合成生物学 ›› 2022, Vol. 3 ›› Issue (1): 66-77.DOI: 10.12211/2096-8280.2021-100
CRISPR/Cas9及其衍生编辑器在衰老研究中的应用进展
龚仕涛, 王宇, 陈宇庭
- 中国科学院深圳先进技术研究院,合成生物学研究所,基因组工程与治疗研究中心,广东 深圳 518055
-
收稿日期:2021-10-27修回日期:2022-02-09出版日期:2022-02-28发布日期:2022-03-14 -
通讯作者:陈宇庭 -
作者简介:龚仕涛 (1994—),男,硕士研究生。研究方向为基因组编辑与基因治疗。E-mail:st.gong@siat.ac.cn陈宇庭 (1992—),男,助理研究员。研究方向为基因编辑与合成生物学。E-mail:chen.yt@siat.ac.cn -
基金资助:国家自然科学基金(32101173)
Advances in application of CRISPR/Cas9 and its derivative editors in aging research
GONG Shitao, WANG Yu, CHEN Yuting
- Center for Genome Engineering and Therapy,Institute of Synthetic Biology,Shenzhen Institute of Advanced Technology,Shenzhen 518055,Guangdong,China
-
Received:2021-10-27Revised:2022-02-09Online:2022-02-28Published:2022-03-14 -
Contact:CHEN Yuting
摘要:
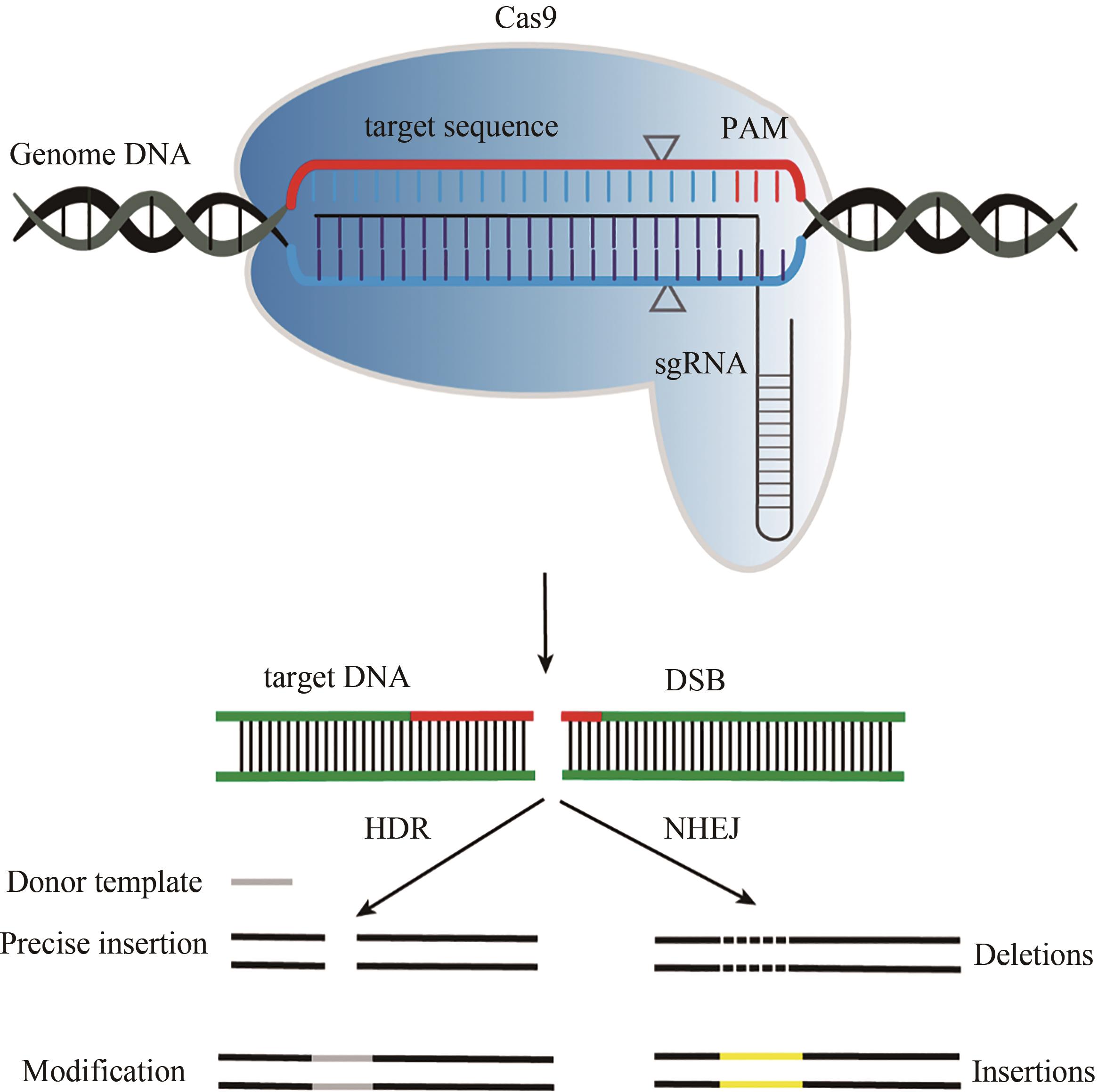
衰老逐渐成为威胁人类健康的重要因素,其中基因突变及其累积是引发衰老的因素之一。基因编辑技术可以纠正或清除错误基因突变,从而具有延缓衰老和治疗衰老相关疾病的潜力。CRISPR/Cas(clustered regularly interspaced short palindromic repeats/CRISPR-associated protein)的基因编辑工具是基于细菌和古细菌获得性免疫系统发明的,已证明可修改多种生物体的基因组。通过对Cas蛋白的定点突变和表达优化改造,可提高CRISPR/Cas系统在靶向位点的编辑效率、保真性及降低其脱靶效应。随后,科学家们发明了许多基于Cas蛋白的编辑器,如碱基编器、引导编辑器、转座/重组酶等。CRISPR/Cas及其衍生的编辑器,可实现多种形式精确的基因修饰,从而满足不同的基因编辑需求。本文主要概述了CRISPR/Cas系统及其种类、Cas9(CRISPR-associated protein 9)蛋白及其变体、基于Cas9蛋白衍生的基因编辑器,并讨论了这些编辑器在衰老及其相关疾病,如早衰综合症、心血管疾病、年龄相关性黄斑变性、神经退行性疾病等方面的应用研究进展。未来,高精确基因编辑器和递送技术的发明,将加速基因编辑技术用于治疗衰老相关疾病和逆转衰老的基础与临床研究,最终实现人类健康衰老。
中图分类号:
引用本文
龚仕涛, 王宇, 陈宇庭. CRISPR/Cas9及其衍生编辑器在衰老研究中的应用进展[J]. 合成生物学, 2022, 3(1): 66-77.
GONG Shitao, WANG Yu, CHEN Yuting. Advances in application of CRISPR/Cas9 and its derivative editors in aging research[J]. Synthetic Biology Journal, 2022, 3(1): 66-77.
| Name | Description of protein variant or mutations | PAM (5′ to 3′) |
|---|---|---|
| SpCas9 | Native Streptococcus pyogenes Cas9 | NGG[ |
| VRER SpCas9 | D1135V, G1218R, R1335E, T1337R | NGCG[ |
| VQR SpCas9 | D1135V, R1335Q, T1337R | NGAN or NGNG[ |
| EQR SpCas9 | D1135E, R1335Q, T1337R | NGAG[ |
| xCas9-3.7 | A262T, R324L, S409I, E480K, E543D, M694I, E1219V | NG, GAA, GAT[ |
| eSpCas9 (1.0) | K810A, K1003A, R1060A | NGG |
| eSpCas9 (1.1) | K848A, K1003A, R1060A | NGG |
| Cas9-HF1 | N497A, R661A, Q695A, Q926A | NGG |
| HypaCas9 | N692A, M694A, Q695A, H698A | NGG |
| evoCas9 | M495V, Y515N, K526E, R661Q | NGG |
| HiFi Cas9 | R691A | NGG |
| ScCas9 | Native Streptococcus canis Cas9 | NNG[ |
| StCas9 | Native Streptococcus thermophilus Cas9 | NNAGAAW[ |
| NmCas9 | Native Neisseria meningitidis Cas9 | NNNNGATT[ |
| SaCas9 | Native Staphylococcus aureus Cas9 | NNGRRT[ |
| CjCas9 | Native Campylobacter jejuni Cas9 | NNNVRYM[ |
| CasX | Phyla Deltaproteobacteria and Planctomycetes | TTCN[ |
| SpG | variants of Streptococcus pyogenes Cas9 | NGN[ |
| SpRY | SpCas9 variant | NHN[ |
| SpCas9 | R1333K, R1335 and T1337N | NRRH, NRCH and NRTH[ |
| SpCas9 | computational models | NNNN[ |
表1 Cas9变体及所识别的PAM序列
Tab. 1 Cas9 variants and identified PAM sequences
| Name | Description of protein variant or mutations | PAM (5′ to 3′) |
|---|---|---|
| SpCas9 | Native Streptococcus pyogenes Cas9 | NGG[ |
| VRER SpCas9 | D1135V, G1218R, R1335E, T1337R | NGCG[ |
| VQR SpCas9 | D1135V, R1335Q, T1337R | NGAN or NGNG[ |
| EQR SpCas9 | D1135E, R1335Q, T1337R | NGAG[ |
| xCas9-3.7 | A262T, R324L, S409I, E480K, E543D, M694I, E1219V | NG, GAA, GAT[ |
| eSpCas9 (1.0) | K810A, K1003A, R1060A | NGG |
| eSpCas9 (1.1) | K848A, K1003A, R1060A | NGG |
| Cas9-HF1 | N497A, R661A, Q695A, Q926A | NGG |
| HypaCas9 | N692A, M694A, Q695A, H698A | NGG |
| evoCas9 | M495V, Y515N, K526E, R661Q | NGG |
| HiFi Cas9 | R691A | NGG |
| ScCas9 | Native Streptococcus canis Cas9 | NNG[ |
| StCas9 | Native Streptococcus thermophilus Cas9 | NNAGAAW[ |
| NmCas9 | Native Neisseria meningitidis Cas9 | NNNNGATT[ |
| SaCas9 | Native Staphylococcus aureus Cas9 | NNGRRT[ |
| CjCas9 | Native Campylobacter jejuni Cas9 | NNNVRYM[ |
| CasX | Phyla Deltaproteobacteria and Planctomycetes | TTCN[ |
| SpG | variants of Streptococcus pyogenes Cas9 | NGN[ |
| SpRY | SpCas9 variant | NHN[ |
| SpCas9 | R1333K, R1335 and T1337N | NRRH, NRCH and NRTH[ |
| SpCas9 | computational models | NNNN[ |
| 1 | PARTRIDGE L, DEELEN J, SLAGBOOM P E. Facing up to the global challenges of ageing[J]. Nature, 2018, 561(7721): 45-56. |
| 2 | KAPAHI P, CHEN D, ROGERS A N, et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging[J]. Cell Metabolism, 2010, 11(6): 453-465. |
| 3 | SHALEM O, SANJANA N E, ZHANG F. High-throughput functional genomics using CRISPR-Cas9[J]. Nature Reviews Genetics, 2015, 16(5): 299-311. |
| 4 | DOMINGUEZ A A, LIM W A, QI L S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation[J]. Nature Reviews Molecular Cell Biology, 2016, 17(1): 5-15. |
| 5 | THAKORE P I, BLACK J B, HILTON I B, et al. Editing the epigenome: technologies for programmable transcription and epigenetic modulation[J]. Nature Methods, 2016, 13(2): 127-137. |
| 6 | PICKAR-OLIVER A, GERSBACH C A. The next generation of CRISPR-Cas technologies and applications[J]. Nature Reviews Molecular Cell Biology, 2019, 20(8): 490-507. |
| 7 | HILLE F, RICHTER H, WONG S P, et al. The biology of CRISPR-Cas: backward and forward[J]. Cell, 2018, 172(6): 1239-1259. |
| 8 | AGARI Y, SAKAMOTO K, TAMAKOSHI M, et al. Transcription profile of thermus thermophilus CRISPR systems after phage infection[J]. Journal of Molecular Biology, 2010, 395(2): 270-281. |
| 9 | GUIDOTTI L G, CHISARI F V. Noncytolytic control of viral infections by the innate and adaptive immune response[J]. Annual Review of Immunology, 2001, 19: 65-91. |
| 10 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712. |
| 11 | DELTCHEVA E, CHYLINSKI K, SHARMA C M, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase Ⅲ[J]. Nature, 2011, 471(7340): 602-607. |
| 12 | OOST J VAN DER, JORE M M, WESTRA E R, et al. CRISPR-based adaptive and heritable immunity in prokaryotes[J]. Trends in Biochemical Sciences, 2009, 34(8): 401-407. |
| 13 | MOUGIAKOS I, BOSMA E F, DE VOS W M, et al. Next generation prokaryotic engineering: the CRISPR-Cas toolkit[J]. Trends in Biotechnology, 2016, 34(7): 575-587. |
| 14 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 15 | BARRANGOU R. RNA-mediated programmable DNA cleavage[J]. Nature Biotechnology, 2012, 30(9): 836-838. |
| 16 | JIANG F G, DOUDNA J A. CRISPR-Cas9 structures and mechanisms[J]. Annual Review of Biophysics, 2017, 46: 505-529. |
| 17 | ZETSCHE B, GOOTENBERG J S, ABUDAYYEH O O, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771. |
| 18 | KOONIN E V, MAKAROVA K S, ZHANG F. Diversity, classification and evolution of CRISPR-Cas systems[J]. Current Opinion in Microbiology, 2017, 37: 67-78. |
| 19 | SHMAKOV S, SMARGON A, SCOTT D, et al. Diversity and evolution of class 2 CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2017, 15(3): 169-182. |
| 20 | CHYLINSKI K, MAKAROVA K S, CHARPENTIER E, et al. Classification and evolution of type Ⅱ CRISPR-Cas systems[J]. Nucleic Acids Research, 2014, 42(10): 6091-6105. |
| 21 | SHMAKOV S, ABUDAYYEH O O, MAKAROVA K S, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems[J]. Molecular Cell, 2015, 60(3): 385-397. |
| 22 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 23 | MALI P, YANG L H, ESVELT K M, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826. |
| 24 | JIANG F G, TAYLOR D W, CHEN J S, et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage[J]. Science, 2016, 351(6275): 867-871. |
| 25 | JINEK M, JIANG F G, TAYLOR D W, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation[J]. Science, 2014, 343(6176): 1247997. |
| 26 | AMITAI G, SOREK R. CRISPR-Cas adaptation: insights into the mechanism of action[J]. Nature Reviews Microbiology, 2016, 14(2): 67-76. |
| 27 | FU Y F, FODEN J A, KHAYTER C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells[J]. Nature Biotechnology, 2013, 31(9): 822-826. |
| 28 | HSU P D, SCOTT D A, WEINSTEIN J A, et al. DNA targeting specificity of RNA-guided Cas9 nucleases[J]. Nature Biotechnology, 2013, 31(9): 827-832. |
| 29 | KLEINSTIVER B P, PREW M S, TSAI S Q, et al. Engineered CRISPR-cas9 nucleases with altered PAM specificities[J]. Nature, 2015, 523(7561): 481-485. |
| 30 | HU J H, MILLER S M, GEURTS M H, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity[J]. Nature, 2018, 556(7699): 57-63. |
| 31 | CHATTERJEE P, JAKIMO N, JACOBSON J M. Minimal PAM specificity of a highly similar SpCas9 ortholog[J]. Science Advances, 2018, 4(10): eaau0766. |
| 32 | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| 33 | DEVEAU H, BARRANGOU R, GARNEAU J E, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus[J]. Journal of Bacteriology, 2008, 190(4): 1390-1400. |
| 34 | ESVELT K M, MALI P, BRAFF J L, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing[J]. Nature Methods, 2013, 10(11): 1116-1121. |
| 35 | HOU Z G, ZHANG Y, PROPSON N E, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(39): 15644-15649. |
| 36 | ZHANG Y, HEIDRICH N, AMPATTU B J, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis [J]. Molecular Cell, 2013, 50(4): 488-503. |
| 37 | RAN F A, CONG L, YAN W X, et al. In vivo genome editing using Staphylococcus aureus Cas9[J]. Nature, 2015, 520(7546): 186-191. |
| 38 | YAMADA M, WATANABE Y, GOOTENBERG J S, et al. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems[J]. Molecular Cell, 2017, 65(6): 1109-1121.e3. |
| 39 | BURSTEIN D, HARRINGTON L B, STRUTT S C, et al. New CRISPR-Cas systems from uncultivated microbes[J]. Nature, 2017, 542(7640): 237-241. |
| 40 | WALTON R T, CHRISTIE K A, WHITTAKER M N, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296. |
| 41 | MILLER S M, WANG T N, RANDOLPH P B, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs[J]. Nature Biotechnology, 2020, 38(4): 471-481. |
| 42 | KIM N, KIM H K, LEE S, et al. Prediction of the sequence-specific cleavage activity of Cas9 variants[J]. Nature Biotechnology, 2020, 38(11): 1328-1336. |
| 43 | KULCSÁR P I, TÁLAS A, HUSZÁR K, et al. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage[J]. Genome Biology, 2017, 18(1): 190. |
| 44 | KLEINSTIVER B P, PATTANAYAK V, PREW M S, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects[J]. Nature, 2016, 529(7587): 490-495. |
| 45 | CHEN J S, DAGDAS Y S, KLEINSTIVER B P, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy[J]. Nature, 2017, 550(7676): 407-410. |
| 46 | CASINI A, OLIVIERI M, PETRIS G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast[J]. Nature Biotechnology, 2018, 36(3): 265-271. |
| 47 | LEE J K, JEONG E, LEE J, et al. Directed evolution of CRISPR-Cas9 to increase its specificity[J]. Nature Communications, 2018, 9: 3048. |
| 48 | WANG M, ZURIS J A, MENG F T, et al. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(11): 2868-2873. |
| 49 | VAKULSKAS C A, DEVER D P, RETTIG G R, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells[J]. Nature Medicine, 2018, 24(8): 1216-1224. |
| 50 | FU Y F, SANDER J D, REYON D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs[J]. Nature Biotechnology, 2014, 32(3): 279-284. |
| 51 | HUANG T P, ZHAO K T, MILLER S M, et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors[J]. Nature Biotechnology, 2019, 37(6): 626-631. |
| 52 | CEBRIAN-SERRANO A, DAVIES B. CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools[J]. Mammalian Genome, 2017, 28(7/8): 247-261. |
| 53 | NISHIMASU H, SHI X, ISHIGURO S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space[J]. Science, 2018, 361(6408): 1259-1262. |
| 54 | CHATTERJEE P, LEE J, NIP L, et al. A Cas9 with PAM recognition for adenine dinucleotides[J]. Nature Communications, 2020, 11: 2474. |
| 55 | CHATTERJEE P, JAKIMO N, LEE J, et al. An engineered ScCas9 with broad PAM range and high specificity and activity[J]. Nature Biotechnology, 2020, 38(10): 1154-1158. |
| 56 | KLEINSTIVER B P, PREW M S, TSAI S Q, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition[J]. Nature Biotechnology, 2015, 33(12): 1293-1298. |
| 57 | HIRANO H, GOOTENBERG J S, HORII T, et al. Structure and engineering of Francisella novicida Cas9[J]. Cell, 2016, 164(5): 950-961. |
| 58 | REES H A, LIU D R. Base editing: precision chemistry on the genome and transcriptome of living cells[J]. Nature Reviews Genetics, 2018, 19(12): 770-788. |
| 59 | ANZALONE A V, KOBLAN L W, LIU D R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature Biotechnology, 2020, 38(7): 824-844. |
| 60 | GRÜNEWALD J, ZHOU R H, LAREAU C A, et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing[J]. Nature Biotechnology, 2020, 38(7): 861-864. |
| 61 | GEHRKE J M, CERVANTES O, CLEMENT M K, et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities[J]. Nature Biotechnology, 2018, 36(10): 977-982. |
| 62 | ZHANG X H, ZHU B Y, CHEN L, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells[J]. Nature Biotechnology, 2020, 38(7): 856-860. |
| 63 | LAPINAITE A, KNOTT G J, PALUMBO C M, et al. DNA capture by a CRISPR-Cas9-guided adenine base editor[J]. Science, 2020, 369(6503): 566-571. |
| 64 | NISHIMASU H, RAN F A, HSU P D, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA[J]. Cell, 2014, 156(5): 935-949. |
| 65 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| 66 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| 67 | LANDRUM M J, LEE J M, BENSON M, et al. ClinVar: public archive of interpretations of clinically relevant variants[J]. Nucleic Acids Research, 2015, 44(D1): D862-D868. |
| 68 | NISHIDA K, ARAZOE T, YACHIE N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): aaf8729. |
| 69 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| 70 | ZAFRA M P, SCHATOFF E M, KATTI A, et al. Optimized base editors enable efficient editing in cells, organoids and mice[J]. Nature Biotechnology, 2018, 36(9): 888-893. |
| 71 | CHEN L W, PARK J E, PAA P, et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins[J]. Nature Communications, 2021, 12: 1384. |
| 72 | ZHANG X H, ZHU B Y, CHEN L, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells[J]. Nature Biotechnology, 2020, 38(7): 856-860. |
| 73 | GRÜNEWALD J, ZHOU R H, LAREAU C A, et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing[J]. Nature Biotechnology, 2020, 38(7): 861-864. |
| 74 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 75 | SÜRÜN D, SCHNEIDER A, MIRCETIC J, et al. Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors[J]. Genes, 2020, 11(5): 511. |
| 76 | PETRI K, ZHANG W T, MA J Y, et al. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells[J]. Nature Biotechnology, 2021: 1-5. |
| 77 | LIU Y, LI X Y, HE S T, et al. Efficient generation of mouse models with the prime editing system[J]. Cell Discovery, 2020, 6: 27. |
| 78 | KIM H K, YU G, PARK J, et al. Predicting the efficiency of prime editing guide RNAs in human cells[J]. Nature Biotechnology, 2021, 39(2): 198-206. |
| 79 | CHEN S P, WANG H H. An engineered cas-transposon system for programmable and site-directed DNA transpositions[J]. The CRISPR Journal, 2019, 2(6): 376-394. |
| 80 | STRECKER J, LADHA A, GARDNER Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases[J]. Science, 2019, 365(6448): 48-53. |
| 81 | CHAIKIND B, BESSEN J L, THOMPSON D B, et al. A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells[J]. Nucleic Acids Research, 2016, 44(20): 9758-9770. |
| 82 | GUILINGER J P, THOMPSON D B, LIU D R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification[J]. Nature Biotechnology, 2014, 32(6): 577-582. |
| 83 | MARUYAMA T, DOUGAN S K, TRUTTMANN M C, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining[J]. Nature Biotechnology, 2015, 33(5): 538-542. |
| 84 | LIN S, STAAHL B T, ALLA R K, et al. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery[J]. eLife, 2014, 3: e04766. |
| 85 | WYSS-CORAY T. Ageing, neurodegeneration and brain rejuvenation[J]. Nature, 2016, 539(7628): 180-186. |
| 86 | HERNANDEZ-SEGURA A, NEHME J, DEMARIA M. Hallmarks of cellular senescence[J]. Trends in Cell Biology, 2018, 28(6): 436-453. |
| 87 | WANG W, ZHENG Y X, SUN S H, et al. A genome-wide CRISPR-based screen identifies KAT7 as a driver of cellular senescence[J]. Science Translational Medicine, 2021, 13(575): eabd2655. |
| 88 | GONZALO S, KREIENKAMP R, ASKJAER P. Hutchinson-Gilford progeria syndrome: a premature aging disease caused by LMNA gene mutations[J]. Ageing Research Reviews, 2017, 33: 18-29. |
| 89 | SANTIAGO-FERNÁNDEZ O, OSORIO F G, QUESADA V, et al. Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome[J]. Nature Medicine, 2019, 25(3): 423-426. |
| 90 | BEYRET E, LIAO H K, YAMAMOTO M, et al. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome[J]. Nature Medicine, 2019, 25(3): 419-422. |
| 91 | KOBLAN L W, ERDOS M R, WILSON C, et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice[J]. Nature, 2021, 589(7843): 608-614. |
| 92 | CARROLL K J, MAKAREWICH C A, MCANALLY J, et al. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(2): 338-343. |
| 93 | XU L, LAU Y S, GAO Y D, et al. Life-long AAV-mediated CRISPR genome editing in dystrophic heart improves cardiomyopathy without causing serious lesions in mdx mice[J]. Molecular Therapy, 2019, 27(8): 1407-1414. |
| 94 | KIM K, PARK S W, KIM J H, et al. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration[J]. Genome Research, 2017, 27(3): 419-426. |
| 95 | LIM C K W, GAPINSKE M, BROOKS A K, et al. Treatment of a mouse model of ALS by in vivo base editing[J]. Molecular Therapy, 2020, 28(4): 1177-1189. |
| 96 | ORTIZ-VIRUMBRALES M, MORENO C L, KRUGLIKOV I, et al. CRISPR/Cas9-Correctable mutation-related molecular and physiological phenotypes in iPSC-derived Alzheimer's PSEN2 N141I neurons[J]. Acta Neuropathologica Communications, 2017, 5(1): 77. |
| 97 | PARK H, OH J, SHIM G, et al. In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nano complexes alleviates deficits in mouse models of Alzheimer's disease[J]. Nature Neuroscience, 2019, 22(4): 524-528. |
| 98 | SUN J, CARLSON-STEVERMER J, DAS U, et al. CRISPR/Cas9 editing of APP C-terminus attenuates β-cleavage and promotes α-cleavage[J]. Nature Communications, 2019, 10: 53. |
| 99 | WANG X L, CAO C W, HUANG J J, et al. One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system[J]. Scientific Reports, 2016, 6: 20620. |
| 100 | VERMILYEA S C, BABINSKI A, TRAN N, et al. In vitro CRISPR/Cas9-directed gene editing to model LRRK2 G2019S Parkinson's disease in common marmosets[J]. Scientific Reports, 2020, 10: 3447. |
| [1] | 林继聪, 邹根, 刘宏民, 魏勇军. CRISPR/Cas基因组编辑技术在丝状真菌次级代谢产物合成中的应用[J]. 合成生物学, 2023, 4(4): 738-755. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||