合成生物学 ›› 2023, Vol. 4 ›› Issue (4): 824-839.DOI: 10.12211/2096-8280.2022-077
蓝细菌CRISPRa系统的开发及其代谢工程应用
王甜甜1,2, 朱虹1, 杨琛1
- 1.中国科学院合成生物学重点实验室,中国科学院分子植物科学卓越创新中心,上海 200032
2.中国科学院大学,北京 100049
-
收稿日期:2022-12-30修回日期:2023-02-20出版日期:2023-08-31发布日期:2023-09-14 -
通讯作者:杨琛 -
作者简介:王甜甜 (1992—),女,博士研究生。研究方向为蓝细菌基因组编辑及萜类化合物代谢工程。E-mail:wangtiantian@cemps.ac.cn朱虹 (1983—),女,硕士研究生。研究方向为微生物代谢工程。E-mail:zhuhong@cemps.ac.cn杨琛 (1974—),女,研究员,博士,博士生导师。研究方向为:①开发代谢流量分析与代谢组分析技术;②研究重要模式及工业微生物代谢网络调控的分子机制,揭示各种调节机制对代谢流量的调控机理,为合理改造细胞代谢、优化微生物生产提供理论依据;③开展光合微生物的代谢工程与合成生物学研究,建立萜类化合物的光合自养细胞工厂。E-mail:cyang@cemps.ac.cn -
基金资助:国家重点研发计划(2021YFA0909702);国家自然科学基金(31925001);中国科学院战略性先导科技专项(XDB27020000)
Development of CRISPRa for metabolic engineering applications in cyanobacteria
WANG Tiantian1,2, ZHU Hong1, YANG Chen1
- 1.CAS Key Laboratory of Synthetic Biology,CAS Center for Excellence in Molecular Plant Sciences,Chinese Academy of Sciences (CAS),Shanghai 200032,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2022-12-30Revised:2023-02-20Online:2023-08-31Published:2023-09-14 -
Contact:YANG Chen
摘要:
蓝细菌是光合作用研究的模式生物之一,也是构建光能自养细胞工厂的良好底盘。然而目前蓝细菌的遗传操作工具仍然较为缺乏且效率较低,开发高效的蓝细菌基因调控工具对于蓝细菌系统与合成生物学研究具有重要意义。本研究在模式蓝细菌聚球藻PCC 7942中开发了CRISPR激活系统,测试了多个转录激活因子,将内源的RNA聚合酶ω-亚基RpoZ与无DNA切割活性的dCas9融合表达,利用高强度启动子表达向导RNA,进而敲除内源rpoZ基因并优化了dCas9-RpoZ的表达及靶向位点。利用建立的CRISPRa系统对重要生物燃料——异戊烯醇的生物合成途径进行了工程改造,该系统不仅能够实现单基因或多基因的高表达,还可以同时对不同基因进行转录激活和抑制,将蓝细菌中异戊烯醇的产量提高了17倍,展示了该系统有望成为构建光能自养细胞工厂的有力工具。
中图分类号:
引用本文
王甜甜, 朱虹, 杨琛. 蓝细菌CRISPRa系统的开发及其代谢工程应用[J]. 合成生物学, 2023, 4(4): 824-839.
WANG Tiantian, ZHU Hong, YANG Chen. Development of CRISPRa for metabolic engineering applications in cyanobacteria[J]. Synthetic Biology Journal, 2023, 4(4): 824-839.
引物 Primer | 5′-3′序列 Sequences 5′-3′ |
|---|---|
| Pendo-F | GAGACACAACGTGGCTTTCCCGCGGCCGCTTACGAAATCATCCTGTGGAGCTTAGTAG |
| Pendo-R | CTATTGAGTATTTCTTATCCATTTTTGCCTCCTAAAATAAAAAGTTTAAATTAAATC |
| dCas9-F | ATGGATAAGAAATACTCAATAGGCTTAG |
| dCas9-R | AATGATTTTCTGGTGGCTCATGGAACCGCCACCGCCGGAACCGCCACCGCCTGCGCAGTCACCTCCTAGCTGACTCAA |
| nudⅠ-F | ATGCGACAACGGACTATTGTATG |
| nudⅠ-R | GATGGGTAATTTCGCTGAGATGCATGGTATATCTCCTTCTTACTAGTCTCTCTCTTGTACATTACAGAAGACCTTTCAAACGTAAC |
| dxs-F | ATGCATCTCAGCGAAATTACCCATC |
| dxs-R | AGCCGGATTAATAATCTGGCTTTTTATATTCTCTTTAAGCCGAAGCAGCACCAAT |
| P18082-F | TTAATTAACCTGCCGAG |
| P18082-R | AGCTCTTCGCCCTTGCTCATGGTACCTTTCTCCTCTTTAATGAATTCGCCTG |
| P18082 upstream-F | ACCGTTTCAGCTGGTGATTTGGATCCGCATGCCCGATCAACGTCTC |
| P18082 upstream -R | ACCTCGGCAGGTTAATTAAGACCGGTATGCCTAATGTA |
| sfgfp -F | ATGAGCAAGGGCGAAGAG |
| sfgfp-R | AGCCAAGCTGGAGACCGTTTAAACTCACTACTTGTACAGTTCATCCATGCCA |
| qPCR-nudI-F | ATGGTGCTTATTTGCT |
| qPCR-nudI-R | TGTTCTCCCAGTTCTT |
| qPCR-dxs-F | ACCCATCCCAACCAGC |
| qPCR-dxs-R | TTCCACCACGCCCAAG |
| qPCR-gpps-F | GCGGGTGGAACGGCTG |
| qPCR-gpps-R | CCTTGTGATTGGTGGG |
| NSⅡ-up-F验证 | ACCTTGCGTCGGTGCTGAGTC |
| dxs-R验证 | TTGCTCAAGCTGAGCAACCGAC |
| NSⅢ-up-F验证 | GATGCACGAGCGTAATGCTCAC |
| NSⅢ-del-R验证 | TCTCGCTAATTGTGGGAGAGGAG |
表3 本实验所用到的引物
Table 3 Primers used in this study
引物 Primer | 5′-3′序列 Sequences 5′-3′ |
|---|---|
| Pendo-F | GAGACACAACGTGGCTTTCCCGCGGCCGCTTACGAAATCATCCTGTGGAGCTTAGTAG |
| Pendo-R | CTATTGAGTATTTCTTATCCATTTTTGCCTCCTAAAATAAAAAGTTTAAATTAAATC |
| dCas9-F | ATGGATAAGAAATACTCAATAGGCTTAG |
| dCas9-R | AATGATTTTCTGGTGGCTCATGGAACCGCCACCGCCGGAACCGCCACCGCCTGCGCAGTCACCTCCTAGCTGACTCAA |
| nudⅠ-F | ATGCGACAACGGACTATTGTATG |
| nudⅠ-R | GATGGGTAATTTCGCTGAGATGCATGGTATATCTCCTTCTTACTAGTCTCTCTCTTGTACATTACAGAAGACCTTTCAAACGTAAC |
| dxs-F | ATGCATCTCAGCGAAATTACCCATC |
| dxs-R | AGCCGGATTAATAATCTGGCTTTTTATATTCTCTTTAAGCCGAAGCAGCACCAAT |
| P18082-F | TTAATTAACCTGCCGAG |
| P18082-R | AGCTCTTCGCCCTTGCTCATGGTACCTTTCTCCTCTTTAATGAATTCGCCTG |
| P18082 upstream-F | ACCGTTTCAGCTGGTGATTTGGATCCGCATGCCCGATCAACGTCTC |
| P18082 upstream -R | ACCTCGGCAGGTTAATTAAGACCGGTATGCCTAATGTA |
| sfgfp -F | ATGAGCAAGGGCGAAGAG |
| sfgfp-R | AGCCAAGCTGGAGACCGTTTAAACTCACTACTTGTACAGTTCATCCATGCCA |
| qPCR-nudI-F | ATGGTGCTTATTTGCT |
| qPCR-nudI-R | TGTTCTCCCAGTTCTT |
| qPCR-dxs-F | ACCCATCCCAACCAGC |
| qPCR-dxs-R | TTCCACCACGCCCAAG |
| qPCR-gpps-F | GCGGGTGGAACGGCTG |
| qPCR-gpps-R | CCTTGTGATTGGTGGG |
| NSⅡ-up-F验证 | ACCTTGCGTCGGTGCTGAGTC |
| dxs-R验证 | TTGCTCAAGCTGAGCAACCGAC |
| NSⅢ-up-F验证 | GATGCACGAGCGTAATGCTCAC |
| NSⅢ-del-R验证 | TCTCGCTAATTGTGGGAGAGGAG |
菌株 Strain | 基因型 Genotype | 来源 Source |
|---|---|---|
| Synechococcus elongatus PCC7942 | Wild type | ATCC |
| PCC 7942-G01 | P18082 sfgfp integrated at NSⅡ | This work |
| PCC 7942-G02 | P18082 sfgfp pcpc sgRNA 106 integrated at NSⅡ | This work |
| PCC 7942-G03 | P18082 sfgfp pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G09 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G10 | Pendo dCas9-CAGGGGSGGGGS-rpoA integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G11 | Pendo dCas9-CAGGGGSGGGGS-rpoD integrated at NSⅡ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G12 | Pendo dCas9-CAGGGGSGGGGS-rpoA NTD integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G13 | Pendo dCas9-CAGGGGSGGGGS-soxS integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G14 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G15 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G16 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G17 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G18 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ | This work |
| PCC 7942-G19 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G27 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G28 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G29 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G30 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G31 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G32 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G33 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G34 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G35 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G36 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G37 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G38 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G39 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G40 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G45 | PJ23108 dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ | This work |
| PCC 7942-S01 | P18082 nudⅠ Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-S02 | P18082 nudⅠ dxs Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-S03 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 nudⅠ Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S04 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S05 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 nudⅠ dxs Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S06 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S07 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-106 integrated at NSⅡ | This work |
| PCC 7942-S12 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 Ptrc crRNA-g1 PJ23119 tracrRNA integrated at NSⅡ | This work |
| PCC 7942-S13 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 Ptrc crRNA-g2 PJ23119 tracrRNA integrated at NSⅡ | This work |
| PCC 7942-S14 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 Ptrc crRNA-106 PJ23119 tracrRNA integrated at NSⅡ | This work |
表1 本实验所用到的菌株
Table 1 Strains used in this study
菌株 Strain | 基因型 Genotype | 来源 Source |
|---|---|---|
| Synechococcus elongatus PCC7942 | Wild type | ATCC |
| PCC 7942-G01 | P18082 sfgfp integrated at NSⅡ | This work |
| PCC 7942-G02 | P18082 sfgfp pcpc sgRNA 106 integrated at NSⅡ | This work |
| PCC 7942-G03 | P18082 sfgfp pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G09 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G10 | Pendo dCas9-CAGGGGSGGGGS-rpoA integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G11 | Pendo dCas9-CAGGGGSGGGGS-rpoD integrated at NSⅡ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G12 | Pendo dCas9-CAGGGGSGGGGS-rpoA NTD integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G13 | Pendo dCas9-CAGGGGSGGGGS-soxS integrated at NSⅢ P18082 sfgfpPcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G14 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G15 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G16 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G17 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G18 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ | This work |
| PCC 7942-G19 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G27 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G28 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G29 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G30 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G31 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G32 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G33 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G34 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G35 | Pendo rpoZ-CAGGGGSGGGGS-dCas9 integrated at rpoZ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G36 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H1 integrated at NSⅡ | This work |
| PCC 7942-G37 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H2 integrated at NSⅡ | This work |
| PCC 7942-G38 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H3 integrated at NSⅡ | This work |
| PCC 7942-G39 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-G40 | Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 sfgfp Pcpc sgRNA H5 integrated at NSⅡ | This work |
| PCC 7942-G45 | PJ23108 dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ | This work |
| PCC 7942-S01 | P18082 nudⅠ Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-S02 | P18082 nudⅠ dxs Pcpc sgRNA H4 integrated at NSⅡ | This work |
| PCC 7942-S03 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 nudⅠ Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S04 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S05 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at NSⅢ P18082 nudⅠ dxs Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S06 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 integrated at NSⅡ | This work |
| PCC 7942-S07 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-106 integrated at NSⅡ | This work |
| PCC 7942-S12 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 Ptrc crRNA-g1 PJ23119 tracrRNA integrated at NSⅡ | This work |
| PCC 7942-S13 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 Ptrc crRNA-g2 PJ23119 tracrRNA integrated at NSⅡ | This work |
| PCC 7942-S14 | Pendo dCas9-CAGGGGSGGGGS-rpoZ integrated at rpoZ P18082 nudⅠ dxs Pcpc sgRNA-H4 Ptrc crRNA-106 PJ23119 tracrRNA integrated at NSⅡ | This work |
质粒 Plasmid | 描述 Description | 来源 Source |
|---|---|---|
| P01 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp | This work |
| P02 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H1 | This work |
| P03 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H2 | This work |
| P04 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H3 | This work |
| P05 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H4 | This work |
| P06 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H5 | This work |
| P07 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA 106 | This work |
| P08 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ Pcpc sgRNA H4 | This work |
| P09 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc sgRNA H4 | This work |
| P10 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc sgRNA 106 | This work |
| P11 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc crRNA g1 PJ23119 tracrRNA | This work |
| P12 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc crRNA g2 PJ23119 tracrRNA | This work |
| P13 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc crRNA 106 PJ23119 tracrRNA | This work |
| P14 | PCL1920; Kanr ; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-soxS | This work |
| P15 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P16 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoD | This work |
| P17 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoA | This work |
| P18 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoA NTD | This work |
| P19 | PCL1920; Kanr; RpoZ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P20 | PCL1920; Kanr;RpoZ targeting; Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P21 | PCL1920; Kanr; RpoZ targeting; Pendo rpoZ-CAGGGGSGGGGS-dCas9 | This work |
| P22 | PCL1920; Kanr; RpoZ targeting; PJ23108 dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P23 | PCL1920; Kanr; RpoZ targeting; Pcpc dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P24 | PCL1920; Kanr; RpoZ targeting; Pcpc dCas9 | This work |
| P25 | PCL1920; Kanr; NSⅢ targeting; Pcpc dCas9-CAGGGGSGGGGS-rpoZ | This work |
表2 本实验所用到的质粒
Table 2 Plasmids used in this study
质粒 Plasmid | 描述 Description | 来源 Source |
|---|---|---|
| P01 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp | This work |
| P02 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H1 | This work |
| P03 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H2 | This work |
| P04 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H3 | This work |
| P05 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H4 | This work |
| P06 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA H5 | This work |
| P07 | PCL1920; Specr; NSⅡ targeting; P18082 sfgfp Pcpc sgRNA 106 | This work |
| P08 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ Pcpc sgRNA H4 | This work |
| P09 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc sgRNA H4 | This work |
| P10 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc sgRNA 106 | This work |
| P11 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc crRNA g1 PJ23119 tracrRNA | This work |
| P12 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc crRNA g2 PJ23119 tracrRNA | This work |
| P13 | PCL1920; Specr; NSⅡ targeting; P18082 nudⅠ dxs Pcpc crRNA 106 PJ23119 tracrRNA | This work |
| P14 | PCL1920; Kanr ; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-soxS | This work |
| P15 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P16 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoD | This work |
| P17 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoA | This work |
| P18 | PCL1920; Kanr; NSⅢ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoA NTD | This work |
| P19 | PCL1920; Kanr; RpoZ targeting; Pendo dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P20 | PCL1920; Kanr;RpoZ targeting; Pendo rpoZ-CAGGGGSGGGGS-dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P21 | PCL1920; Kanr; RpoZ targeting; Pendo rpoZ-CAGGGGSGGGGS-dCas9 | This work |
| P22 | PCL1920; Kanr; RpoZ targeting; PJ23108 dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P23 | PCL1920; Kanr; RpoZ targeting; Pcpc dCas9-CAGGGGSGGGGS-rpoZ | This work |
| P24 | PCL1920; Kanr; RpoZ targeting; Pcpc dCas9 | This work |
| P25 | PCL1920; Kanr; NSⅢ targeting; Pcpc dCas9-CAGGGGSGGGGS-rpoZ | This work |
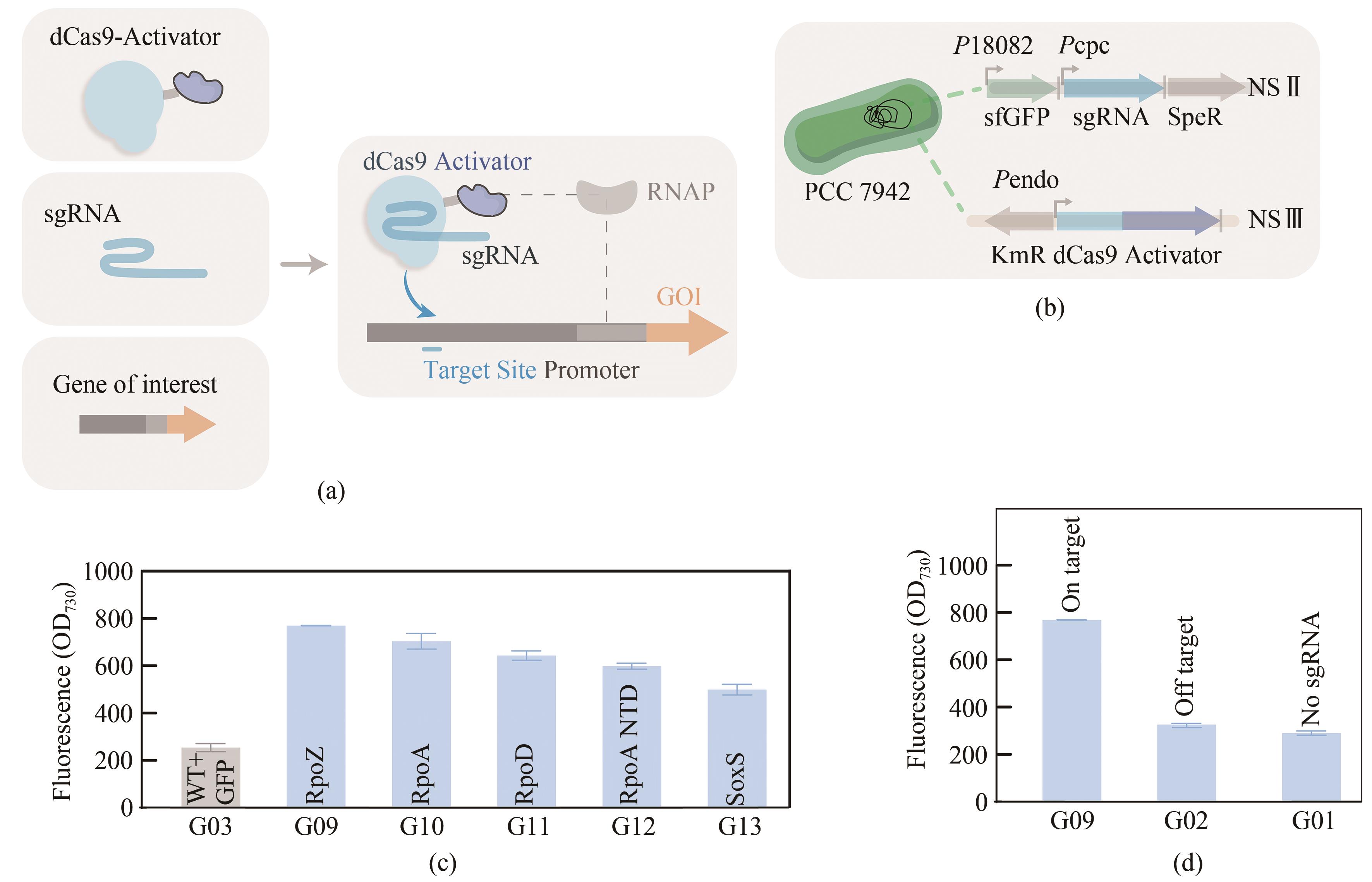
图1 在PCC 7942中配置CRISPRa(a)CRISPRa系统的组成元件;(b)dCas9与转录激活因子的基因模块整合至PCC 7942中性位点NSⅢ,报告基因sfgfp与sgRNA整合至PCC 7942中性位点NSⅡ;(c)表达不同转录激活因子的菌株的荧光信号值;(d)以无sgRNA及脱靶RNA构建的菌株G01与G02的荧光信号值
Fig. 1 Configuration of CRISPRa in PCC7942(a) Components of the CRISPRa system. (b) The expression cassette encoding dCas9 anda transcriptional activator fusion protein was inserted into the NSⅢ site of PCC7942, and the report genes sfgfp and sgRNA were integrated into the NSⅡ site of PCC7942. (c) The fluorescence intensities of strains G03, G09, G10, G11, G12, and G13 with different transcriptional activators. (d) The fluorescence signal level of G01(control strain without sgRNA) and G02(control strain with an off target sgRNA 106).
靶点 TargetSites | DNA序列(5′→3′) DNA Sequences(5′→3′) | Target Strand | Distance toTSS/bp |
|---|---|---|---|
| H1 | ATGTAACACCGTGCGTGTTG | NT | -216 |
| H2 | GAAGATCCGGCCTGCAGCCA | NT | -236 |
| H3 | GGCTCGAGTCGACAGTTCAT | NT | -273 |
| H4 | CTACGGAACTCTTGTGCGTA | T | -327 |
| H5 | GCAAAAGCTCATTTCTGAAG | T | -397 |
| g1 | CACTCAAAGGATAGACGGGA | NT | +48 |
| g2 | TCCCATTGCGCTAAGCCCTA | NT | +960 |
表4 本研究用到的CRISPRa系统靶位点序列
Table 4 Targeting sequences for the CRISPRa system used in this study
靶点 TargetSites | DNA序列(5′→3′) DNA Sequences(5′→3′) | Target Strand | Distance toTSS/bp |
|---|---|---|---|
| H1 | ATGTAACACCGTGCGTGTTG | NT | -216 |
| H2 | GAAGATCCGGCCTGCAGCCA | NT | -236 |
| H3 | GGCTCGAGTCGACAGTTCAT | NT | -273 |
| H4 | CTACGGAACTCTTGTGCGTA | T | -327 |
| H5 | GCAAAAGCTCATTTCTGAAG | T | -397 |
| g1 | CACTCAAAGGATAGACGGGA | NT | +48 |
| g2 | TCCCATTGCGCTAAGCCCTA | NT | +960 |
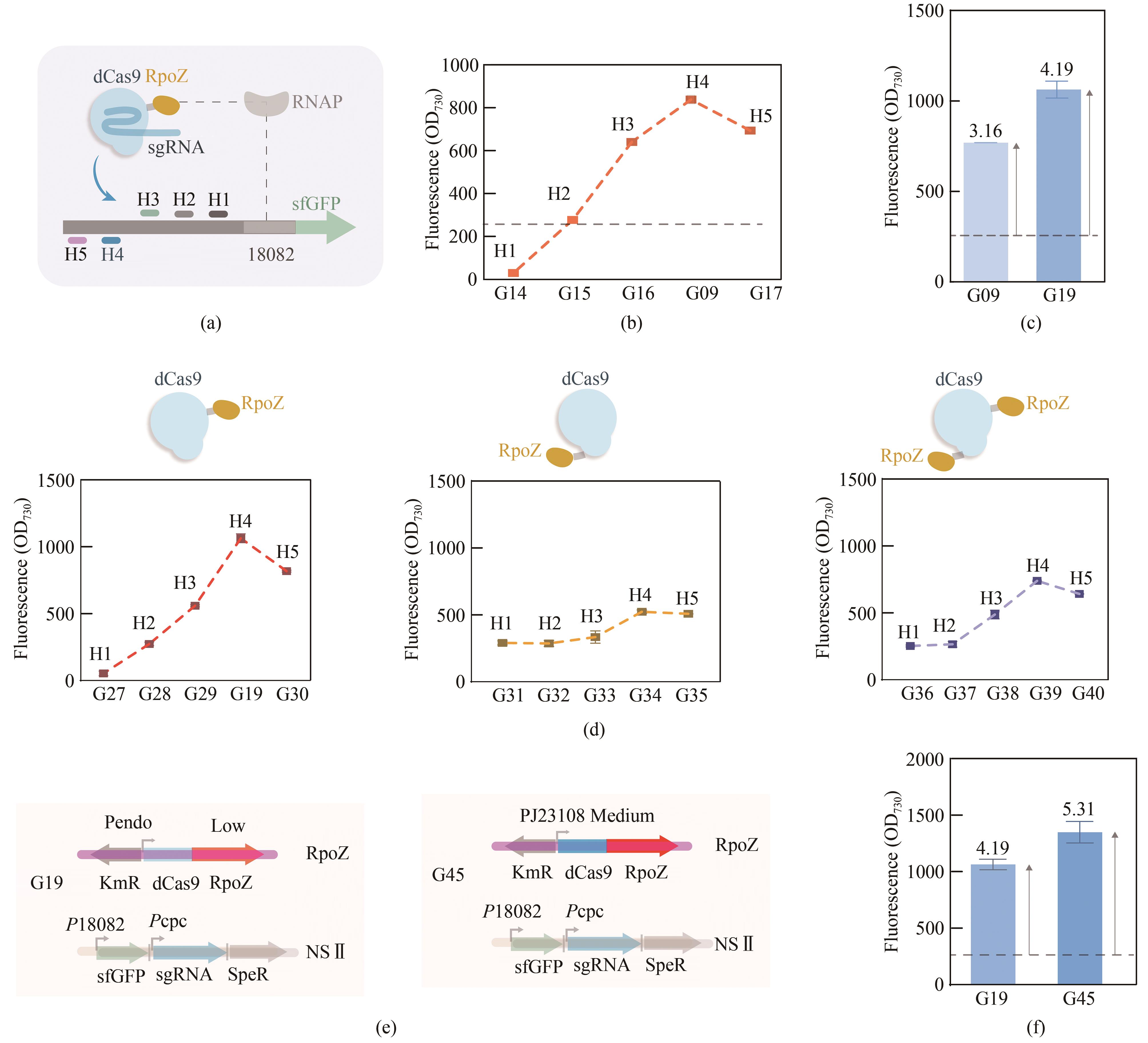
图2 CRISPRa系统的优化(a)目标基因上游不同靶向位点的位置示意图;(b)CRISPRa靶向不同位点的菌株的荧光信号值;(c)内源rpoZ被敲除的G19菌株的荧光信号值;(d)dCas9与RpoZ融合表达顺序的改变对于CRISPRa激活效果的影响;(e)以强启动子PJ23108替换弱启动子Pendo,构建dCas9-RpoZ表达增强的菌株G45;(f)菌株G45的荧光信号值
Fig. 2 Optimization of the CRISPRa system(a)Scheme of different targeting sites for CRISPRa. (b) The fluorescence intensities of strains G09, G14, G15,G16, and G17 with different targeting sites. (c) The fluorescence intensity of G19 strain with genetic knockout of endogenous rpoZ. (d) Effect of different orientations of the dCas9 and RpoZ fusion on CRISPRa performance. (e) Expression of dCas9-RpoZ was increased in G45 strain by replacing the promoter Pendo with PJ23108. (f) The fluorescence intensity of G45 strain.
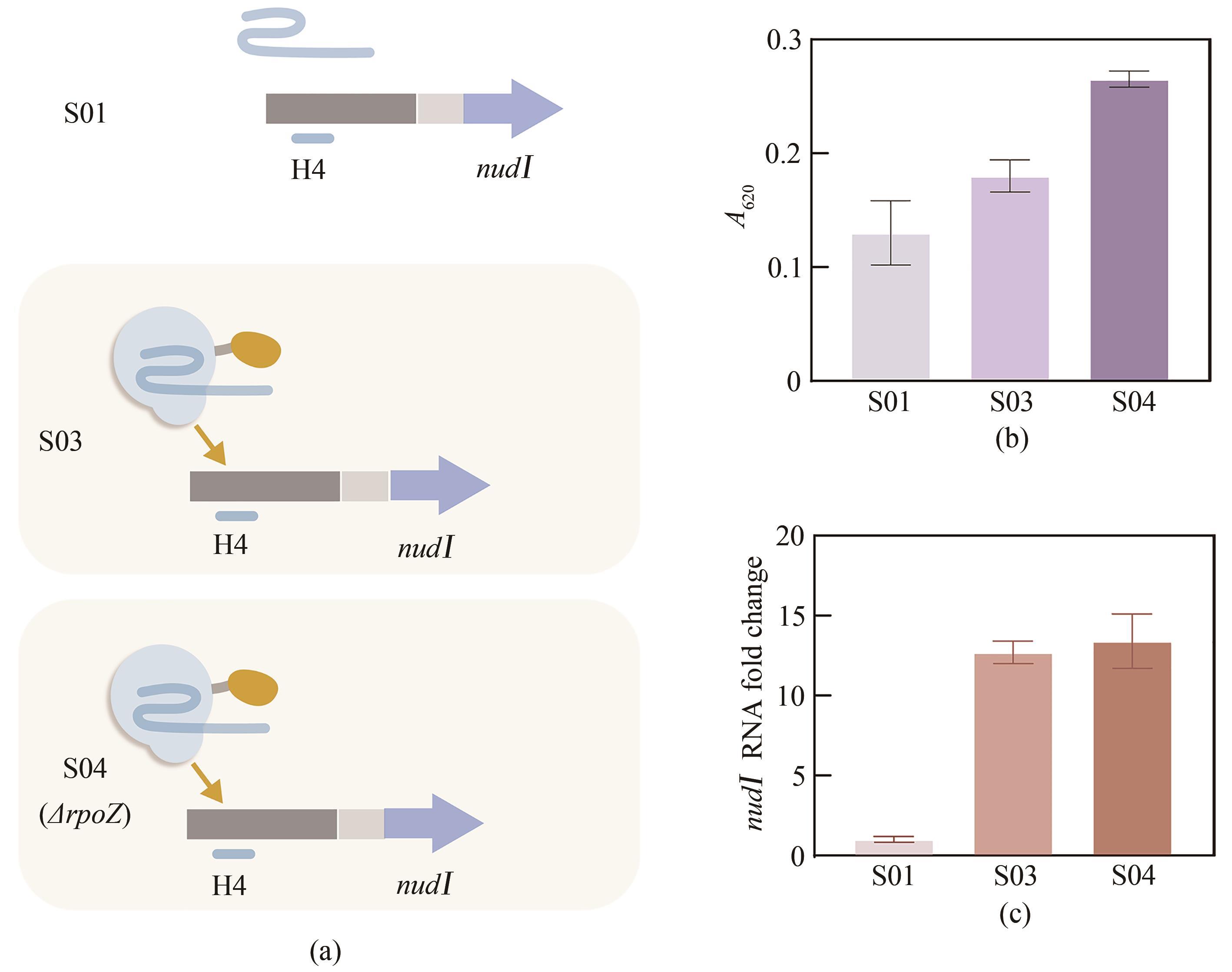
图3 CRISPRa激活nudⅠ(a)sgRNA(靶位点H4)与nudⅠ基因整合至PCC 7942中性位点NSⅡ,dCas9-RpoZ基因模块整合至PCC 7942中性位点NSⅢ或替换内源rpoZ基因,分别得到菌株S03和S04,S01为没有dCas9-RpoZ模块的对照菌株;(b)菌株S01、S03和S04培养96 h后利用显色反应检测的上清液中异戊烯醇含量;(c)菌株S01、S03和S04培养72 h后nudⅠ的转录水平
Fig. 3 Activation of nudⅠ by CRISPRa(a) Scheme of nudⅠ activation by CRISPRa. The sgRNA H4 and nudⅠ were inserted into the NSⅡ site. The DNA fragment encoding dCas9-RpoZ was inserted into the NSⅢ site or to replace endogenous rpoZ, generating strains S03 and S04, respectively. The control strain S01 lacked dCas9-RpoZ. (b) Isopentenol production by strains S01, S03, and S04. Cells were cultivated for 96 h, and isopentenol in the culture supernatant was detected by a colorimetric assay. (c) Transcriptional level of nudⅠ in strains S01, S03, and S04. RNA was isolated from cellscultivated for 72 h.

图4 CRISPRa同时激活nudⅠ与dxs(a)dxs编码的脱氧木酮糖-5-磷酸合成酶催化MEP途径的第一步反应;(b)sgRNA(靶位点H4)、nudⅠ及dxs整合至PCC 7942中性位点NSⅡ,dCas9-RpoZ基因模块整合至PCC 7942中性位点NSⅢ或替换内源rpoZ基因,分别得到菌株S05和S06,S02为没有dCas9-RpoZ模块的对照菌株,S07为整合了脱靶RNA(off target)的菌株;(c)菌株S02、S05、S06、S07培养96 h后利用显色反应检测的上清液中异戊烯醇含量;(d)菌株S02、S05、S06、S07培养72 h后nudⅠ与dxs的转录水平
Fig. 4 Simultaneous activation of nudⅠ and dxs by CRISPRa(a) 1-Deoxy-D-xylulose 5-phosphate synthase encoded by dxs catalyzes the first reaction of the MEP pathway. (b)Scheme of nudⅠ and dxs activation by CRISPRa. The sgRNA H4, nudⅠ, and dxs were inserted into the NSⅡ site. The DNA fragment encoding dCas9-RpoZ was inserted into the NSⅢ site or to replace endogenous rpoZ, generating strains S05 and S06, respectively. S02 (without dCas9-RpoZ) and S07 (with an off target sgRNA 106) are the control strains. (c) Isopentenol production by strains S02, S05, S06, and S07.Cells were cultivated for 96 h, and isopentenol in the culture supernatant was detected by a colorimetric assay. (f) Transcriptional levels of nudI and dxs in strains S02, S05, S06, and S07. RNA was isolated from cells cultivated for 72 h.

图5 CRISPRa 同时激活nudI与dxs并抑制gpps我们检测了上述菌株中nudⅠ、dxs及gpps的转录水平以及菌株的生长与异戊烯醇的生成。结果显示与菌株S06相比,菌株S13中gpps的转录水平降低了85%,而菌株S12和S14中gpps的表达水平没有明显改变;菌株S13中nudI与dxs的转录水平与菌株S06相比降低了25%~30%,但是与对照菌株S02相比仍然提高了5.33倍和2.09倍[图5(d)]。菌株S13的异戊烯醇产量明显高于菌株S06,在光照培养第8天达到最高值[图5(e)]。菌株S13的生长明显变慢[图5(f)],可能是由于gpps基因受到强烈抑制而导致光合作用相关的叶绿素和类胡萝卜素合成下降。除了显色法以外,我们还利用气相色谱检测了菌株的异戊烯醇生成。结果显示菌株S13的异戊烯醇产量达到22.1 mg/L,相比菌株S06和S02分别提高了2倍和17倍[图5(g)]。因此,该CRISPRa系统能够激活nudⅠ与dxs的转录并同时抑制gpps基因的转录,大幅提高了蓝细菌中异戊烯醇的合成。(a)gpps编码的香叶基焦磷酸合成酶是影响异戊烯醇合成的竞争途径上的关键酶;(b)gpps(Synpcc7942_0776)基因在PCC 7942基因组上的位置示意图;(c)在S06菌株gpps所在操纵子的转录起始位点以及gpps基因起始密码子附近各选了一个靶向位点(g1与g2),分别得到菌株S12和S13,S14为整合了脱靶RNA的对照菌株;(d)菌株S12、S13、S14及S02、S06在OD730约为0.7时gpps、nudⅠ和dxs的转录水平;(e)菌株S06、S12、S13与S14培养4~8天时利用显色反应检测的上清液中异戊烯醇含量;(f)野生型菌株WT与S02、S06、S12、S13、S14菌株的生长曲线;(g)菌株S02、S06与S13在OD730约为2.0时利用气相色谱检测的培养上清液中异戊烯醇的含量
Fig. 5 Simultaneous activation of nudⅠ and dxs and repression of gpps by CRISPRa(a) gpps catalyzes a key competing reaction for isopentenol biosynthesis; (b) Location of the gpps gene and the targeting sites g1 and g2 on the PCC 7942 genome; (c) The g1 and g2 sites of gpps in strain S06 were targeted, generating strains S12 and S13, respectively. S14 (with an off target sgRNA 106) is a control strain. (d) Transcriptional levels of gpps, nudⅠ and dxs in strains S02, S06, S12, S13, and S14. RNA was isolated from cells grown to OD730 of about 0.7. (e) Isopentenol production by strains S02, S06, S13, and S14. Isopentenol in the culture supernatant was detected by a colorimetric assay. (f) Isopentenol production by strains S02, S06, and S13. Culture supernatants were collected at OD730 of about 2.0, and isopentenol was detected by gas chromatography.
| 1 | GAO X, GAO F, LIU D, et al. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2 [J]. Energy & Environmental Science, 2016, 9(4): 1400-1411. |
| 2 | NI J, TAO F, XU P, et al. Engineering cyanobacteria for photosynthetic production of C3 platform chemicals and terpenoids from CO2 [J]. Advancesin Experimental Medicine and Biology, 2018, 1080: 239-259. |
| 3 | ROUSSOU S, ALBERGATI A, LIANG F Y, et al. Engineered cyanobacteria with additional overexpression of selected Calvin-Benson-Bassham enzymes show further increased ethanol production[J]. Metabolic Engineering Communications, 2021, 12: e00161. |
| 4 | KOBAYASHI S, ATSUMI S, IKEBUKURO K, et al. Light-induced production of isobutanol and 3-methyl-1-butanol by metabolically engineered cyanobacteria[J]. Microbial Cell Factories, 2022, 21(1): 7. |
| 5 | WANG B, ECKERT C, MANESS P C, et al. A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803[J]. ACS Synthetic Biology, 2018, 7(1): 276-286. |
| 6 | CHI X T, ZHANG S S, SUN H L, et al. Adopting a theophylline-responsive riboswitch for flexible regulation and understanding of glycogen metabolism in Synechococcus elongatusPCC 7942[J]. Frontiersin Microbiology, 2019, 10: 551. |
| 7 | CAICEDO-BURBANO P, SMIT T, PINEDA HERNÁNDEZ H, et al. Construction of fully segregated genomic libraries in polyploid organisms such as Synechocystis sp. PCC 6803[J]. ACS Synthetic Biology, 2020, 9(10): 2632-2638. |
| 8 | BEHLE A, SAAKE P, GERMANN A T, et al. Comparative dose-response analysis of inducible promoters in cyanobacteria[J]. ACS Synthetic Biology, 2020, 9(4): 843-855. |
| 9 | YUNUS I S, ANFELT J, SPORRE E, et al. Synthetic metabolic pathways for conversion of CO2 into secreted short-to medium-chain hydrocarbons using cyanobacteria[J]. Metabolic Engineering, 2022, 72: 14-23. |
| 10 | WENDT K E, UNGERER J, COBB R E, et al. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973[J]. Microbial Cell Factories, 2016, 15(1): 115. |
| 11 | SENGUPTA A, PRITAM P, JAISWAL D, et al. Photosynthetic co-production of succinate and ethylene in a fast-growing cyanobacterium, Synechococcus elongatus PCC 11801[J]. Metabolites, 2020, 10(6): 250. |
| 12 | LI H, SHEN C R, HUANG C H, et al. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production[J]. Metabolic Engineering, 2016, 38: 293-302. |
| 13 | YAO L, SHABESTARY K, BJÖRK S M, et al. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp. PCC 6803 for enhanced industrial phenotypes[J]. Nature Communications, 2020, 11(1): 1666. |
| 14 | CHOI S Y, WOO H M. CRISPRi-dCas12a: A dCas12a-mediated CRISPR interference for repression of multiple genes and metabolic engineering in cyanobacteria[J]. ACS Synthetic Biology, 2020, 9(9): 2351-2361. |
| 15 | HU J H, MILLER S M, GEURTS M H, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity[J]. Nature, 2018, 556(7699): 57-63. |
| 16 | LU Z H, YANG S H, YUAN X, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis [J]. Nucleic Acids Research, 2019, 47(7): e40. |
| 17 | LIU Y, WAN X Y, WANG B J. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria[J]. Nature Communications, 2019, 10: 3693. |
| 18 | BIKARD D, JIANG W Y, SAMAI P, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system[J]. Nucleic Acids Research, 2013, 41(15): 7429-7437. |
| 19 | YU L J, SU W, FEY P D, et al. Yield improvement of the anti-MRSA antibiotics WAP-8294A by CRISPR/dCas9 combined with refactoring self-protection genes in Lysobacter enzymogenes OH11[J]. ACS Synthetic Biology, 2018, 7(1): 258-266. |
| 20 | PENG R, WANG Y, FENG W W, et al. CRISPR/dCas9-mediated transcriptional improvement of the biosynthetic gene cluster for the epothilone production in Myxococcus xanthus [J]. Microbial Cell Factories, 2018, 17(1): 15. |
| 21 | DONG C, FONTANA J, PATEL A, et al. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria[J]. Nature Communications, 2018, 9: 2489. |
| 22 | NIU F X, HUANG Y B, JI L N, et al. Genomic and transcriptional changes in response to pinene tolerance and overproduction in evolved Escherichia coli [J]. Syntheticand Systems Biotechnology, 2019, 4(3): 113-119. |
| 23 | FONTANA J, DONG C, KIATTISEWEE C, et al. Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements[J]. Nature Communications, 2020, 11(1): 1618. |
| 24 | HO H I, FANG J R, CHEUNG J, et al. Programmable CRISPR-Cas transcriptional activation in bacteria[J]. MolecularSystemsBiology, 2020, 16(7): e9427. |
| 25 | WITHERS S T, GOTTLIEB S S, LIEU B, et al. Identification of isopentenol biosynthetic genes from Bacillus subtilisby a screening method based on isoprenoid precursor toxicity[J]. Appliedand Environmental Microbiology, 2007, 73(19): 6277-6283. |
| 26 | FOO J L, JENSEN H M, DAHL R H, et al. Improving microbial biogasoline production in Escherichia coli using tolerance engineering[J]. mBio, 2014, 5(6): e01932. |
| 27 | GEORGE K W, THOMPSON M G, KANG A, et al. Metabolic engineering for the high-yield production of isoprenoid-based C5 alcohols in E. coli [J]. ScientificReports, 2015, 5: 11128. |
| 28 | KANG A, GEORGE K W, WANG G, et al. Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production[J]. MetabolicEngineering, 2016, 34: 25-35. |
| 29 | TIAN T, KANG J W, KANG A, et al. Redirecting metabolic flux via combinatorial multiplex CRISPRi-mediated repression for isopentenol production in Escherichia coli [J]. ACS SyntheticBiology, 2019, 8(2): 391-402. |
| 30 | CHOU H H, KEASLING J D. Synthetic pathway for production of five-carbon alcohols from isopentenyl diphosphate[J]. Applied and Environmental Microbiology, 2012, 78(22): 7849-7855. |
| 31 | RIPPKA R, DERUELLES J, WATERBURY J B, etal. Generic assignments, strain histories and properties of pure cultures of cyanobacteria[J]. Microbiology, 1979, 111(1): 1-61. |
| 32 | PÉDELACQ J D, CABANTOUS S, TRAN T, et al. Engineering and characterization of a superfolder green fluorescent protein[J]. NatureBiotechnology, 2006, 24(1): 79-88. |
| 33 | LINSHIZ G, JENSEN E, STAWSKI N, et al. End-to-end automated microfluidic platform for synthetic biology: from design to functional analysis[J]. Journal of Biological Engineering, 2016, 10: 3. |
| 34 | ORLOVA I, NAGEGOWDA D A, KISH C M, et al. The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of tobacco geranylgeranyl diphosphate synthase in planta[J]. The Plant Cell, 2009, 21(12): 4002-4017. |
| 35 | GILBERT L A, HORLBECK M A, ADAMSON B, et al. Genome-scale CRISPR-mediated control of gene repression and activation[J]. Cell, 2014, 159(3): 647-661. |
| 36 | KONERMANN S, BRIGHAM M D, TREVINO A E, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex[J]. Nature, 2015, 517(7536): 583-588. |
| 37 | JOUNG J, KONERMANN S, GOOTENBERG J S, et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening[J]. NatureProtocols, 2017, 12(4): 828-863. |
| 38 | HORLBECK M A, GILBERT L A, VILLALTA J E, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation[J]. eLife, 2016, 5: e19760. |
| 39 | WANG G C, CHOW R D, BAI Z G, et al. Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity[J]. Nature Immunology, 2019, 20(11): 1494-1505. |
| 40 | VAN DER WEYDEN L, OFFORD V, TURNER G, et al. Membrane protein regulators of melanoma pulmonary colonization identified using a CRISPRa screen and spontaneous metastasis assay in mice[J]. G3 Genes Genomes Genetics, 2021, 11(7): jkab157. |
| 41 | SIEPE D H, HENNEBERG L T, WILSON S C, et al. Identification of orphan ligand-receptor relationships using a cell-based CRISPRa enrichment screening platform[J]. eLife, 2022, 11: e81398. |
| [1] | 孙绘梨, 崔金玉, 栾国栋, 吕雪峰. 面向高效光驱固碳产醇的蓝细菌合成生物技术研究进展[J]. 合成生物学, 2023, 4(6): 1161-1177. |
| [2] | 董正鑫, 孙韬, 陈磊, 张卫文. 调控工程在光合蓝细菌中的应用[J]. 合成生物学, 2022, 3(5): 966-984. |
| [3] | 陶飞, 孙韬, 王钰, 魏婷, 倪俊, 许平. “双碳”背景下聚球藻底盘研究的挑战与机遇[J]. 合成生物学, 2022, 3(5): 932-952. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
