合成生物学 ›› 2021, Vol. 2 ›› Issue (3): 428-443.DOI: 10.12211/2096-8280.2021-023
• 研究论文 • 上一篇
细胞内大片段DNA数据存储的多RS码交织编码
陈为刚1,2, 葛奇1, 王盼盼1, 韩明哲2,3, 郭健1
- 1.天津大学微电子学院,天津 300072
2.教育部合成生物学前沿科学中心,天津大学,天津 300072
3.天津大学化工学院,天津 300072
-
收稿日期:2021-02-09修回日期:2021-03-28出版日期:2021-06-30发布日期:2021-07-14 -
通讯作者:陈为刚 -
作者简介:陈为刚 (1980—),男,博士,副教授。研究方向为DNA数据存储、信息论与编码理论。 E-mail:chenwg@tju.edu.cn
Multiple interleaved RS codes for data storage using up to Mb-scale synthetic DNA in living cells
CHEN Weigang1,2, GE Qi1, WANG Panpan1, HAN Mingzhe2,3, GUO Jian1
- 1.School of Microelectronics,Tianjin University,Tianjin 300072,China
2.Frontiers Science Center for Synthetic Biology (MOE),Tianjin University,Tianjin 300072,China
3.School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
-
Received:2021-02-09Revised:2021-03-28Online:2021-06-30Published:2021-07-14 -
Contact:CHEN Weigang
摘要:
合成DNA作为潜在的数字信息存储介质,存储密度高,可用时间久,有望成为未来数据存储的重要选项。然而,DNA的合成与测序读出往往造成碱基的多种错误,无法满足数据存储的可靠性要求,而保证可靠性的编码方案往往效率较低。针对该问题,提出了一种面向酿酒酵母内大片段DNA数据存储的高效率编码方法。数据编码通过多个极高码率的里德-所罗门(RS)码的码字交织构建数据DNA单元,将其与酵母的自主复制序列(ARS)交替镶嵌,构成酵母人工染色体序列;数据读出时,利用二代高通量测序,组合了读段从头(de novo)组装、ARS导引例,用20×二代测序数据可无错恢复原始数据。该编码方法不仅能实现数据可靠存储,实现的DNA数据部分逻辑密度为1.973 bit/bp,即使考虑生物单元开销,总体逻辑密度仍达到1.947 bit/bp。该设计流程可支持Kb到Mb不同长度的DNA的编码,为大片段DNA数据存储的“湿”实验提供灵活的实验前验证与评估。
中图分类号:
引用本文
陈为刚, 葛奇, 王盼盼, 韩明哲, 郭健. 细胞内大片段DNA数据存储的多RS码交织编码[J]. 合成生物学, 2021, 2(3): 428-443.
CHEN Weigang, GE Qi, WANG Panpan, HAN Mingzhe, GUO Jian. Multiple interleaved RS codes for data storage using up to Mb-scale synthetic DNA in living cells[J]. Synthetic Biology Journal, 2021, 2(3): 428-443.
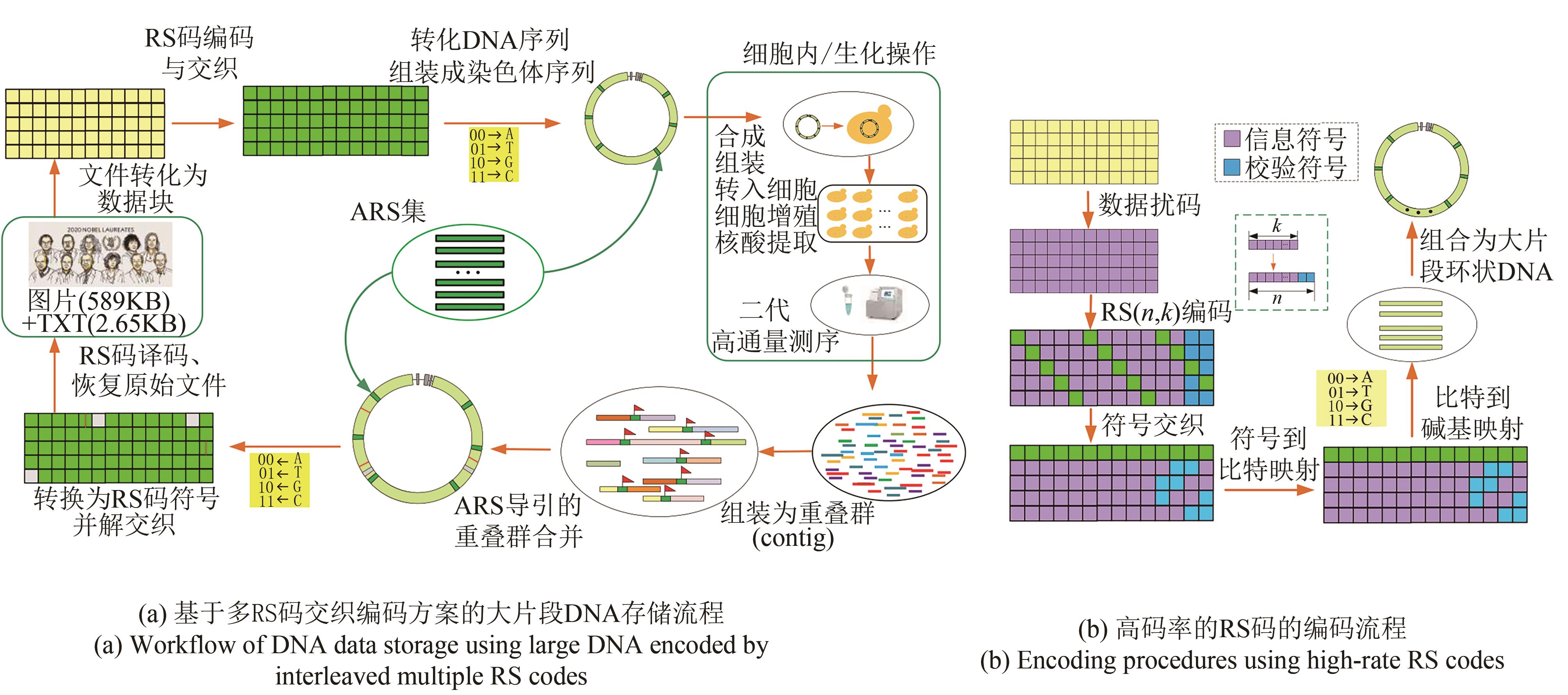
图1 面向大片段DNA数据存储的高码率RS码编码方法[(a) Workflow of DNA data storage with large DNA fragments encoded by interleaved multiple RS codes. The workflow consists of three steps. First, digital data for a picture and a text file was converted into DNA sequences by interleaved multiple RS codes. Second, the artificial chromosome was assembled from multiple DNA sequences with ARS to stabilize the assembly and replication. Third, with the high-throughput sequencing, data readout uses short read assembly based on the de Bruijn graphs, ARS guided contig combination and RS erasure and error correction to achieve reliable data recovery. (b) Encoding procedures using high code rate RS codes. The encoding procedures include data scrambling, RS encoding, symbol interleaving, symbol-to-bit mapping, bit-to-base mapping and combining with ARSs to form a ring chromosome]
Fig. 1 Encoding scheme using a very high code rate RS codes for data storage with large DNA
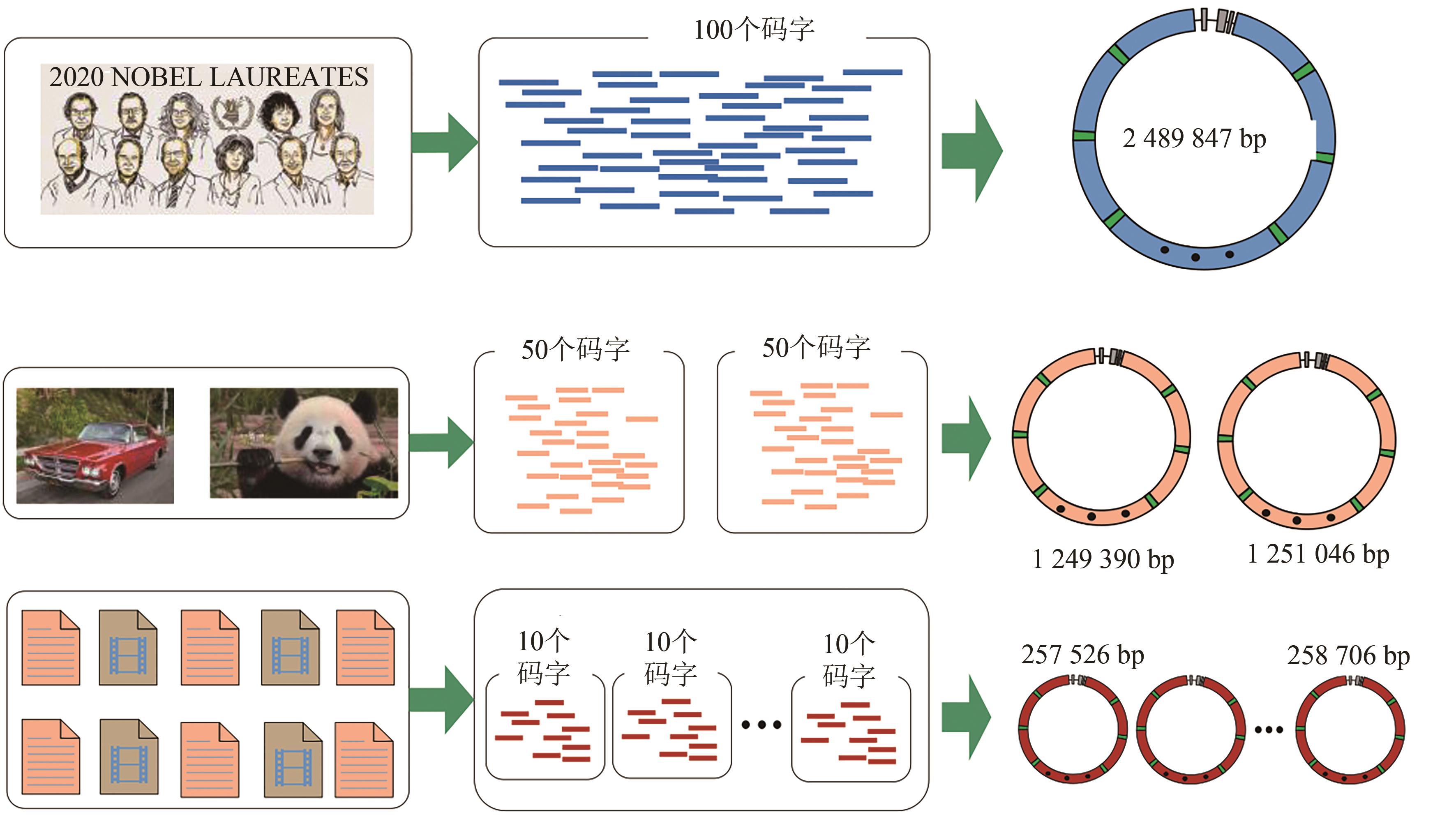
图2 不同数量数据段组合构建不同长度的大片段DNA(Three examples are shown in details. In example 1, the original file was converted into the artificial chromosome with a length of 2 489 847 bp. In example 2, two picture files were converted into two smaller artificial chromosomes. In example 3, ten small files were converted into ten independent small artificial chromosomes)
Fig. 2 Building of variant-length large DNA integrating different number of data blocks
| 主要结果 | 总体逻辑密度 (包含引物或载体骨架) /bit·bp-1 | 数据部分逻辑密度(不包含引物或载体骨架) /bit·bp-1 |
|---|---|---|
| Church等[ | 0.60 | 0.83 |
| Goldman等[ | 0.19 | 0.29 |
| Grass等[ | 0.83 | 1.16 |
| Bornholt等[ | 0.57 | 0.85 |
| Erlich等[ | 1.19 | 1.57 |
| Blawat等[ | 0.89 | 1.08 |
| Organick等[ | 0.81 | 1.10 |
| Chen 等[ | 1.19 | 1.24 |
| Ping 等[ | 1.32 | 1.88 |
| 本工作(单段DNA,2 489 847 p) | 1.947 | 1.973 |
表1 不同编码方法的碱基逻辑密度比较
Tab. 1 Base logical density using different encoding schemes
| 主要结果 | 总体逻辑密度 (包含引物或载体骨架) /bit·bp-1 | 数据部分逻辑密度(不包含引物或载体骨架) /bit·bp-1 |
|---|---|---|
| Church等[ | 0.60 | 0.83 |
| Goldman等[ | 0.19 | 0.29 |
| Grass等[ | 0.83 | 1.16 |
| Bornholt等[ | 0.57 | 0.85 |
| Erlich等[ | 1.19 | 1.57 |
| Blawat等[ | 0.89 | 1.08 |
| Organick等[ | 0.81 | 1.10 |
| Chen 等[ | 1.19 | 1.24 |
| Ping 等[ | 1.32 | 1.88 |
| 本工作(单段DNA,2 489 847 p) | 1.947 | 1.973 |
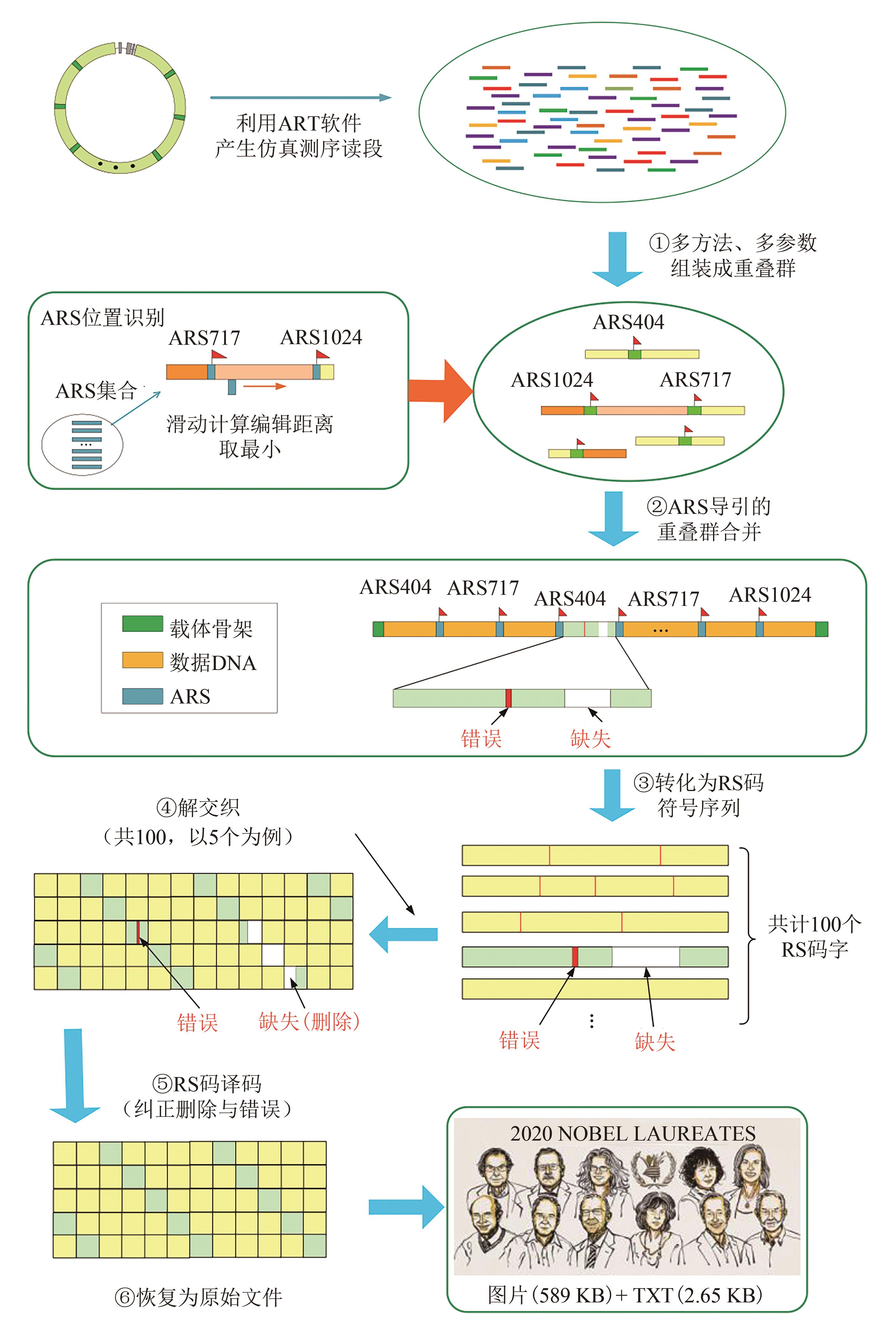
图3 基于短读段从头组装、ARS引导的多重叠群合并、RS码纠错纠删的数据恢复流程
Fig. 3 Data readout processes (The process includes de novo assembly from short reads using multiple programs with multiple k-mers, ARS navigated combination of multiple contigs, converting into symbol sequences of RS code, deinterleaving, and RS erasure and error correction)
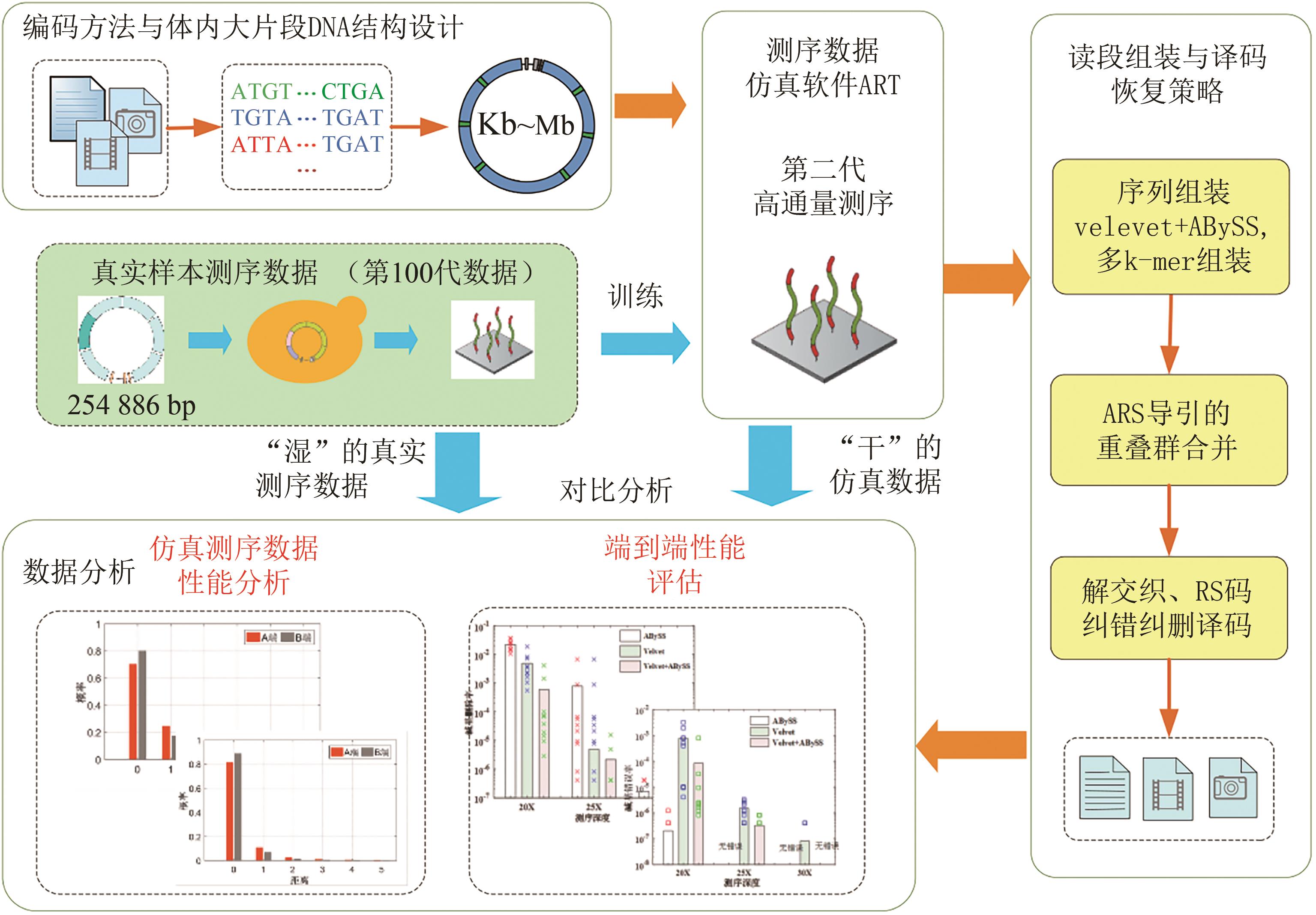
图4 基于计算机仿真的编码大片段DNA体内存储验证流程(Sequencing reads from real large DNA chunks were used to train the parameters of high-throughput sequencing and end-to-end performance verification were performed. And reads assembly and decoding recovery schemes include contig assembly, ARS navigated combination of multiple contigs, deinterleaving and RS erasure and error correction)
Fig. 4 Verification procedures using computer simulation for proposed encoding method and construction scheme of large DNA chunks in living cells
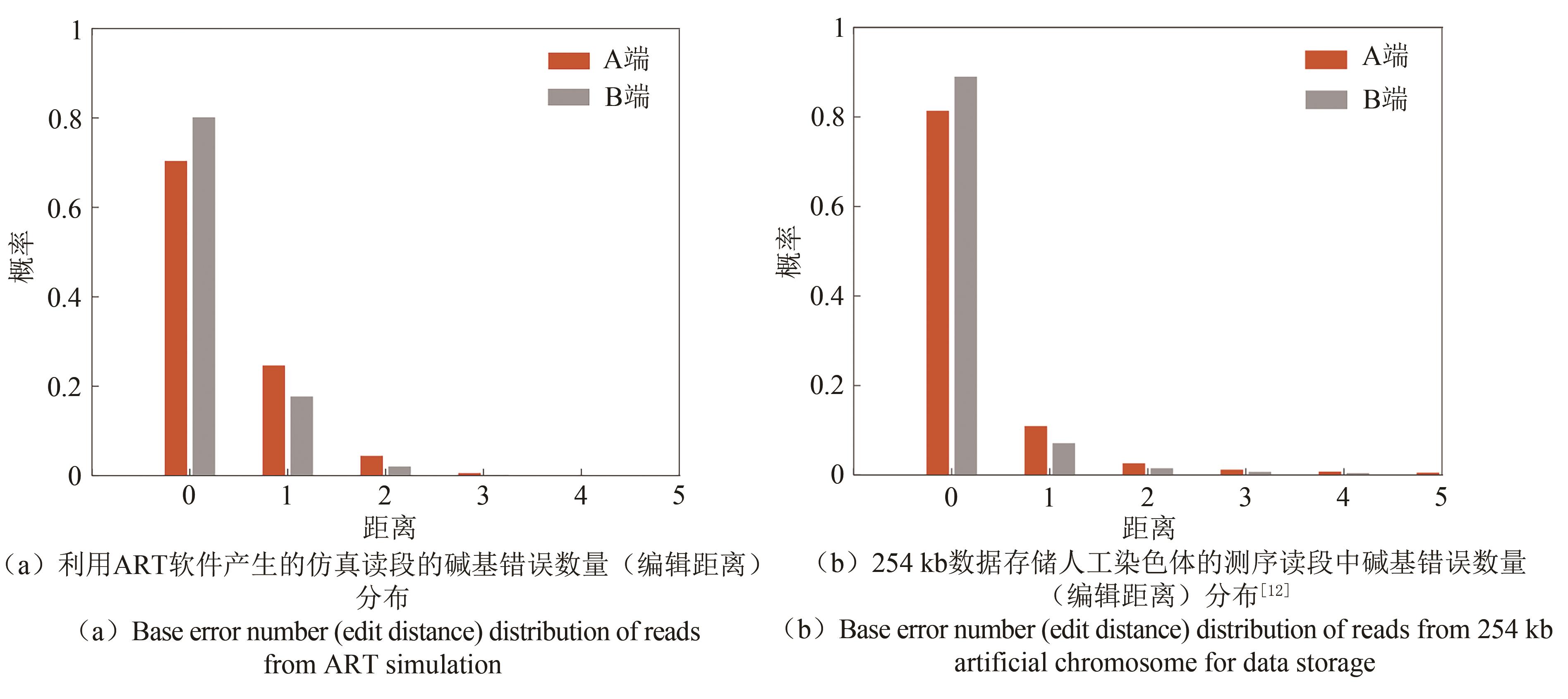
图5 仿真读段与实际测序读段的碱基错误数量(编辑距离)分布(Red columns represent sequencing data from one end. Grey columns represent sequencing data from the other end. The probability of reads with errors from ART simulation is about 20%. And the quality of reads from ART simulation is slightly less good than real sequencing reads, which can well illustrate the error correction ability of the proposed method)
Fig. 5 Base error number (edit distance) distribution in simulation and real sequencing reads.
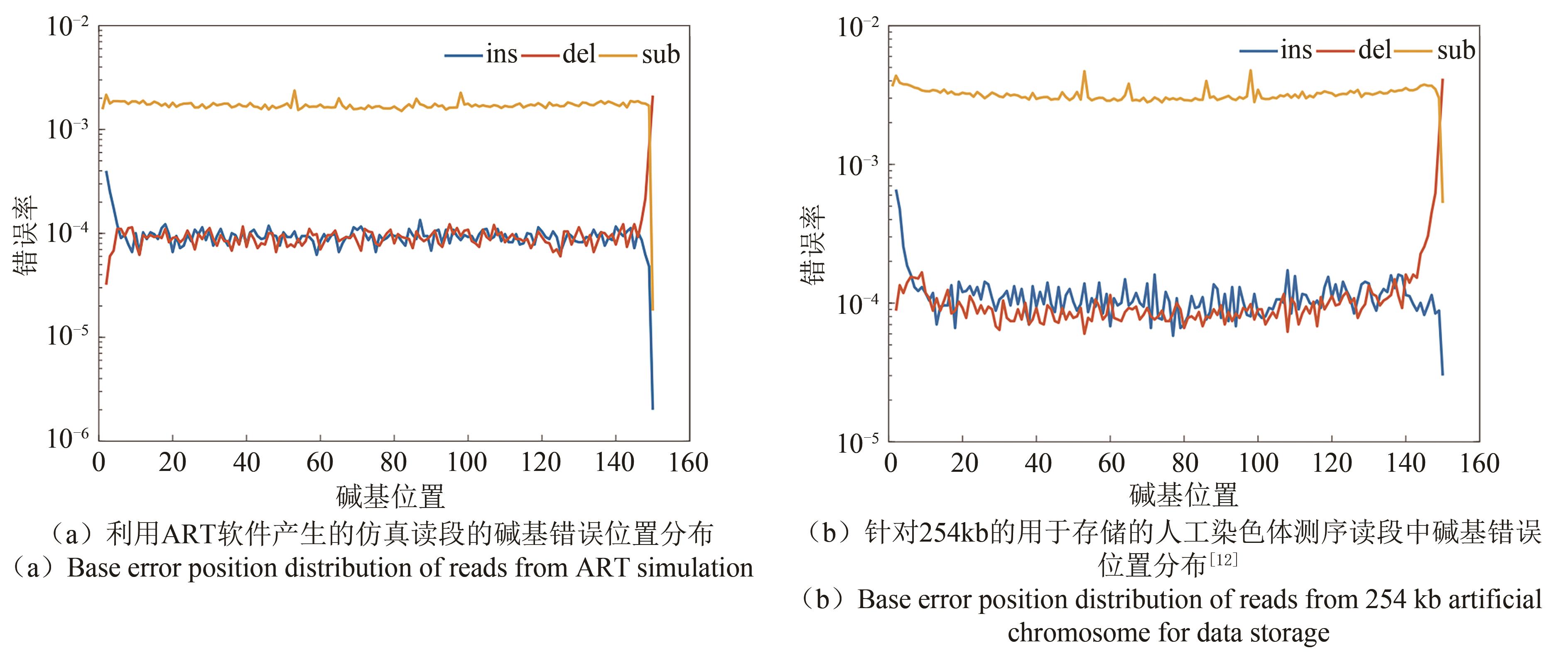
图6 仿真与实测读段的碱基错误随着位置变化情况(The second-generation sequencing reads contain insertion, deletion and substitution errors, and the error rate is about 10-4~10-3. Furthermore, the error rate of insertions and deletions is significantly lower than that of substitutions. Reads from ART simulation are very consistent with the characteristics of the real second-generation sequencing reads. It is verified that using simulation method to generate sequencing reads is feasible)
Fig. 6 Base error position distribution in simulation and real sequencing.
测序 覆盖度 | 组装软件 | k-mer | 非交织RS码不能恢复数据段数量以及缺失与错误(缺失数量+错误数量) | 解交织后RS码译码 |
|---|---|---|---|---|
| 20× | Velvet | {25,27,29,31,33} | 实验1:2个(2705+0,2450+7711) | 成功 |
| 实验2:3个(3525+0,673+1913,5194+0) | 成功 | |||
| 实验3:5个(6779+0,11003+0, 45+1855,9296+0,18612+0) | 失败 | |||
| 实验4:1个(5498+0) | 成功 | |||
| 实验5:1个(17306+0) | 成功 | |||
| 实验6:1个(8387+0) | 成功 | |||
| 实验7:1个(1134+0) | 成功 | |||
| 实验8:3个(2668+16,8320+0,9298+919) | 成功 | |||
| 实验9:1个(2466+1213) | 成功 | |||
| 实验10:2个(3657+0,0+4712) | 成功 | |||
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验2(3525+1,1+1905) | 成功 | |
| 实验5(10041+0) | 成功 | |||
| 25× | Velvet | 25 | 实验6(937+0) | 成功 |
| 实验9(10620+1) | 成功 | |||
| 27 | 实验2(13604+1) | 成功 | ||
| 29 | 实验8(21488+0,3373+0) | 成功 | ||
| 实验10(13023+17) | 成功 | |||
| 31 | 实验2(1692+0) | 成功 | ||
| 实验7(1105+0) | 成功 | |||
| 实验8(3373+0) | 成功 | |||
| 33 | 实验2(1696+0) | 成功 | ||
| 实验10(1854+1986) | 成功 | |||
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1(16950+0) | 成功 | |
| 实验8(2151+0) | 成功 | |||
| Velvet+ ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| 30× | Velvet | 27 | 实验1~7, 9,10均无大片段错误 实验8(1292+0) | 成功 |
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 |
表2 采用交织多个RS码的数据恢复分析
Tab. 2 Data recovery analysis using interleaved multiple RS codes
测序 覆盖度 | 组装软件 | k-mer | 非交织RS码不能恢复数据段数量以及缺失与错误(缺失数量+错误数量) | 解交织后RS码译码 |
|---|---|---|---|---|
| 20× | Velvet | {25,27,29,31,33} | 实验1:2个(2705+0,2450+7711) | 成功 |
| 实验2:3个(3525+0,673+1913,5194+0) | 成功 | |||
| 实验3:5个(6779+0,11003+0, 45+1855,9296+0,18612+0) | 失败 | |||
| 实验4:1个(5498+0) | 成功 | |||
| 实验5:1个(17306+0) | 成功 | |||
| 实验6:1个(8387+0) | 成功 | |||
| 实验7:1个(1134+0) | 成功 | |||
| 实验8:3个(2668+16,8320+0,9298+919) | 成功 | |||
| 实验9:1个(2466+1213) | 成功 | |||
| 实验10:2个(3657+0,0+4712) | 成功 | |||
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验2(3525+1,1+1905) | 成功 | |
| 实验5(10041+0) | 成功 | |||
| 25× | Velvet | 25 | 实验6(937+0) | 成功 |
| 实验9(10620+1) | 成功 | |||
| 27 | 实验2(13604+1) | 成功 | ||
| 29 | 实验8(21488+0,3373+0) | 成功 | ||
| 实验10(13023+17) | 成功 | |||
| 31 | 实验2(1692+0) | 成功 | ||
| 实验7(1105+0) | 成功 | |||
| 实验8(3373+0) | 成功 | |||
| 33 | 实验2(1696+0) | 成功 | ||
| 实验10(1854+1986) | 成功 | |||
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1(16950+0) | 成功 | |
| 实验8(2151+0) | 成功 | |||
| Velvet+ ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| 30× | Velvet | 27 | 实验1~7, 9,10均无大片段错误 实验8(1292+0) | 成功 |
| {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | ||
| ABySS | {25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 | |
| Velvet+ABySS | V{25,27,29,31,33} A{25,27,29,31,33} | 实验1~10均无大片段错误 | 成功 |

图7 不同测序覆盖度与组装方法,在ARS识别与重叠群合并后,数据DNA部分错误分布('×' and '□' represent base erasure probability and base error probability of 10 experiments respectively. The histogram represents the average of ten experiments. When the sequencing depth increases, the performance of different assembly methods improves, and the number of residual errors and erasures decreases rapidly)
Fig. 7 Base error distribution of the data DNA after ARS identification and contig merging using different assembly methods and sequencing coverage
| 1 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA [J]. Science, 2012, 337(6102): 1628. |
| 2 | GOLDMAN N, BERTONE P, CHEN S Y, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA [J]. Nature, 2013, 494(7435): 77-80. |
| 3 | MEISER L C, ANTKOWIAK P L, KOCH J, et al. Reading and writing digital data in DNA [J]. Nature Protocols, 2020, 15(1): 86-101. |
| 4 | DONG Y M, SUN F J, PING Z, et al. DNA storage: research landscape and future prospects [J]. National Science Review, 2020, 7(6): 1092-1107. |
| 5 | PING Z, MA D Z, HUANG X L, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage [J]. Gigascience, 2019, 8(6): giz075. |
| 6 | 董一名, 孙法家, 武瑞君, 等. DNA数字信息存储的研究进展[J]. 合成生物学, 2021, 2(3): 323-334. |
| DONG Yiming, SUN Fajia, WU Ruijun, et al. Research progress on DNA molecules for digital information storage [J]. Synthetic Biology Journal, 2021, 2(3): 323-334. | |
| 7 | 韩明哲,陈为刚, 宋理富, 等. DNA信息存储:生命系统与信息系统的桥梁 [J]. 合成生物学, 2021.2(3): 309-322. |
| HAN Mingzhe, CHEN Weigang, SONG Lifu,et al. DNA information storage:bridging biological and digital world [J]. Synthetic Biology Journal, 2021, 2(3):309-322. | |
| 8 | Semiconductor Industry Association (ISA), Semiconductor Research Corporation (SRC). The decadal plan for semiconductors [R/OL]. 2021. . |
| 9 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. Toward a DNA-based archival storage system [J]. IEEE Micro, 2017, 37(3): 98-104. |
| 10 | ERLICH Y, ZIELINSKI D. DNA fountain enables a robust and efficient storage architecture [J]. Science, 2017, 355(6328): 950-954. |
| 11 | YAZDI TABATABAEI HOSSEIN S M, GABRYS R, MILENKOVIC O. Portable and error-free DNA-based data storage [J]. Scientific Reports, 2017, 7(1): 5011. |
| 12 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage [J]. Nature Biotechnology, 2018, 36(3): 242-248. |
| 13 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes [J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555. |
| 14 | BLAWAT M, GAEDKE K, HUETTER I, et al. Forward error correction for DNA data storage [J]. Procedia Computer Science, 2016, 80: 1011-1022. |
| 15 | 陈为刚, 黄刚, 李炳志, 等. 音视频文件的DNA信息存储 [J]. 中国科学: 生命科学, 2020, 50(1): 81-85. |
| CHEN Weigang, HUANG Gang, LI Bingzhi, et al. DNA information storage for audio and video files [J]. SCIENTIA SINICA Vitae, 2020, 50(1): 81-85. | |
| 16 | PING Z, CHEN S, HUANG X, et al. Towards practical and robust DNA-based data archiving by codec system named Yin-Yang [EB/OL]. [2021-05-27]. . |
| 17 | PRESS W H, HAWKINS J A, JONES S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints [J]. Proceedings of the National Academy of Science of United States of America, 2020, 117(31): 18489-18496. |
| 18 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications [J]. Nature Methods, 2014, 11(5): 499-507. |
| 19 | CHEN W G, HAN M Z, ZHOU J T, et al. An artificial chromosome for data storage, [J] National Science Review, 2021, 8(5): nwab028. |
| 20 | DAVIS J. Microvenus [J]. Art Journal, 1996, 55(1): 70-74. |
| 21 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria [J]. Nature, 2017, 547(7663): 345-349. |
| 22 | HAO M, QIAO H Y, GAO Y, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale [J]. Communications Biology, 2020, 3(1): 416-424. |
| 23 | NGUYEN H H, PARK J, PARK S J, et al. Long-term stability and integrity of plasmid-based DNA data storage [J]. Polymers, 2018, 10(1): 28. |
| 24 | YIM S S, MCBEE R M, SONG A M, et al. Robust direct digital-to-biological data storage in living cells [J]. Nature Chemical Biology, 2021,17(3):246-253. |
| 25 | POSTMA E D, DASHKO S, BREEMEN L V, et al. A supernumerary designer chromosome for modular in vivo pathway assembly in Saccharomyces cerevisiae [J]. Nucleic Acids Research, 2021, 49(3): 1769-1783. |
| 26 | SONG L F, ZENG A P. Orthogonal information encoding in living cells with high error-tolerance, safety, and fidelity [J]. ACS Synthetic Biology, 2018, 7(3): 866-874. |
| 27 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome [J]. Science, 2010, 329(5987): 52-56. |
| 28 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X [J]. Science, 2017, 355(6329): eaaf4706. |
| 29 | XIE Z X, LI B Z, MITCHELL L A, et al. "Perfect" designer chromosome V and behavior of a ring derivative [J]. Science, 2017, 355(6329): eaaf4704. |
| 30 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome [J]. Science, 2017, 355(6329): eaaf4791. |
| 31 | FREDENS J, WANG K H, TORRE D D L, et al. Total synthesis of Escherichia coli with a recoded genome [J]. Nature, 2019, 569(7757): 514-518. |
| 32 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING Mingzhu, LI Bingzhi, WANG Ying, et al. Significant research progress in synthetic biology [J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 33 | 彭凯, 逯晓云, 程健, 等. DNA合成、组装与纠错技术研究进展[J]. 合成生物学, 2020, 1(6): 697-708. |
| PENG Kai, LU Xiaoyun, CHENG Jian, et al. Advances in technologies for de novo DNA synthesis,assembly and error correction [J]. Synthetic Biology Journal, 2020, 1(6): 697-708. | |
| 34 | 刘晓, 王慧媛, 熊燕, 等. 基因合成与基因组编辑[J]. 中国细胞生物学学报, 2019, 41(11): 2072-2082. |
| LIU Xiao, WANG Huiyuan, XIONG Yan, et al. Progress in gene synthesis and genome editing [J]. Chinese Journal of Cell Biology, 2019, 41(11): 2072-2082. | |
| 35 | 罗周卿, 戴俊彪. 合成基因组学:设计与合成的艺术[J]. 生物工程学报, 2017, 33(3): 331-342. |
| LUO Zhouqing, DAI Junbiao. Synthetic genomics: the art of design and synthesis [J]. Chinese Journal of Biotechnology 2017, 33(3): 331-342. | |
| 36 | 王会, 戴俊彪, 罗周卿. 基因组的"读-改-写"技术[J]. 合成生物学, 2020, 1(5): 503-515. |
| WANG Hui, DAI Junbiao, LUO Zhouqing. Reading, editing, and writing techniques for genome research [J]. Synthetic Biology Journal, 2020, 1(5): 503-515. | |
| 37 | KARAS B J, JABLANOVIC J, SUN L J, et al. Direct transfer of whole genomes from bacteria to yeast [J]. Nature Methods, 2013, 10(5): 410-412. |
| 38 | 朱雪龙. 应用信息论基础[M]. 北京: 清华大学出版社, 2001. |
| ZHU X L.Fundamentals of applied information theory[M]. Beijing: Tsinghua University Press, 2001. | |
| 39 | LIN S, COSTELLO D J. Error control coding [M]. London:Pearson Education Inc, 2004. |
| 40 | REED I S, SOLOMON G. Polynomial codes over certain finite fields [J]. Journal of the Society for Industrial & Applied Mathematics, 1960, 8(2): 300-304. |
| 41 | MATSUI H, MITA S. A new encoding and decoding system of Reed-Solomon codes for HDD [J]. IEEE Transactions on Magnetics, 2009, 45(10): 3757-3760. |
| 42 | RIGGLE C M, MCCARTHY S G. Design of error correction systems for disk drives [J]. IEEE Transactions on Magnetics, 1998, 34(4): 2362-2371. |
| 43 | LEE Joohyun, LEE Jaejin, PARK T. Error control scheme for high-speed DVD systems [J]. IEEE Transactions on Consumer Electronics, 2005, 51(4): 1197-1203. |
| 44 | SONG M A, KUO S Y, LAN I F. A low complexity design of Reed Solomon code algorithm for advanced RAID system [J]. IEEE Transactions on Consumer Electronics, 2007, 53(2): 265-273. |
| 45 | IM S, SHIN D. Flash-Aware RAID techniques for dependable and High-Performance flash memory SSD [J]. Computers IEEE Transactions on Computers, 2011, 60(1): 80-92. |
| 46 | HUANG J Z, LIANG X H, QIN X, et al. Scale-RS: an efficient scaling scheme for RS-Coded storage clusters [J]. IEEE Transactions on Parallel & Distributed Systems, 2015, 26(6): 1704-1717. |
| 47 | CHEN W G, WANG T, HAN C C, et al. Erasure-correction-enhanced iterative decoding for LDPC-RS product codes [J]. China Communications, 2021, 18(1): 49-60. |
| 48 | SIOW C C, NIEDUSZYNSKA S R, MÜLLER C A, et al. OriDB, the DNA replication origin database updated and extended [J]. Nucleic Acids Research, 2012, 40(D1): 682-686. |
| 49 | LOMAN N J, MISRA R V, DALLMAN T J, et al. Performance comparison of benchtop high-throughput sequencing platforms [J]. Nature Biotechnology, 2012, 30(5): 434-439. |
| 50 | MARDIS E R. Next-generation DNA sequencing methods [J]. Annual Review of Genomics and Human Genetics, 2008, 9(1): 387-402. |
| 51 | SHENDURE J, JI H. Next-generation DNA sequencing [J]. Nature Biotechnology, 2008, 26(10): 1135-1145. |
| 52 | METZKER M L. Sequencing technologies—the next generation [J]. Nature Reviews Genetics, 2010, 11(1): 31-46. |
| 53 | COMPEAU P E C, PEVZNER P A, TESLER G. How to apply de Bruijn graphs to genome assembly [J]. Nature Biotechnology, 2011, 29(11): 987. |
| 54 | ZERBINO D R, BIRNEY E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs [J]. Genome Research, 2008, 18(5): 821-829. |
| 55 | SIMPSON J T, WONG K, JACKMAN S D, et al. ABySS: a parallel assembler for short read sequence data [J]. Genome Research, 2009, 19(6): 1117-1123. |
| 56 | HUANG W C, LI L P, MYERS J R, et al. ART: a next-generation sequencing read simulator [J]. Bioinformatics, 2012, 28: 593-594. |
| 57 | CHEN W G, WANG L X, HAN M Z, et al. Sequencing barcode construction and identification methods based on block error-correction codes [J]. Science China Life Sciences, 2020,63(10):1580-1592. |
| 58 | CHEN W G, WANG P P, WANG L X, et al. Low-complexity and highly robust barcodes for error-rich single molecular sequencing [J]. 3 Biotech, 2021, 11(2): 1-11. |
| 59 | 格林 M R, 萨姆布鲁克 J. 分子克隆实验指南 [M]. 贺福初, 陈薇, 杨晓明, 译. 4版. 北京: 科学出版社, 2013: 226-227. |
| GREEN M R, SAMBROOK J. Molecular cloning: a laboratory manual [M]. HE F C, CHEN W, YANG X M, trans. 4th ed. Beijing: Science Press, 2013: 226-227. | |
| 60 | DUNNEN J D, GROOTSCHOLTEN P M, DAUWERSE J, et al. Reconstruction of the 2.4 Mb human DMD-gene by homologous YAC recombination [J]. Human Molecular Genetics, 1992, 1(1): 19-28. |
| [1] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
