合成生物学 ›› 2022, Vol. 3 ›› Issue (1): 78-97.DOI: 10.12211/2096-8280.2021-006
哺乳动物合成基因组学研究进展
何博, 付宗恒, 吴毅, 赵广荣
- 天津大学化工学院,系统生物工程教育部重点实验室,教育部合成生物学前沿科学中心,天津 300072
-
收稿日期:2021-01-13修回日期:2021-11-27出版日期:2022-02-28发布日期:2022-03-14 -
通讯作者:吴毅,赵广荣 -
作者简介:何博 (1995—),女,博士研究生。研究方向为合成基因组学。E-mail:hebo@tju.edu.cn吴毅 (1989—),男,博士,研究员,博士生导师。研究方向为合成基因组学。E-mail:yi.wu@tju.edu.cn赵广荣 (1966—),男,博士,教授,博士生导师。研究方向为合成生物学。E-mail:grzhao@tju.edu.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2019YFA0903800)
Research progress of synthetic mammalian genomics
HE Bo, FU Zongheng, WU Yi, ZHAO Guangrong
- Frontiers Science Center for Synthetic Biology,Key Laboratory of Systems Bioengineering (Ministry of Education),School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
-
Received:2021-01-13Revised:2021-11-27Online:2022-02-28Published:2022-03-14 -
Contact:WU Yi, ZHAO Guangrong
摘要:
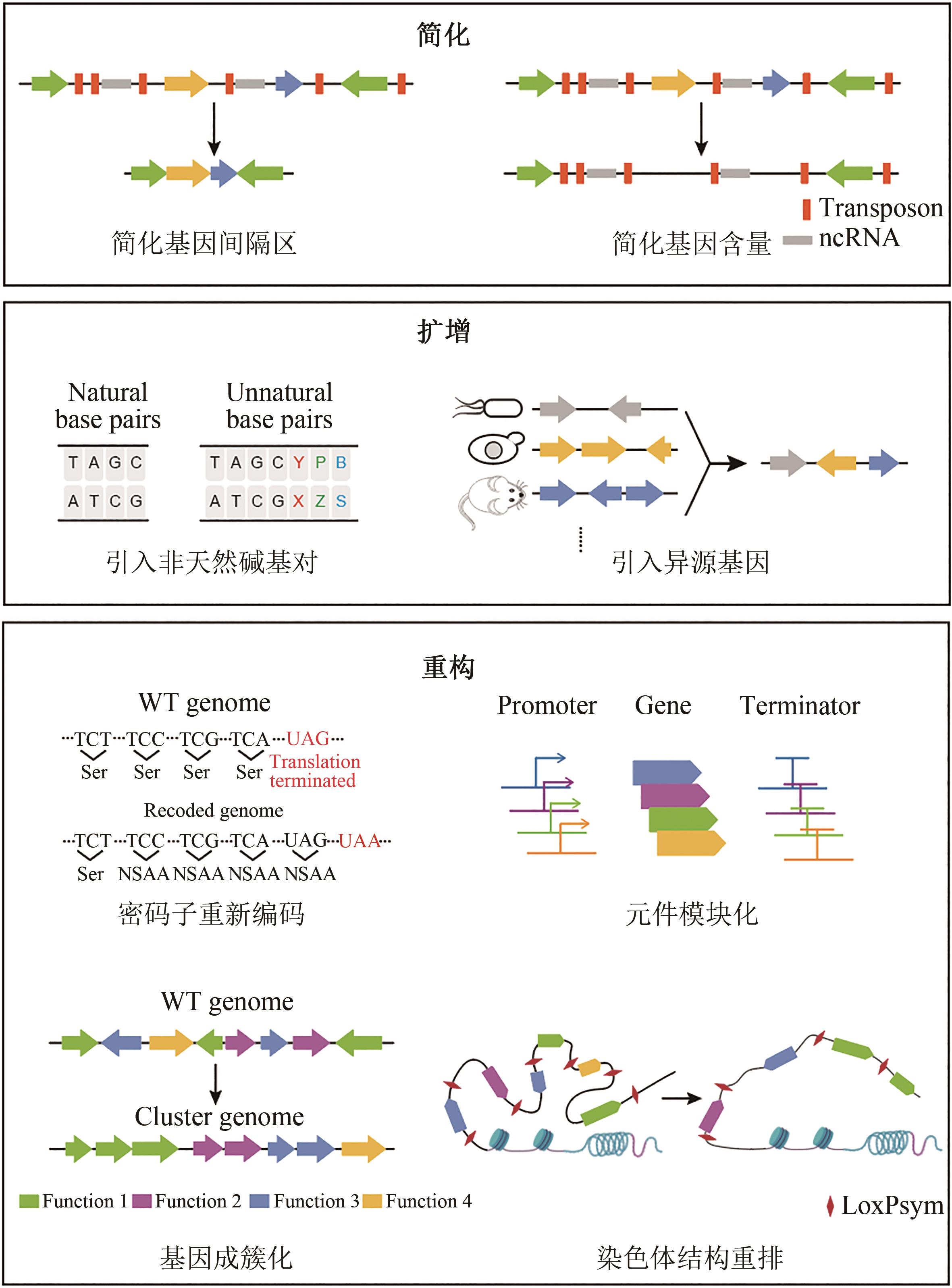
合成基因组学通过设计与合成大片段DNA序列,开展基因组尺度的工程化改造或从头合成,从而揭示基因型和表型的关联,并构建预定功能的生物。正如在大肠杆菌、酿酒酵母等低等模式生物中合成基因组学的实践,对哺乳动物基因组的大片段DNA设计再造也必然会加深对更为复杂的动物基因组的理解并加强对基因组的功能重塑。面对生命健康领域存在的重大挑战,设计合成哺乳动物大片段DNA为其提供了新的思路和解决方案,特别是在染色体疾病模型构建、人源化免疫系统等方面展示出独特的应用潜力。然而,目前大片段DNA在哺乳动物细胞中的设计和操纵仍是一个巨大的挑战,面临着高等哺乳动物基因组注释不完善、复杂序列组装困难、穿梭载体通用性差、大片段DNA转移低效等问题。本文围绕设计-组装-转移的技术路线,评述哺乳动物合成基因组学领域的重要研究进展,详细介绍重要的技术突破,并展望哺乳动物合成基因组学在医药健康领域的应用。
中图分类号:
引用本文
何博, 付宗恒, 吴毅, 赵广荣. 哺乳动物合成基因组学研究进展[J]. 合成生物学, 2022, 3(1): 78-97.
HE Bo, FU Zongheng, WU Yi, ZHAO Guangrong. Research progress of synthetic mammalian genomics[J]. Synthetic Biology Journal, 2022, 3(1): 78-97.
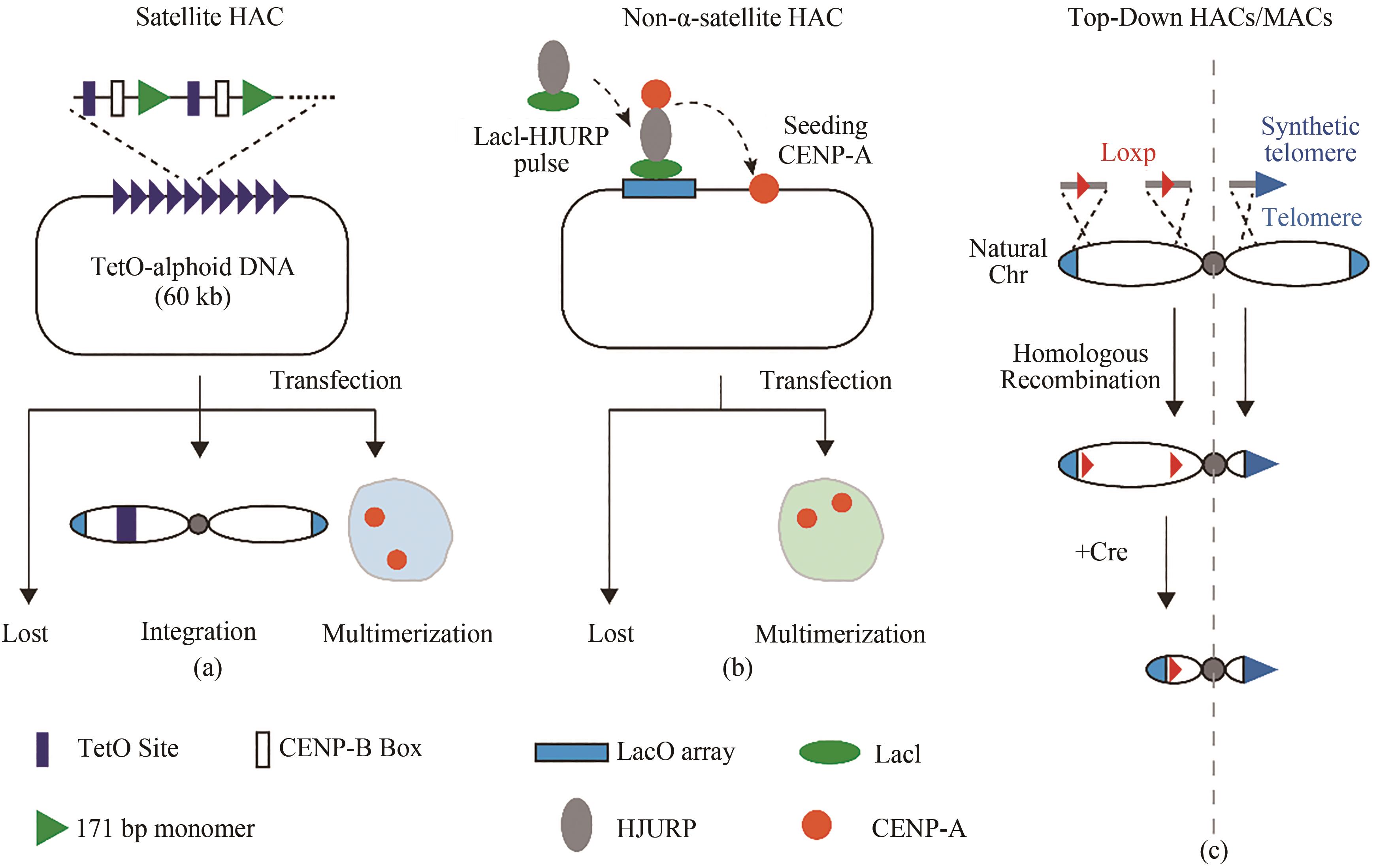
图2 合成型哺乳动物染色体着丝粒区域(a)基于卫星(satellite)重复序列自下而上构建的着丝粒区域;(b)基于非天然重复序列自下而上构建的着丝粒区域;(c)自上而下截短的人工染色体
Fig. 2 Centromeric regions of synthetic mammalian chromosomes(a) Bottom-up centromere region based on satellite repeat sequence; (b) Bottom-up centromere region based on non-natural repetitive sequences; (c) Top-down artificial chromosomes from truncated natural chromosomes
| 方法 | 转移介质 | 最大承载量 | 目的细胞 | 存在缺点 | 文献 |
|---|---|---|---|---|---|
| 脂质体转染 | FuGENE 6 | 200 kb | HT1080 | 脂质材料对细胞有毒性 | [ |
| Lipofection | 400 kb | HT1080 | [ | ||
| 电转 | 缓冲溶液 | 200 kb | mESc | 需要昂贵细胞电转仪 | [ |
| 显微注射 | 缓冲溶液 | 590 kb | CHO | 显微操作通量低 | [ |
| 病毒载体 | HSV-1 | 152 kb | hESc | 操作步骤烦琐、存在生物安全问题 | [ |
| Epstein-barr virus | 330 kb | Raji | [ | ||
| MMCT | 微细胞 | Mb级 | HT1080 | 操作步骤烦琐 | [ |
| 微细胞 | Mb级 | Hela | |||
| Eco-MMCT | 微细胞 | Mb级 | mESc | ||
| 酵母原生质体-细胞融合 | 酵母原生质体 | 1.1 Mb | HEK293 | 操作步骤烦琐 | [ |
| 100 kb | HeLa | [ | |||
| 1.4 Mb | mES |
表1 哺乳动物细胞大片段DNA的转移方法
Tab. 1 Method for delivering large fragments of DNA to mammalian cells
| 方法 | 转移介质 | 最大承载量 | 目的细胞 | 存在缺点 | 文献 |
|---|---|---|---|---|---|
| 脂质体转染 | FuGENE 6 | 200 kb | HT1080 | 脂质材料对细胞有毒性 | [ |
| Lipofection | 400 kb | HT1080 | [ | ||
| 电转 | 缓冲溶液 | 200 kb | mESc | 需要昂贵细胞电转仪 | [ |
| 显微注射 | 缓冲溶液 | 590 kb | CHO | 显微操作通量低 | [ |
| 病毒载体 | HSV-1 | 152 kb | hESc | 操作步骤烦琐、存在生物安全问题 | [ |
| Epstein-barr virus | 330 kb | Raji | [ | ||
| MMCT | 微细胞 | Mb级 | HT1080 | 操作步骤烦琐 | [ |
| 微细胞 | Mb级 | Hela | |||
| Eco-MMCT | 微细胞 | Mb级 | mESc | ||
| 酵母原生质体-细胞融合 | 酵母原生质体 | 1.1 Mb | HEK293 | 操作步骤烦琐 | [ |
| 100 kb | HeLa | [ | |||
| 1.4 Mb | mES |
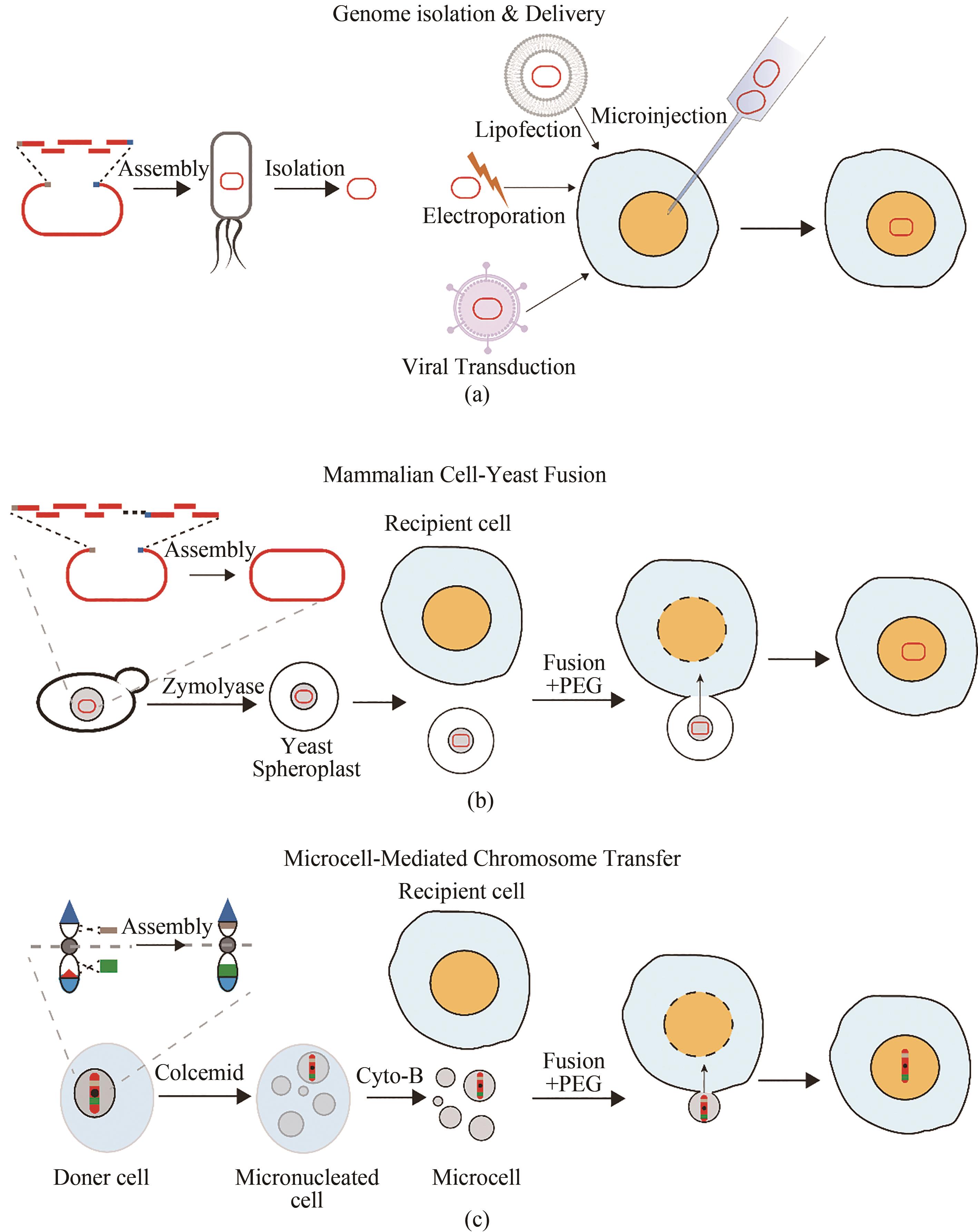
图3 哺乳动物细胞大片段DNA的转移方法(a)由酿酒酵母组装的DNA分子在大肠杆菌中富集后提取纯化到体外并通过显微注射、脂质体转染、电穿孔转移或病毒介导的转染等方法转移到宿主细胞中;(b)由酿酒酵母组装的DNA分子通过PEG介导的酵母原生质体与哺乳动物细胞融合转移到宿主细胞中;(c)在供体细胞中通过自上而下构建的人工染色体以微细胞介导的染色体转移方式转移到宿主细胞
Fig. 3 Method for delivering large fragments of DNA to mammalian cells(a) DNA molecules assembled by S.cerevisiae are enriched in E. coli, extracted and purified in vitro and transferred to host cells by microinjection, lipofection, electroporation, or virus-mediated transduction; (b) DNA molecules assembled by S.cerevisiae are transferred to host cells through the PEG-mediated fusion of yeast protoplasts and mammalian cells; (c) Top-down artificial chromosomes constructed in donor cell are transferred to host cell through the microcell-mediated chromosome transfer
| 1 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 2 | BOEKE J D, CHURCH G, HESSEL A, et al. The genome project-write[J]. Science, 2016, 353(6295): 126-127. |
| 3 | BAKER M. Synthetic genomes: the next step for the synthetic genome[J]. Nature, 2011, 473(7347): 403, 405-403, 408. |
| 4 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 5 | HUTCHISON C A, CHUANG R Y, NOSKOV V N, et al. Design and synthesis of a minimal bacterial genome[J]. Science, 2016, 351(6280): aad6253. |
| 6 | WANG K H, FREDENS J, BRUNNER S F, et al. Defining synonymous codon compression schemes by genome recoding[J]. Nature, 2016, 539(7627): 59-64. |
| 7 | FREDENS J, WANG K H, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| 8 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329): eaaf4791. |
| 9 | MERCY G, MOZZICONACCI J, SCOLARI V F, et al. 3D organization of synthetic and scrambled chromosomes[J]. Science, 2017, 355(6329): eaaf4597. |
| 10 | XIE Z X, LI B Z, MITCHELL L A, et al. “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 11 | MITCHELL L A, WANG A, STRACQUADANIO G, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond[J]. Science, 2017, 355(6329): eaaf4831. |
| 12 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 13 | ZHANG W M, ZHAO G H, LUO Z Q, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329): eaaf3981. |
| 14 | OSTROV N, BEAL J, ELLIS T, et al. Technological challenges and milestones for writing genomes[J]. Science, 2019, 366(6463): 310-312. |
| 15 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363(6429): 884-887. |
| 16 | OSTROV N, LANDON M, GUELL M, et al. Design, synthesis, and testing toward a 57-codon genome[J]. Science, 2016, 353(6301): 819-822. |
| 17 | HOCHREIN L, MITCHELL L A, SCHULZ K, et al. L-SCRaMbLE as a tool for light-controlled Cre-mediated recombination in yeast[J]. Nature Communications, 2018, 9: 1931.doi:10.1038/s41467-017-02208-6 . |
| 18 | LUO Z Q, WANG L H, WANG Y, et al. Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES[J]. Nature Communications, 2018, 9: 1930.doi:10.1038/s41467-017-00866-y . |
| 19 | LIU W, LUO Z Q, WANG Y, et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods[J]. Nature Communications, 2018, 9: 1936.doi:10.1038/s41467-018-04254-0 . |
| 20 | WU Y, ZHU R Y, MITCHELL L A, et al. In vitro DNA SCRaMbLE[J]. Nature Communications, 2018, 9(1): 1935.doi:10.1038/s41467-018-03743-6 . |
| 21 | SHEN M J, WU Y, YANG K, et al. Heterozygous diploid and inter species SCRaMbLEing[J]. Nature Communications, 2018, 9(1): 1934.doi:10.1038/s41467-018-04157-0 . |
| 22 | JIA B, WU Y, LI B Z, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9: 1933.doi:10.1038/s41467-018-03084-4 . |
| 23 | BLOUNT B A, GOWERS G O F, HO J C H, et al. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome[J]. Nature Communications, 2018, 9: 1932.doi:10.1038/s41467-018-03143-w . |
| 24 | JIANG S, LI H, HONG H, et al. Spatial density of open chromatin: an effective metric for the functional characterization of topologically associated domains[J]. Briefings in Bioinformatics, 2020, 22(3): bbaa210. |
| 25 | MALYSHEV D A, DHAMI K, LAVERGNE T, et al. A semi-synthetic organism with an expanded genetic alphabet[J]. Nature, 2014, 509(7500): 385-388. |
| 26 | LUO X Z, REITER M A, D'ESPAUX L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast[J]. Nature, 2019, 567(7746): 123-126. |
| 27 | ROBERTSON W E, FUNKE L F H, DE LA TORRE D, et al. Creating custom synthetic genomes in Escherichia coli with REXER and GENESIS[J]. Nature Protocols, 2021, 16(5): 2345-2380. |
| 28 | ROBERTSON W E, FUNKE L F H, DE LA TORRE D, et al. Sense codon reassignment enables viral resistance and encoded polymer synthesis[J]. Science, 2021, 372(6546): 1057-1062. |
| 29 | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G, et al. Design of a synthetic yeast genome[J]. Science, 2017, 355(6329): 1040-1044. |
| 30 | KANNAN K, GIBSON D G. Yeast genome, by design[J]. Science, 2017, 355(6329): 1024-1025. |
| 31 | CONSORTIUM E P. The ENCODE (ENCyclopedia Of DNA Elements) Project[J]. Science, 2004, 306(5696): 636-640. |
| 32 | QU H Z, FANG X D. A brief review on the human encyclopedia of DNA elements (ENCODE) project[J]. Genomics, Proteomics & Bioinformatics, 2013, 11(3): 135-141. |
| 33 | CONSORTIUM E P, SNYDER M P, GINGERAS T R, et al. Perspectives on encode[J]. Nature, 2020, 583(7818): 693-698. |
| 34 | LEE C M, BARBER G P, CASPER J, et al. UCSC Genome Browser enters 20th year[J]. Nucleic Acids Research, 2020, 48(D1): D756-D761. |
| 35 | KARCZEWSKI K J, FRANCIOLI L C, TIAO G, et al. The mutational constraint spectrum quantified from variation in 141, 456 humans[J]. Nature, 2020, 581(7809): 434-443. |
| 36 | TAM V, PATEL N, TURCOTTE M, et al. Benefits and limitations of genome-wide association studies[J]. Nature Reviews Genetics, 2019, 20(8): 467-484. |
| 37 | LANDER E S, LINTON L M, BIRREN B, et al. Initial sequencing and analysis of the human genome[J]. Nature, 2001, 409(6822): 860-921. |
| 38 | VENTER J C, ADAMS M D, MYERS E W, et al. The sequence of the human genome[J]. Science, 2001, 291(5507): 1304-1351. |
| 39 | International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome[J]. Nature, 2004, 431(7011): 931-945. |
| 40 | LOGSDON G A, VOLLGER M R, HSIEH P, et al. The structure, function and evolution of a complete human chromosome 8[J]. Nature, 2021, 593(7857): 101-107. |
| 41 | CARNINCI P, KASUKAWA T, KATAYAMA S, et al. The transcriptional landscape of the mammalian genome[J]. Science, 2005, 309(5740): 1559-1563. |
| 42 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 43 | TUNYASUVUNAKOOL K, ADLER J, WU Z, et al. Highly accurate protein structure prediction for the human proteome[J]. Nature, 2021, 596(7873): 590-596. |
| 44 | HAYDEN K E. Human centromere genomics: now it’s personal[J]. Chromosome Research, 2012, 20(5): 621-633. |
| 45 | FUKAGAWA T, EARNSHAW W C. The centromere: chromatin foundation for the kinetochore machinery[J]. Developmental Cell, 2014, 30(5): 496-508. |
| 46 | LOGSDON G A, GAMBOGI C W, LISKOVYKH M A, et al. Human artificial chromosomes that bypass centromeric DNA[J]. Cell, 2019, 178(3): 624-639. |
| 47 | EBERSOLE T A, ROSS A, CLARK E, et al. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats[J]. Human Molecular Genetics, 2000, 9(11): 1623-1631. |
| 48 | GRIMES B R, RHOADES A A, WILLARD H F. α-Satellite DNA and vector composition influence rates of human artificial chromosome formation[J]. Molecular Therapy, 2002, 5(6): 798-805. |
| 49 | IKENO M, GRIMES B, OKAZAKI T, et al. Construction of YAC-based mammalian artificial chromosomes[J]. Nature Biotechnology, 1998, 16(5): 431-439. |
| 50 | MEJÍA J E, ALAZAMI A, WILLMOTT A, et al. Efficiency of de novo centromere formation in human artificial chromosomes[J]. Genomics, 2002, 79(3): 297-304. |
| 51 | OHZEKI J I, BERGMANN J H, KOUPRINA N, et al. Breaking the HAC Barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly[J]. The EMBO Journal, 2012, 31(10): 2391-2402. |
| 52 | OKADA T, OHZEKI J I, NAKANO M, et al. CENP-B controls centromere formation depending on the chromatin context[J]. Cell, 2007, 131(7): 1287-1300. |
| 53 | HARRINGTON J J, BOKKELEN G VAN, MAYS R W, et al. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes[J]. Nature Genetics, 1997, 15(4): 345-355. |
| 54 | IIDA Y, KIM J H, KAZUKI Y, et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression[J]. DNA Research, 2010, 17(5): 293-301. |
| 55 | KOUPRINA N, PETROV N, MOLINA O, et al. Human artificial chromosome with regulated centromere: a tool for genome and cancer studies[J]. ACS Synthetic Biology, 2018, 7(9): 1974-1989. |
| 56 | KOUPRINA N, SAMOSHKIN A, ERLIANDRI I, et al. Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere[J]. ACS Synthetic Biology, 2012, 1(12): 590-601. |
| 57 | KIM J H, KONONENKO A, ERLIANDRI I, et al. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(50): 20048-20053. |
| 58 | PESENTI E, LISKOVYKH M, OKAZAKI K, et al. Analysis of complex DNA rearrangements during early stages of HAC formation[J]. ACS Synthetic Biology, 2020, 9(12): 3267-3287. |
| 59 | BARNETT M A, BUCKLE V J, EVANS E P, et al. Telomere directed fragmentation of mammalian chromosomes[J]. Nucleic Acids Research, 1993, 21(1): 27-36. |
| 60 | BROWN D M, GLASS J I. Technology used to build and transfer mammalian chromosomes[J]. Experimental Cell Research, 2020, 388(2): 111851. |
| 61 | LAJOIE M J, ROVNER A J, GOODMAN D B, et al. Genomically recoded organisms expand biological functions[J]. Science, 2013, 342(6156): 357-360. |
| 62 | KLAWITTER S, FUCHS N V, UPTON K R, et al. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells[J]. Nature Communications, 2016, 7: 10286. |
| 63 | LEE E C, LIANG Q, ALI H, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery[J]. Nature Biotechnology, 2014, 32(4): 356-363. |
| 64 | MULLER H, ANNALURU N, SCHWERZMANN J W, et al. Assembling large DNA segments in yeast[J]. Methods in Molecular Biology, 2012, 852: 133-150. |
| 65 | JUHAS M, AJIOKA J W. High molecular weight DNA assembly in vivo for synthetic biology applications[J]. Critical Reviews in Biotechnology, 2017, 37(3): 277-286. |
| 66 | ITAYA M, TSUGE K, KOIZUMI M, et al. Combining two genomes in one cell: stable cloning of the Synechocystis PCC6803 genome in the Bacillus subtilis 168 genome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(44): 15971-15976. |
| 67 | ITAYA M, FUJITA K, KUROKI A, et al. Bottom-up genome assembly using the Bacillus subtilis genome vector[J]. Nature Methods, 2008, 5(1): 41-43. |
| 68 | ZHANG Y M, BUCHHOLZ F, MUYRERS J P P, et al. A new logic for DNA engineering using recombination in Escherichia coli [J]. Nature Genetics, 1998, 20(2): 123-128. |
| 69 | FU J, BIAN X Y, HU S, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting[J]. Nature Biotechnology, 2012, 30(5): 440-446. |
| 70 | GIBSON D G, BENDERS G A, AXELROD K C, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(51): 20404-20409. |
| 71 | ZHOU J T, WU R H, XUE X L, et al. CasHRA (Cas9-facilitated Homologous Recombination Assembly) method of constructing megabase-sized DNA[J]. Nucleic Acids Research, 2016, 44(14): e124. |
| 72 | MITCHELL L A, MCCULLOCH L H, PINGLAY S, et al. De novo assembly and delivery to mouse cells of a 101 kb functional human gene[J]. Genetics, 2021, 218(1). . |
| 73 | POSTMA E D, DASHKO S, BREEMEN L VAN, et al. A supernumerary designer chromosome for modular in vivo pathway assembly in Saccharomyces cerevisiae [J]. Nucleic Acids Research, 2021, 49(3): 1769-1783. |
| 74 | SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature, 2018, 560(7718): 331-335. |
| 75 | TAGWERKER C, DUPONT C L, KARAS B J, et al. Sequence analysis of a complete 1.66 Mb Prochlorococcus marinus MED4 genome cloned in yeast[J]. Nucleic Acids Research, 2012, 40(20): 10375-10383. |
| 76 | KARAS B J, TAGWERKER C, YONEMOTO I T, et al. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid[J]. ACS Synthetic Biology, 2012, 1(1): 22-28. |
| 77 | 王培霞, 马渊, 吴毅. 大DNA体内组装技术进展[J]. 生物加工过程, 2019, 17(1): 15-22. |
| WANG Peixia, MA Yuan, WU Yi. Advances in large DNA in vivo assembly[J]. Chinese Journal of Bioprocess Engineering, 2019, 17(1): 15-22. | |
| 78 | NOSKOV V N, KARAS B J, YOUNG L, et al. Assembly of large, high G+C bacterial DNA fragments in yeast[J]. ACS Synthetic Biology, 2012, 1(7): 267-273. |
| 79 | KAZUKI Y, HOSHIYA H, TAKIGUCHI M, et al. Refined human artificial chromosome vectors for gene therapy and animal transgenesis[J]. Gene Therapy, 2011, 18(4): 384-393. |
| 80 | KUROIWA Y, TOMIZUKA K, SHINOHARA T, et al. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts[J]. Nature Biotechnology, 2000, 18(10): 1086-1090. |
| 81 | KUROIWA Y, KASINATHAN P, CHOI Y J, et al. Cloned transchromosomic calves producing human immunoglobulin[J]. Nature Biotechnology, 2002, 20(9): 889-894. |
| 82 | SANO A, MATSUSHITA H, WU H, et al. Physiological level production of antigen-specific human immunoglobulin in cloned transchromosomic cattle[J]. PLoS One, 2013, 8(10): e78119. |
| 83 | BUERSTEDDE J M, TAKEDA S. Increased ratio of targeted to random integration after transfection of chicken B cell lines[J]. Cell, 1991, 67(1): 179-188. |
| 84 | WILLBANKS A, LEARY M, GREENSHIELDS M, et al. The evolution of epigenetics: from prokaryotes to humans and its biological consequences[J]. Genetics & Epigenetics, 2016, 8: 25-36. |
| 85 | LARTIGUE C, VASHEE S, ALGIRE M A, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast[J]. Science, 2009, 325(5948): 1693-1696. |
| 86 | MEJÍA J E, WILLMOTT A, LEVY E, et al. Functional complementation of a genetic deficiency with human artificial chromosomes[J]. The American Journal of Human Genetics, 2001, 69(2): 315-326. |
| 87 | SHI J F, MA Y F, ZHU J, et al. A review on electroporation-based intracellular delivery[J]. Molecules, 2018, 23(11): 3044. |
| 88 | STEWART M P, LANGER R, JENSEN K F. Intracellular delivery by membrane disruption: Mechanisms, strategies, and concepts[J]. Chemical Reviews, 2018, 118(16): 7409-7531. |
| 89 | GNIRKE A, HUXLEY C, PETERSON K, et al. Microinjection of intact 200- to 500-kb fragments of YAC DNA into mammalian cells[J]. Genomics, 1993, 15(3): 659-667. |
| 90 | WADE-MARTINS R, SMITH E R, TYMINSKI E, et al. An infectious transfer and expression system for genomic DNA loci in human and mouse cells[J]. Nature Biotechnology, 2001, 19(11): 1067-1070. |
| 91 | MORALLI D, MONACO Z L. Developing de novo human artificial chromosomes in embryonic stem cells using HSV-1 amplicon technology[J]. Chromosome Research, 2015, 23(1): 105-110. |
| 92 | SUN T Q, FENSTERMACHER D A, VOS J M H. Human artificial episomal chromosomes for cloning large DNA fragments in human cells[J]. Nature Genetics, 1994, 8(1): 33-41. |
| 93 | SUZUKI T, KAZUKI Y, HARA T, et al. Current advances in microcell-mediated chromosome transfer technology and its applications[J]. Experimental Cell Research, 2020, 390(1): 111915. |
| 94 | BROWN D M, CHAN Y A, DESAI P J, et al. Efficient size-independent chromosome delivery from yeast to cultured cell lines[J]. Nucleic Acids Research, 2016, 45(7): e50. |
| 95 | LI L P, BLANKENSTEIN T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast-embryonic stem cell fusion[J]. Nature Protocols, 2013, 8(8): 1567-1582. |
| 96 | MARSCHALL P, MALIK N, LARIN Z. Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine[J]. Gene Therapy, 1999, 6(9): 1634-1637. |
| 97 | MACDONALD L E, KAROW M, STEVENS S, et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(14): 5147-5152. |
| 98 | MURPHY A J, MACDONALD L E, STEVENS S, et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(14): 5153-5158. |
| 99 | LARTIGUE C, GLASS J I, ALPEROVICH N, et al. Genome transplantation in bacteria: changing one species to another[J]. Science, 2007, 317(5838): 632-638. |
| 100 | MONTOLIU L. Purification of yeast artificial chromosome DNA for microinjection using pulsed-field gel electrophoresis and ultrafiltration[J]. Cold Spring Harbor Protocols, 2018. doi:10.1101/pdb.prot093948 |
| 101 | MONTOLIU L. Large-scale preparation of yeast agarose plugs to isolate yeast artificial chromosome DNA[J]. Cold Spring Harbor Protocols, 2018. DOI: 10.1101/pdb.prot093955 |
| 102 | LEE J T, JAENISCH R. A method for high efficiency YAC lipofection into murine embryonic stem cells[J]. Nucleic Acids Research, 1996, 24(24): 5054-5055. |
| 103 | DOHERTY A M, FISHER E M. Microcell-mediated chromosome transfer (MMCT): small cells with huge potential[J]. Mammalian Genome, 2003, 14(9): 583-592. |
| 104 | SINENKO S A, PONOMARTSEV S V, TOMILIN A N. Human artificial chromosomes for pluripotent stem cell-based tissue replacement therapy[J]. Experimental Cell Research, 2020, 389(1): 111882. |
| 105 | LISKOVYKH M, LEE N C, LARIONOV V, et al. Moving toward a higher efficiency of microcell-mediated chromosome transfer[J]. Molecular Therapy-Methods & Clinical Development, 2016, 3: 16043. |
| 106 | SUZUKI T, KAZUKI Y, OSHIMURA M, et al. Highly efficient transfer of chromosomes to a broad range of target cells using Chinese hamster ovary cells expressing murine leukemia virus-derived envelope proteins[J]. PLoS One, 2016, 11(6): e0157187. |
| 107 | JAKOBOVITS A, MOORE A L, GREEN L L, et al. Germ-line transmission and expression of a human-derived yeast artificial chromosome[J]. Nature, 1993, 362(6417): 255-258. |
| 108 | LI L P, LAMPERT J C, CHEN X J, et al. Transgenic mice with a diverse human T cell antigen receptor repertoire[J]. Nature Medicine, 2010, 16(9): 1029-1034. |
| 109 | SUZUKI N, ITOU T, HASEGAWA Y, et al. Cell to cell transfer of the chromatin-packaged human β-globin gene cluster[J]. Nucleic Acids Research, 2009, 38(5): e33. |
| 110 | O'DOHERTY A, RUF S, MULLIGAN C, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes[J]. Science, 2005, 309(5743): 2033-2037. |
| 111 | KAZUKI Y, HIRATSUKA M, TAKIGUCHI M, et al. Complete genetic correction of iPS cells from Duchenne muscular dystrophy[J]. Molecular Therapy, 2010, 18(2): 386-393. |
| 112 | TAYLOR L D, CARMACK C E, HUSZAR D, et al. Human immunoglobulin transgenes undergo rearrangement, somatic mutation and class switching in mice that lack endogenous IgM[J]. International Immunology, 1994, 6(4): 579-591. |
| 113 | GREEN L L, HARDY M C, MAYNARD-CURRIE C E, et al. Antigen-specific human monoclonal antibodies from mice engineered with human Ig heavy and light chain YACs[J]. Nature Genetics, 1994, 7(1): 13-21. |
| 114 | LONBERG N, TAYLOR L D, HARDING F A, et al. Antigen-specific human antibodies from mice comprising four distinct genetic modifications[J]. Nature, 1994, 368(6474): 856-859. |
| 115 | SCHWARTZBERG P L, GOFF S P, ROBERTSON E J. Germ-line transmission of a c-abl mutation produced by targeted gene disruption in ES cells[J]. Science, 1989, 246(4931): 799-803. |
| 116 | ZIJLSTRA M, LI E, SAJJADI F, et al. Germ-line transmission of a disrupted beta 2-microglobulin gene produced by homologous recombination in embryonic stem cells[J]. Nature, 1989, 342(6248): 435-438. |
| 117 | MANSOUR S L, THOMAS K R, CAPECCHI M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes[J]. Nature, 1988, 336(6197): 348-352. |
| 118 | BRÜGGEMANN M, CASKEY H M, TEALE C, et al. A repertoire of monoclonal antibodies with human heavy chains from transgenic mice[J]. Proceedings of the National Academy of Sciences of the United States of America, 1989, 86(17): 6709-6713. |
| 119 | XU J L, XU K, JUNG S, et al. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants[J]. Nature, 2021, 595(7866): 278-282. |
| 120 | MATSUSHITA H, SANO A, WU H, et al. Species-specific chromosome engineering greatly improves fully human polyclonal antibody production profile in cattle[J]. PLoS One, 2015, 10(6): e0130699. |
| 121 | GARDNER C L, SUN C Q, LUKE T, et al. Antibody preparations from human transchromosomic cows exhibit prophylactic and therapeutic efficacy against venezuelan equine encephalitis virus[J]. Journal of Virology, 2017, 91(14): e00226. |
| 122 | LUKE T, WU H, ZHAO J C, et al. Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS-CoV in vivo [J]. Science Translational Medicine, 2016, 8(326): 326ra21. |
| 123 | BEIGEL J H, VOELL J, KUMAR P, et al. Safety and tolerability of a novel, polyclonal human anti-MERS coronavirus antibody produced from transchromosomic cattle: a phase 1 randomised, double-blind, single-dose-escalation study[J]. The Lancet Infectious Diseases, 2018, 18(4): 410-418. |
| 124 | NIU D, WEI H J, LIN L, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9[J]. Science, 2017, 357(6357): 1303-1307. |
| 125 | YUE Y N, XU W H, KAN Y N, et al. Extensive germline genome engineering in pigs[J]. Nature Biomedical Engineering, 2021, 5(2): 134-143. |
| 126 | KAZUKI Y, KOBAYASHI K, AUEVIRIYAVIT S, et al. Trans-chromosomic mice containing a human CYP3A cluster for prediction of xenobiotic metabolism in humans[J]. Human Molecular Genetics, 2012, 22(3): 578-592. |
| 127 | UNO N, ABE S, OSHIMURA M, et al. Combinations of chromosome transfer and genome editing for the development of cell/animal models of human disease and humanized animal models[J]. Journal of Human Genetics, 2018, 63(2): 145-156. |
| 128 | KAZUKI Y, KOBAYASHI K, HIRABAYASHI M, et al. Humanized UGT2 and CYP3A transchromosomic rats for improved prediction of human drug metabolism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(8): 3072-3081. |
| [1] | 汪君仪, 武晓乐, 曹月阳, 李炳志. 基因组设计与合成:从复写到理性设计[J]. 合成生物学, 2021, 2(2): 247-255. |
| [2] | 李祎, 林振泉, 刘子鹤. 酿酒酵母适应性实验室进化工具的最新进展[J]. 合成生物学, 2021, 2(2): 287-301. |
| [3] | 王会, 戴俊彪, 罗周卿. 基因组的“读-改-写”技术[J]. 合成生物学, 2020, 1(5): 503-515. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||