J]. Journal of Biotechnology, 2011, 155(3): 312-319. |
| 128 | RALEIRAS P, KHANNA N, MIRANDA H, et al. Turning around the electron flow in an uptake hydrogenase. EPR spectroscopy and in vivo activity of a designed mutant in HupSL from Nostoc punctiforme [J]. Energy & Environmental Science, 2016, 9(2): 581-594. |
| 129 | SCHINK B, SCHLEGEL H G. The membrane-bound hydrogenase of Alcaligenes eutrophus.Ⅰ. Solubilization, purification, and biochemical properties[J]. Biochimica et Biophysica Acta, 1979, 567(2): 315-324. |
| 130 | LENZ O, GLEICHE A, STRACK A, et al. Requirements for heterologous production of a complex metalloenzyme: the membrane-bound [NiFe] hydrogenase[J]. Journal of Bacteriology, 2005, 187(18): 6590-6595. |
| 131 | BUHRKE T, LENZ O, KRAUSS N, et al. Oxygen tolerance of the H2-sensing [NiFe] hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site[J]. Journal of Biological Chemistry, 2005, 280(25): 23791-23796. |
| 132 | SCHIFFELS J, PINKENBURG O, SCHELDEN M, et al. An innovative cloning platform enables large-scale production and maturation of an oxygen-tolerant [NiFe]-hydrogenase from Cupriavidus necator in Escherichia coli [J]. PLoS One, 2013, 8(7): e68812. |
| 133 | PORTHUN A, BERNHARD M, FRIEDRICH B. Expression of a functional NAD-reducing [NiFe] hydrogenase from the gram-positive Rhodococcus opacus in the Gram-negative Ralstonia eutropha [J]. Archives of Microbiology, 2002, 177(2): 159-166. |
| 134 | WELLS M A, MERCER J, MOTT R A, et al. Engineering a non-native hydrogen production pathway into Escherichia coli via a cyanobacterial [NiFe] hydrogenase[J]. Metabolic Engineering, 2011, 13(4): 445-453. |
| 135 | PREISSLER J, WAHLEFELD S, LORENT C, et al. Enzymatic and spectroscopic properties of a thermostable [NiFe]- hydrogenase performing H2-driven NAD+-reduction in the presence of O2 [J]. Biochimica et Biophysica Acta Bioenergetics, 2018, 1859(1): 8-18. |
| 136 | RIECKENBERG F, GÖTZ K, HILTERHAUS L, et al. Strategies for reliable and improved large-scale production of Pyrococcus furiosus with integrated purification of hydrogenaseⅠ[J]. Bioprocess and Biosystems Engineering, 2014, 37(12): 2475-2482. |
| 137 | SOBOH B, LINDENSTRAUSS U, GRANICH C, et al. [NiFe]-hydrogenase maturation in vitro: analysis of the roles of the HybG and HypD accessory proteins 1[J]. Biochemical Journal, 2014, 464(2): 169-177. |
| 138 | PAGNIER A, BALCI B, SHEPARD E M, et al. [FeFe]- hydrogenase in vitro maturation[J]. Angewandte Chemie International Edition, 2022, 61(49): e202212074. |
| 139 | KÖHLER V, WILSON Y M, DÜRRENBERGER M, et al. Synthetic cascades are enabled by combining biocatalysts with artificial metalloenzymes[J]. Nature Chemistry, 2013, 5(2): 93-99. |
| 140 | MAITY B, TAHER M, MAZUMDAR S, et al. Artificial metalloenzymes based on protein assembly[J]. Coordination Chemistry Reviews, 2022, 469: 214593. |
| 141 | ONODA A, HAYASHI T. Artificial hydrogenase: biomimetic approaches controlling active molecular catalysts[J]. Current Opinion in Chemical Biology, 2015, 25: 133-140. |
| 142 | VOLBEDA A, CHARON M H, PIRAS C, et al. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas [J]. Nature, 1995, 373(6515): 580-587. |
| 143 | DARAOSHEH A Q, ABUL-FUTOUH H, MURAKAMI N, et al. Novel [FeFe]-hydrogenase mimics: unexpected course of the reaction of ferrocenyl α-thienyl thioketone with Fe3(CO)12 [J]. Materials, 2022, 15(8): 2867. |
| 144 | LAND H, SENGER M, BERGGREN G, et al. Current state of [FeFe]-hydrogenase research: biodiversity and spectroscopic investigations[J]. ACS Catalysis, 2020, 10(13): 7069-7086. |
| 145 | 石婷, 宋展, 宋世怡, 等. 体外生物转化(ivBT):生物制造的新前沿 [J]. 合成生物学, 2024, 5(6):1437-1460. |
| SHI T, SONG Z, SONG S Y, et al. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6):1437-1460. | |
| 146 | KIM J E, KIM E J, CHEN H, et al. Advanced water splitting for green hydrogen gas production through complete oxidation of starch by in vitro metabolic engineering[J]. Metabolic Engineering, 2017, 44: 246-252. |
| 147 | BOMMARIUS A S, PAYE M F. Stabilizing biocatalysts[J]. Chemical Society Reviews, 2013, 42(15): 6534-6565. |
| 148 | ZHANG Y H P. Production of biocommodities and bioelectricity by cell-free synthetic enzymatic pathway biotransformations: challenges and opportunities[J]. Biotechnology and Bioengineering, 2010, 105(4): 663-677. |
| 149 | ROLLIN J A, MARTIN DEL CAMPO J, MYUNG S, et al. High-yield hydrogen production from biomass by in vitro metabolic engineering: mixed sugars coutilization and kinetic modeling[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(16): 4964-4969. |
| 150 | 雷航彬, 何宁, 李斐煊,等. 氢化酶固定化研究进展 [J]. 合成生物学, 2024, 5(6):1485-1497. |
| LEI H B, HE N, LI F X, et al. Advance in the immobilization of hydrogenases [J]. Synthetic Biology Journal, 2024, 5(6):1485-1497. | |
| 151 | ROSANO G L, MORALES E S, CECCARELLI E A. New tools for recombinant protein production in Escherichia coli: a 5-year update[J]. Protein Science, 2019, 28(8): 1412-1422. |
| 152 | ZHOU Y L, LU Z H, WANG X, et al. Genetic engineering modification and fermentation optimization for extracellular production of recombinant proteins using Escherichia coli [J]. Applied Microbiology and Biotechnology, 2018, 102(4): 1545-1556. |
| 153 | LIU J H, LI H L, ZHAO G R, et al. Redox cofactor engineering in industrial microorganisms: strategies, recent applications and future directions[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(5): 313-327. |
| 154 | COVARRUBIAS A J, PERRONE R, GROZIO A, et al. NAD+ metabolism and its roles in cellular processes during ageing[J]. Nature Reviews Molecular Cell Biology, 2021, 22(2): 119-141. |
| 155 | ESTRELLA M A, DU J, CHEN L, et al. The metabolites NADP+ and NADPH are the targets of the circadian protein Nocturnin (Curled)[J]. Nature Communications, 2019, 10(1): 2367. |
| 156 | HOLM A K, BLANK L M, OLDIGES M, et al. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli [J]. Journal of Biological Chemistry, 2010, 285(23): 17498-17506. |
| 157 | ANDERSON B M, KAPLAN N O. Enzymatic studies with analogues of diphosphopyridine nucleotide[J]. Journal of Biological Chemistry, 1959, 234(5): 1226-1232. |
| 158 | LIU Y X, LI Q, WANG L, et al. Engineering D-lactate dehydrogenase to favor an non-natural cofactor nicotinamide cytosine dinucleotide[J]. ChemBioChem, 2020, 21(14): 1972-1975. |
| 159 | GUO X J, LIU Y X, WANG Q, et al. Non-natural cofactor and formate-driven reductive carboxylation of pyruvate[J]. Angewandte Chemie International Edition, 2020, 59(8): 3143-3146. |
| 160 | NOWAK C, PICK A, LOMMES P, et al. Enzymatic reduction of nicotinamide biomimetic cofactors using an engineered glucose dehydrogenase: providing a regeneration system for artificial cofactors[J]. ACS Catalysis, 2017, 7(8): 5202-5208. |
| 161 | YOU C, HUANG R, WEI X L, et al. Protein engineering of oxidoreductases utilizing nicotinamide-based coenzymes, with applications in synthetic biology[J]. Synthetic and Systems Biotechnology, 2017, 2(3): 208-218. |
| 162 | BLACK W B, ZHANG L Y, MAK W S, et al. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis[J]. Nature Chemical Biology, 2020, 16(1): 87-94. |
| 163 | HUANG R, CHEN H, UPP D M, et al. A high-throughput method for directed evolution of NAD(P)+-dependent dehydrogenases for the reduction of biomimetic nicotinamide analogues[J]. ACS Catalysis, 2019, 9(12): 11709-11719. |
| 164 | ZACHOS I, NOWAK C, SIEBER V. Biomimetic cofactors and methods for their recycling[J]. Current Opinion in Chemical Biology, 2019, 49: 59-66. |
| 165 | JI D B, WANG L, HOU S H, et al. Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide[J]. Journal of the American Chemical Society, 2011, 133(51): 20857-20862. |
| 166 | SHOJI S, YAMAJI T, MAKINO H, et al. Metabolic design for selective production of nicotinamide mononucleotide from glucose and nicotinamide[J]. Metabolic Engineering, 2021, 65: 167-177. |
| 167 | ZHANG P, YULY J L, LUBNER C E, et al. Electron bifurcation: thermodynamics and kinetics of two-electron brokering in biological redox chemistry[J]. Accounts of Chemical Research, 2017, 50(9): 2410-2417. |
| 168 | KANAI T, ITO S, IMANAKA T. Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1[J]. Journal of Bacteriology, 2003, 185(5): 1705-1711. |
| [1] | 刘庠诗, 吴奕禄, 詹鹏, 黄天灏, 蔡的, 秦培勇. 醇脱氢酶的研究进展及其催化增值生物基呋喃化合物前景展望[J]. 合成生物学, 2023, 4(6): 1122-1139. |
| [2] | 刘伟松, 张坤城, 崔会娟, 朱之光, 张以恒, 张玲玲. 电能辅助二氧化碳生物转化[J]. 合成生物学, 2023, 4(6): 1191-1222. |
| [3] | 王俊婷, 郭潇佳, 李青, 万里, 赵宗保. 创制非天然辅酶偏好型甲醇脱氢酶[J]. 合成生物学, 2021, 2(4): 651-661. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||