合成生物学 ›› 2022, Vol. 3 ›› Issue (1): 155-167.DOI: 10.12211/2096-8280.2021-074
功能性菌群构建的研究进展
黄佳城1,2, 张瑷珲1,2, 付友思1,2, 方柏山1,2
- 1.厦门大学化学化工学院,福建 厦门 361005
2.厦门市合成生物技术重点实验室,福建 厦门 361005
-
收稿日期:2021-07-12修回日期:2021-11-25出版日期:2022-02-28发布日期:2022-03-14 -
通讯作者:方柏山 -
作者简介:黄佳城 (1997—),男,硕士研究生。研究方向为人工智能在合成生物学与宏基因组上的应用等。E-mail:chonpcaacpnohc@gmail.com方柏山 (1957—),男,教授。研究方向为合成生物学与生物分子机器;定向进化与生物催化;生物技术过程开发与优化等。E-mail:fbs@xmu.edu.cn -
基金资助:国家自然科学基金(21978245);博士后创新人才支持计划(BX20200197)
Research progress in construction of functional microbial communities
HUANG Jiacheng1,2, ZHANG Aihui1,2, FU Yousi1,2, FANG Baishan1,2
- 1.College of Chemistry and Chemical Engineering,Xiamen University,Xiamen 361005,Fujian,China
2.The Key Laboratory for Synthetic Biotechnology of Xiamen City,Xiamen 361005,Fujian,China
-
Received:2021-07-12Revised:2021-11-25Online:2022-02-28Published:2022-03-14 -
Contact:FANG Baishan
摘要:
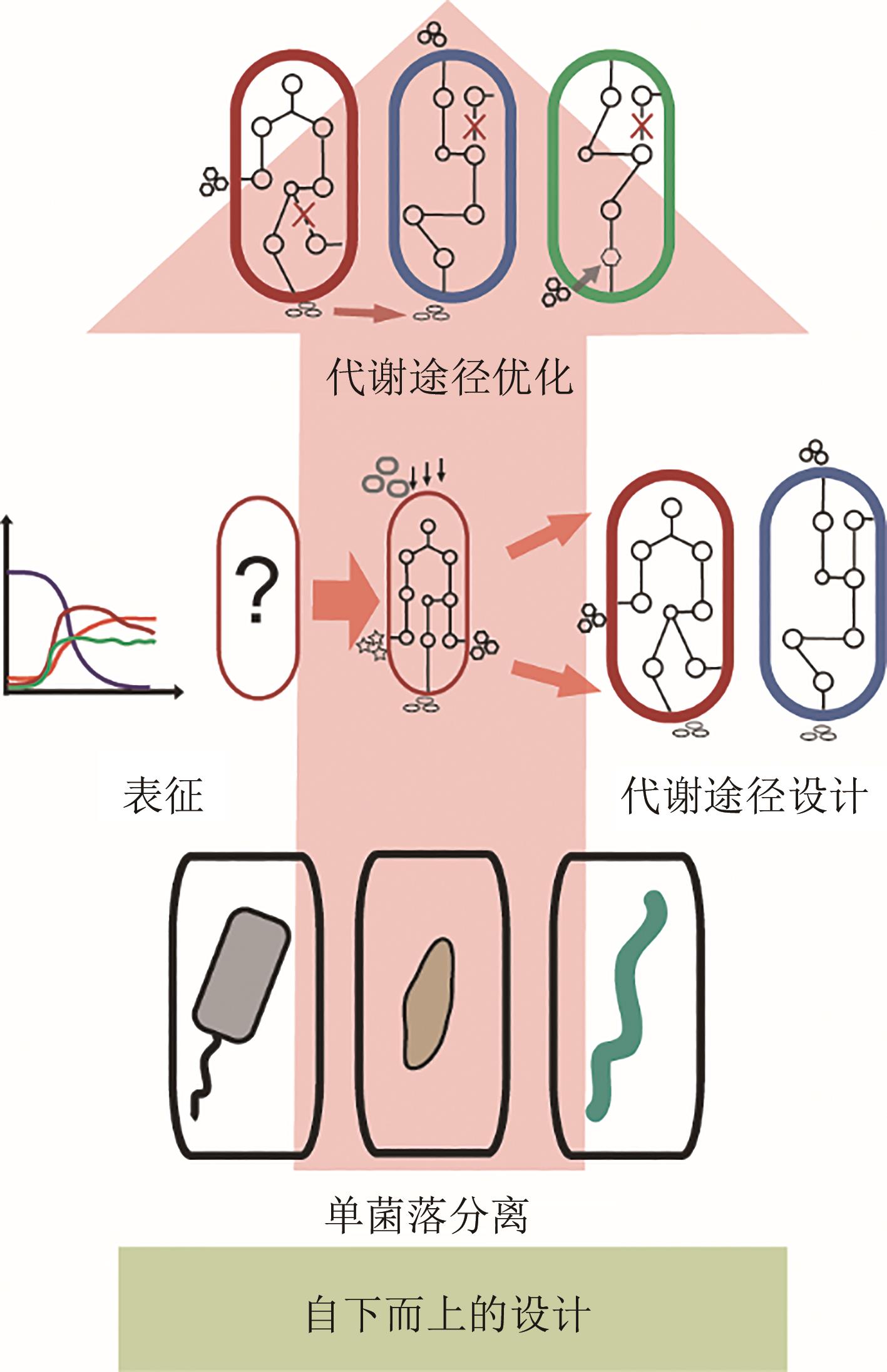
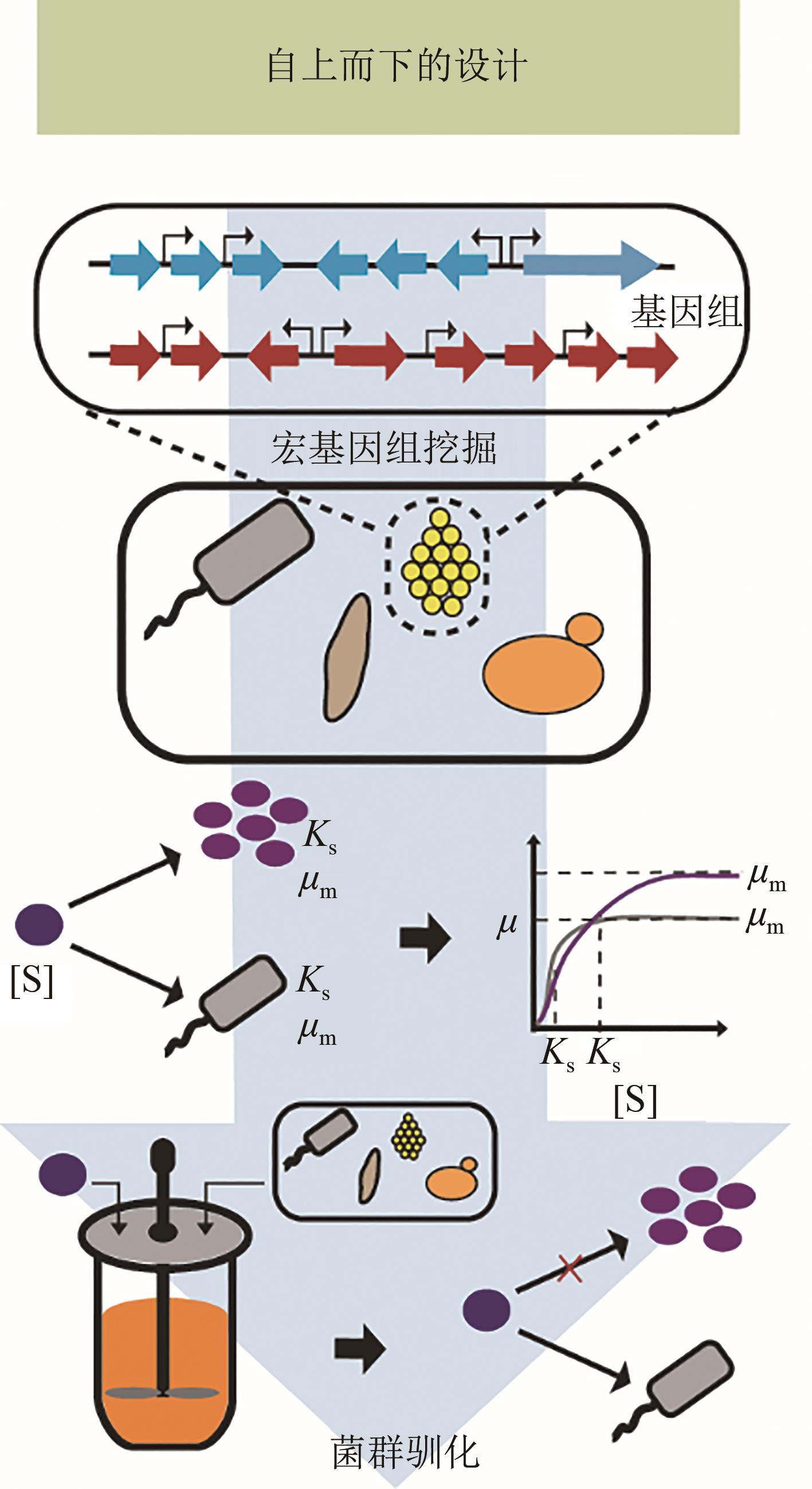
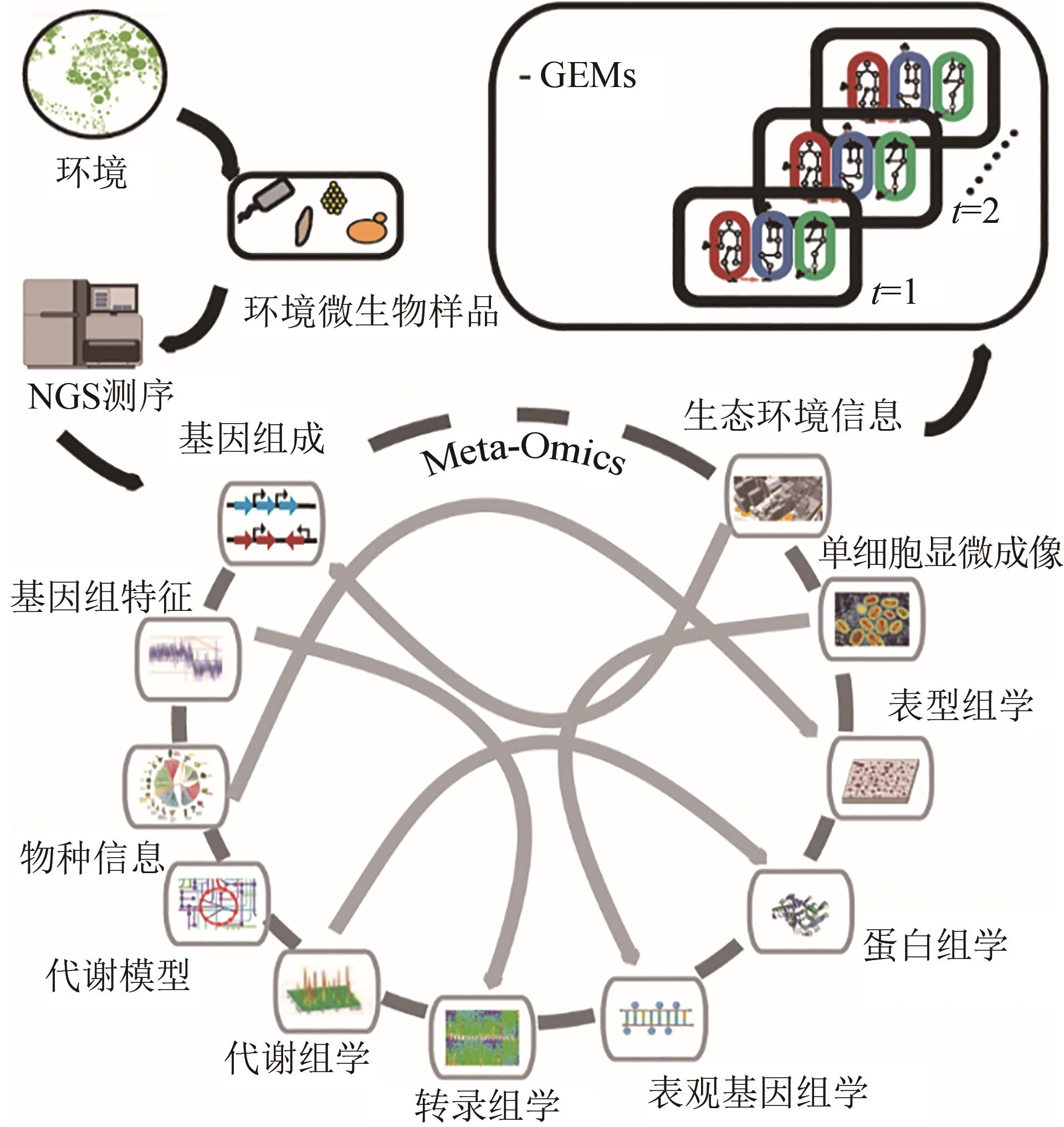
功能性菌群构建作为一个新兴的研究方向,随着合成生物学、微生物组学技术的发展,逐渐成为研究热点。本文将从以下4个方面介绍功能性菌群的研究进展。第一,功能性菌群研究的初衷及其相对于单一生物体工程的优势和设计难点;第二,功能性菌群研究中自下而上(bottom-up)和自上而下(top-down)的设计策略;第三,功能性菌群的分析工具,包括“宏组学”和“多组学联用”手段以及相关的数据处理流程和软件。第四,分别从设计策略和分析工具方面出发,总结了功能性菌群构建过程中的主要挑战,并展望了未来以“智能设计”为核心的发展方向:①利用可解释的时空数据模型解析区域范围内功能性菌群的时空变化关系;②结合图神经网络与多模态学习方法建立多组学群落分析流程;③通过强化学习设计功能性菌群内分布式代谢回路。
中图分类号:
引用本文
黄佳城, 张瑷珲, 付友思, 方柏山. 功能性菌群构建的研究进展[J]. 合成生物学, 2022, 3(1): 155-167.
HUANG Jiacheng, ZHANG Aihui, FU Yousi, FANG Baishan. Research progress in construction of functional microbial communities[J]. Synthetic Biology Journal, 2022, 3(1): 155-167.
| 软件名 | 对应处理的组学数据类型 | 功能 |
|---|---|---|
| QIIME[ | 扩增子测序 | 用于质量控制、OTU处理、物种分类、系统发育重建、可视化的扩增子数据处理流程 |
| bioBakery[ | 宏基因组/扩增子测序 | 基于比对的宏基因组数据处理流程 |
| metaWRAP[ | 宏基因组 | 基于比对的宏基因组数据处理流程 |
| MetaQUBIC[ | 宏基因组/宏转录组 | 基于双聚类的功能基因分类软件 |
| MetaTrans[ | 宏转录组 | 从RNA-Seq数据分析微生物群落结构和功能的数据处理流程 |
| SAMSA[ | 宏转录组 | 用于分析肠道微生物组数据的软件,重点关注样本中的生物体特异性活动或功能活动 |
| MG-RAST[ | 宏基因组/宏转录组 | 通过比较蛋白质和核苷酸数据库产生宏基因组序列功能分类的软件 |
| IdentiPy[ | 宏蛋白组 | 肽识别、搜索、验证、蛋白质推断和量化的数据处理流程 |
| Trans-Proteomic Pipeline[ | 宏蛋白组 | 大规模可重复的定量MS蛋白质组学数据处理软件 |
| compleXView[ | 宏蛋白组 | 根据蛋白质组数据计算丰度、再现性和特异性的度量以推断PPI的网络服务器 |
| Pathos[ | 宏代谢组 | 分析质谱数据并显示代谢物和代谢途径的网络服务器 |
| MetaboAnalyst[ | 宏代谢组/多组学整合 | 用于集成代谢组学数据分析、解释和与其他组学数据集成的网络服务器 |
| Netome[ | 宏代谢组/多组学整合 | 用于处理和分析代谢组学数据以及探索与其他组学数据和元数据关联的工具 |
表1 宏组学数据的主要处理软件
Tab. 1 Major software for processing meta-omics data
| 软件名 | 对应处理的组学数据类型 | 功能 |
|---|---|---|
| QIIME[ | 扩增子测序 | 用于质量控制、OTU处理、物种分类、系统发育重建、可视化的扩增子数据处理流程 |
| bioBakery[ | 宏基因组/扩增子测序 | 基于比对的宏基因组数据处理流程 |
| metaWRAP[ | 宏基因组 | 基于比对的宏基因组数据处理流程 |
| MetaQUBIC[ | 宏基因组/宏转录组 | 基于双聚类的功能基因分类软件 |
| MetaTrans[ | 宏转录组 | 从RNA-Seq数据分析微生物群落结构和功能的数据处理流程 |
| SAMSA[ | 宏转录组 | 用于分析肠道微生物组数据的软件,重点关注样本中的生物体特异性活动或功能活动 |
| MG-RAST[ | 宏基因组/宏转录组 | 通过比较蛋白质和核苷酸数据库产生宏基因组序列功能分类的软件 |
| IdentiPy[ | 宏蛋白组 | 肽识别、搜索、验证、蛋白质推断和量化的数据处理流程 |
| Trans-Proteomic Pipeline[ | 宏蛋白组 | 大规模可重复的定量MS蛋白质组学数据处理软件 |
| compleXView[ | 宏蛋白组 | 根据蛋白质组数据计算丰度、再现性和特异性的度量以推断PPI的网络服务器 |
| Pathos[ | 宏代谢组 | 分析质谱数据并显示代谢物和代谢途径的网络服务器 |
| MetaboAnalyst[ | 宏代谢组/多组学整合 | 用于集成代谢组学数据分析、解释和与其他组学数据集成的网络服务器 |
| Netome[ | 宏代谢组/多组学整合 | 用于处理和分析代谢组学数据以及探索与其他组学数据和元数据关联的工具 |
| 1 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010—2020[J]. Nature Communications, 2020, 11: 5174. |
| 2 | PADDON C J, KEASLING J D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development[J]. Nature Reviews Microbiology, 2014, 12(5): 355-367. |
| 3 | CALERO P, NIKEL P I. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms[J]. Microbial Biotechnology, 2019, 12(1): 98-124. |
| 4 | NIELSEN J, KEASLING J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. |
| 5 | WU G, YAN Q, JONES J A, et al. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications[J]. Trends in Biotechnology, 2016, 34(8): 652-664. |
| 6 | SHOU W Y, RAM S, VILAR J M G. Synthetic cooperation in engineered yeast populations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6): 1877-1882. |
| 7 | BRENNER K, ARNOLD F H. Self-organization, layered structure, and aggregation enhance persistence of a synthetic biofilm consortium[J]. PLoS One, 2011, 6(2): e16791. |
| 8 | MCCARTY N S, LEDESMA-AMARO R. Synthetic biology tools to engineer microbial communities for biotechnology[J]. Trends in Biotechnology, 2019, 37(2): 181-197. |
| 9 | STENUIT B, AGATHOS S N. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology[J]. Current Opinion in Biotechnology, 2015, 33: 305-317. |
| 10 | TSOI R, WU F L, ZHANG C, et al. Metabolic division of labor in microbial systems[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2526-2531. |
| 11 | BASSLER B L, LOSICK R. Bacterially speaking[J]. Cell, 2006, 125(2): 237-246. |
| 12 | ZHANG H R, PEREIRA B, LI Z J, et al. Engineering Escherichia coli coculture systems for the production of biochemical products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(27): 8266-8271. |
| 13 | XIA T, EITEMAN M A, ALTMAN E. Simultaneous utilization of glucose, xylose and Arabinose in the presence of acetate by a consortium of Escherichia coli strains[J]. Microbial Cell Factories, 2012, 11: 77. |
| 14 | VERHOEVEN M D, DE VALK S C, DARAN J M G, et al. Fermentation of glucose-xylose-arabinose mixtures by a synthetic consortium of single-sugar-fermenting Saccharomyces cerevisiae strains[J]. FEMS Yeast Research, 2018, 18(8): foy075. |
| 15 | MINTY J J, SINGER M E, SCHOLZ S A, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(36): 14592-14597. |
| 16 | GILBERT E S, WALKER A W, KEASLING J D. A constructed microbial consortium for biodegradation of the organophosphorus insecticide parathion[J]. Applied Microbiology and Biotechnology, 2003, 61(1): 77-81. |
| 17 | BOKINSKY G, PERALTA-YAHYA P P, GEORGE A, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(50): 19949-19954. |
| 18 | TSAI S L, GOYAL G, CHEN W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production[J]. Applied and Environmental Microbiology, 2010, 76(22): 7514-7520. |
| 19 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4): 377-383. |
| 20 | MINAMI H, KIM J S, IKEZAWA N, et al. Microbial production of plant benzylisoquinoline alkaloids[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(21): 7393-7398. |
| 21 | JONES J A, VERNACCHIO V R, COLLINS S M, et al. Complete biosynthesis of anthocyanins using E. coli polycultures[J]. mBio, 2017, 8(3): e00621-e00617. |
| 22 | VILLARREAL F, CONTRERAS-LLANO L E, CHAVEZ M, et al. Synthetic microbial consortia enable rapid assembly of pure translation machinery[J]. Nature Chemical Biology, 2018, 14(1): 29-35. |
| 23 | BAYER T S, WIDMAIER D M, TEMME K, et al. Synthesis of methyl halides from biomass using engineered microbes[J]. Journal of the American Chemical Society, 2009, 131(18): 6508-6515. |
| 24 | WANG E X, DING M Z, MA Q, et al. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation[J]. Microbial Cell Factories, 2016, 15: 21. |
| 25 | CHEN A Y, DENG Z T, BILLINGS A N, et al. Synthesis and patterning of tunable multiscale materials with engineered cells[J]. Nature Materials, 2014, 13(5): 515-523. |
| 26 | GILBERT C, HOWARTH M, HARWOOD C R, et al. Extracellular self-assembly of functional and tunable protein conjugates from Bacillus subtilis [J]. ACS Synthetic Biology, 2017, 6(6): 957-967. |
| 27 | HONG S H, HEGDE M, KIM J, et al. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device[J]. Nature Communications, 2012, 3: 613. |
| 28 | URRIOS A, MACIA J, MANZONI R, et al. A synthetic multicellular memory device[J]. ACS Synthetic Biology, 2016, 5(8): 862-873. |
| 29 | MACIA J, MANZONI R, CONDE N, et al. Implementation of complex biological logic circuits using spatially distributed multicellular consortia[J]. PLoS Computational Biology, 2016, 12(2): e1004685. |
| 30 | REGOT S, MACIA J, CONDE N, et al. Distributed biological computation with multicellular engineered networks[J]. Nature, 2011, 469(7329): 207-211. |
| 31 | TAMSIR A, TABOR J J, VOIGT C A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'[J]. Nature, 2011, 469(7329): 212-215. |
| 32 | BOEHM C R, GRANT P K, HASELOFF J. Programmed hierarchical patterning of bacterial populations[J]. Nature Communications, 2018, 9: 776. |
| 33 | BASU S, GERCHMAN Y, COLLINS C H, et al. A synthetic multicellular system for programmed pattern formation[J]. Nature, 2005, 434(7037): 1130-1134. |
| 34 | XIU Y, JANG S, JONES J A, et al. Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures[J]. Biotechnology and Bioengineering, 2017, 114(10): 2235-2244. |
| 35 | MEYER A, PELLAUX R, POTOT S, et al. Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors[J]. Nature Chemistry, 2015, 7(8): 673-678. |
| 36 | HUGENHOLTZ P, TYSON G W. Metagenomics[J]. Nature, 2008, 455(7212): 481-483. |
| 37 | RUSSELL S, NORVIG P. Artificial intelligence: a modern approach[M]. 2nd ed. Prentice Hall, 2003. |
| 38 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12): 725-741. |
| 39 | MILLER M B, BASSLER B L. Quorum sensing in bacteria[J]. Annual Review of Microbiology, 2001, 55: 165-199. |
| 40 | SCOTT S R, HASTY J. Quorum sensing communication modules for microbial consortia[J]. ACS Synthetic Biology, 2016, 5(9): 969-977. |
| 41 | MIANO A, LIAO M J, HASTY J. Inducible cell-to-cell signaling for tunable dynamics in microbial communities[J]. Nature Communications, 2020, 11: 1193. |
| 42 | DU P, ZHAO H W, ZHANG H Q, et al. De novo design of an intercellular signaling toolbox for multi-channel cell-cell communication and biological computation[J]. Nature Communications, 2020, 11: 4226. |
| 43 | KONG W T, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14(8): 821-829. |
| 44 | LÓPEZ-MAURY L, MARGUERAT S, BÄHLER J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation[J]. Nature Reviews Genetics, 2008, 9(8): 583-593. |
| 45 | ZENGLER K, ZARAMELA L S. The social network of microorganisms—how auxotrophies shape complex communities[J]. Nature Reviews Microbiology, 2018, 16(6): 383-390. |
| 46 | MEE M T, COLLINS J J, CHURCH G M, et al. Syntrophic exchange in synthetic microbial communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(20): E2149-E2156. |
| 47 | WEGNER A, MEISER J, WEINDL D, et al. How metabolites modulate metabolic flux[J]. Current Opinion in Biotechnology, 2015, 34: 16-22. |
| 48 | SCOTT S R, DIN M O, BITTIHN P, et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis[J]. Nature Microbiology, 2017, 2: 17083. |
| 49 | ELIAS S, BANIN E. Multi-species biofilms: living with friendly neighbors[J]. FEMS Microbiology Reviews, 2012, 36(5): 990-1004. |
| 50 | GLASS D S, RIEDEL-KRUSE I H. A synthetic bacterial cell-cell adhesion toolbox for programming multicellular morphologies and patterns[J]. Cell, 2018, 174(3): 649-658.e16. |
| 51 | ALPHENAAR P A, VISSER A, LETTINGA G. The effect of liquid upward velocity and hydraulic retention time on granulation in UASB reactors treating wastewater with a high sulphate content[J]. Bioresource Technology, 1993, 43(3): 249-258. |
| 52 | LIU Y, TAY J H. The essential role of hydrodynamic shear force in the formation of biofilm and granular sludge[J]. Water Research, 2002, 36(7): 1653-1665. |
| 53 | BATSTONE D J, PUYOL D, FLORES-ALSINA X, et al. Mathematical modelling of anaerobic digestion processes: applications and future needs[J]. Reviews in Environmental Science and Bio/Technology, 2015, 14(4): 595-613. |
| 54 | PICIOREANU C, KREFT J U, LOOSDRECHT M C M VAN. Particle-based multidimensional multi species biofilm model[J]. Applied and Environmental Microbiology, 2004, 70(5): 3024-3040. |
| 55 | BROWN C T, HUG L A, THOMAS B C, et al. Unusual biology across a group comprising more than 15% of domain Bacteria[J]. Nature, 2015, 523(7559): 208-211. |
| 56 | HOVER B M, KIM S H, KATZ M, et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens[J]. Nature Microbiology, 2018, 3(4): 415-422. |
| 57 | MARSHALL J R, YAO P Y, MONTGOMERY S L, et al. Screening and characterization of a diverse panel of metagenomic imine reductases for biocatalytic reductive amination[J]. Nature Chemistry, 2021, 13(2): 140-148. |
| 58 | BURSTEIN D, HARRINGTON L B, STRUTT S C, et al. New CRISPR-Cas systems from uncultivated microbes[J]. Nature, 2017, 542(7640): 237-241. |
| 59 | JOHNS N I, GOMES A L C, YIM S S, et al. Metagenomic mining of regulatory elements enables programmable species-selective gene expression[J]. Nature Methods, 2018, 15(5): 323-329. |
| 60 | RONDA C, CHEN S P, CABRAL V, et al. Metagenomic engineering of the mammalian gut microbiome in situ [J]. Nature Methods, 2019, 16(2): 167-170. |
| 61 | LAM K N, SPANOGIANNOPOULOS P, SOTO-PEREZ P, et al. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome[J]. Cell Reports, 2021, 37(5): 109930. |
| 62 | ZHAO L P, ZHANG F, DING X Y, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes[J]. Science, 2018, 359(6380): 1151-1156. |
| 63 | DREKONJA D, REICH J, GEZAHEGN S, et al. Fecal microbiota transplantation for clostridium difficile infection: a systematic review[J]. Annals of Internal Medicine, 2015, 162(9): 630-638. |
| 64 | SHETH R U, CABRAL V, CHEN S P, et al. Manipulating bacterial communities by in situ microbiome engineering[J]. Trends in Genetics, 2016, 32(4): 189-200. |
| 65 | KNIGHT R, VRBANAC A, TAYLOR B C, et al. Best practices for analysing microbiomes[J]. Nature Reviews Microbiology, 2018, 16(7): 410-422. |
| 66 | QUINCE C, WALKER A W, SIMPSON J T, et al. Shotgun metagenomics, from sampling to analysis[J]. Nature Biotechnology, 2017, 35(9): 833-844. |
| 67 | WILMES P, BOND P L. Metaproteomics: studying functional gene expression in microbial ecosystems[J]. Trends in Microbiology, 2006, 14(2): 92-97. |
| 68 | NICHOLSON J K. Global systems biology, personalized medicine and molecular epidemiology[J]. Molecular Systems Biology, 2006, 2: 52. |
| 69 | PACE N R, STAHL D A, LANE D J, et al. The analysis of natural microbial populations by ribosomal RNA sequences[M]// Marshall K C. Advances in microbial ecology. Springer, 1986: 1-55. |
| 70 | GROUP N I H H W, PETERSON J, GARGES S, et al. The NIH human microbiome project[J]. Genome Research, 2009, 19(12): 2317-2323. |
| 71 | WERNER J J, KNIGHTS D, GARCIA M L, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(10): 4158-4163. |
| 72 | HEROLD M, MARTÍNEZ ARBAS S, NARAYANASAMY S, et al. Integration of time-series meta-omics data reveals how microbial ecosystems respond to disturbance[J]. Nature Communications, 2020, 11: 5281. |
| 73 | YU K, YI S, LI B, et al. An integrated meta-omics approach reveals substrates involved in synergistic interactions in a bisphenol A (BPA)-degrading microbial community[J]. Microbiome, 2019, 7(1): 16. |
| 74 | SEGATA N, BOERNIGEN D, TICKLE T L, et al. Computational meta'omics for microbial community studies[J]. Molecular Systems Biology, 2013, 9: 666. |
| 75 | CAPORASO J G, KUCZYNSKI J, STOMBAUGH J, et al. QIIME allows analysis of high-throughput community sequencing data[J]. Nature Methods, 2010, 7(5): 335-336. |
| 76 | BOLYEN E, RIDEOUT J R, DILLON M R, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2[J]. Nature Biotechnology, 2019, 37(8): 852-857. |
| 77 | MCIVER L J, ABU-ALI G, FRANZOSA E A, et al. bioBakery: a meta'omic analysis environment[J]. Bioinformatics, 2017, 34(7): 1235-1237. |
| 78 | URITSKIY G V, DIRUGGIERO J, TAYLOR J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis[J]. Microbiome, 2018, 6(1): 158. |
| 79 | MA A J, SUN M X, MCDERMAID A, et al. MetaQUBIC: a computational pipeline for gene-level functional profiling of metagenome and metatranscriptome[J]. Bioinformatics, 2019, 35(21): 4474-4477. |
| 80 | MARTINEZ X, POZUELO M, PASCAL V, et al. MetaTrans: an open-source pipeline for metatranscriptomics[J]. Scientific Reports, 2016, 6: 26447. |
| 81 | WESTREICH S T, KORF I, MILLS D A, et al. SAMSA: a comprehensive metatranscriptome analysis pipeline[J]. BMC Bioinformatics, 2016, 17(1): 399. |
| 82 | MEYER F, PAARMANN D, D'SOUZA M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes[J]. BMC Bioinformatics, 2008, 9: 386. |
| 83 | LEVITSKY L I, IVANOV M V, LOBAS A A, et al. IdentiPy: an extensible search engine for protein identification in shotgun proteomics[J]. Journal of Proteome Research, 2018, 17(7): 2249-2255. |
| 84 | DEUTSCH E W, MENDOZA L, SHTEYNBERG D, et al. A guided tour of the Trans-Proteomic Pipeline[J]. Proteomics, 2010, 10(6): 1150-1159. |
| 85 | SOLIS-MEZARINO V, HERZOG F. compleXView: a server for the interpretation of protein abundance and connectivity information to identify protein complexes[J]. Nucleic Acids Research, 2017, 45(W1): W276-W284. |
| 86 | LEADER D P, BURGESS K, CREEK D, et al. Pathos: a web facility that uses metabolic maps to display experimental changes in metabolites identified by mass spectrometry[J]. Rapid Communications in Mass Spectrometry, 2011, 25(22): 3422-3426. |
| 87 | CHONG J, SOUFAN O, LI C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis[J]. Nucleic Acids Research, 2018, 46(W1): W486-W494. |
| 88 | RAHNAVARD A, HITCHCOCK D, PACHECO J A, et al. Netome: a computational framework for metabolite profiling and omics network analysis[E/OL]. BioRxiv, 2018: 443903.. |
| 89 | CALLAHAN B J, MCMURDIE P J, ROSEN M J, et al. DADA2: High-resolution sample inference from Illumina amplicon data[J]. Nature Methods, 2016, 13(7): 581-583. |
| 90 | KneadData. BioBakery 2017, |
| 91 | BEGHINI F, MCIVER L J, BLANCO-MÍGUEZ A, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3[J]. eLife, 2021, 10: e65088. |
| 92 | FRANZOSA E A, MCIVER L J, RAHNAVARD G, et al. Species-level functional profiling of metagenomes and metatranscriptomes[J]. Nature Methods, 2018, 15(11): 962-968. |
| 93 | TRUONG D T, TETT A, PASOLLI E, et al. Microbial strain-level population structure and genetic diversity from metagenomes[J]. Genome Research, 2017, 27(4): 626-638. |
| 94 | LI D H, LUO R B, LIU C M, et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices[J]. Methods, 2016, 102: 3-11. |
| 95 | NURK S, MELESHKO D, KOROBEYNIKOV A, et al. metaSPAdes: a new versatile metagenomic assembler[J]. Genome Research, 2017, 27(5): 824-834. |
| 96 | LU J, BREITWIESER F P, THIELEN P, et al. Bracken: estimating species abundance in metagenomics data[J]. PeerJ Computer Science, 2017, 3: e104. |
| 97 | ALNEBERG J, BJARNASON B S, DE BRUIJN I, et al. Binning metagenomic contigs by coverage and composition[J]. Nature Methods, 2014, 11(11): 1144-1146. |
| 98 | WU Y W, SIMMONS B A, SINGER S W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets[J]. Bioinformatics, 2015, 32(4): 605-607. |
| 99 | KANG D D, FROULA J, EGAN R, et al. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities[J]. PeerJ, 2015, 3: e1165. |
| 100 | MACHADO D, ANDREJEV S, TRAMONTANO M, et al. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities[J]. Nucleic Acids Research, 2018, 46(15): 7542-7553. |
| 101 | ZORRILLA F, BURIC F, PATIL K R, et al. metaGEM: reconstruction of genome scale metabolic models directly from metagenomes[J]. Nucleic Acids Research, 2021, 49(21): e126. |
| 102 | GLAZ B, WANG C, HURLEY M, et al. Artificial intelligence in synthetic biology, defense cyber, and aeromechanical design[R]. CCDC Army Research Laboratory Adelphi United States, 2020. |
| 103 | SOUEIDAN H, NIKOLSKI M. Machine learning for metagenomics: methods and tools[E/OL]. arXiv:1510.06621, 2015.. |
| 104 | THOMPSON L R, SANDERS J G, MCDONALD D, et al. A communal catalogue reveals Earth's multiscale microbial diversity[J]. Nature, 2017, 551(7681): 457-463. |
| 105 | WANG S Z, CAO J N, YU P. Deep learning for spatio-temporal data mining: a survey[J]. IEEE Transactions on Knowledge and Data Engineering, 2020. doi: 10.1109/TKDE.2020.3025580 . |
| 106 | MONTAVON G, SAMEK W, MÜLLER K R. Methods for interpreting and understanding deep neural networks[J]. Digital Signal Processing, 2018, 73: 1-15. |
| 107 | YANG J Y, ANISHCHENKO I, PARK H, et al. Improved protein structure prediction using predicted interresidue orientations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(3): 1496-1503. |
| 108 | ANDERSON C. Google's AI tool DeepVariant promises significantly fewer genome errors[J]. Clinical OMICs, 2018, 5(1): 33. |
| 109 | LIU Z Q, MA A J, MATHÉ E, et al. Network analyses in microbiome based on high-throughput multi-omics data[J]. Briefings in Bioinformatics, 2020, 22(2): 1639-1655. |
| 110 | ZHOU J, CUI G Q, HU S D, et al. Graph neural networks: a review of methods and applications[J]. AI Open, 2020, 1: 57-81. |
| 111 | RAMACHANDRAM D, TAYLOR G W. Deep multimodal learning: a survey on recent advances and trends[J]. IEEE Signal Processing Magazine, 2017, 34(6): 96-108. |
| 112 | LIN G M, WARDEN-ROTHMAN R, VOIGT C A. Retrosynthetic design of metabolic pathways to chemicals not found in nature[J]. Current Opinion in Systems Biology, 2019, 14: 82-107. |
| 113 | NGUYEN T T, NGUYEN N D, NAHAVANDI S. Deep reinforcement learning for multiagent systems: a review of challenges, solutions, and applications[J]. IEEE Transactions on Cybernetics, 2020, 50(9): 3826-3839. |
| [1] | 王也, 王昊晨, 晏明皓, 胡冠华, 汪小我. 生物分子序列的人工智能设计[J]. 合成生物学, 2021, 2(1): 1-14. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||