• 研究论文 •
大肠杆菌中吲哚介导的双向趋化行为的调控与分析
汪昆昆1, 陈泓宇2, 张安栋1, 卢晓云1, 谭丹1
- 1.西安交通大学生命科学与技术学院,生物医学信息工程教育部重点实验室,陕西 西安 710049
2.清华大学生命科学学院,北京 100084
-
收稿日期:2025-04-29修回日期:2025-07-31出版日期:2025-08-01 -
通讯作者:谭丹 -
作者简介:汪昆昆 (1999—),男,硕士研究生。研究方向为基于嗜盐微生物的合成生物学。E-mail:3122113016@stu.xjtu.edu.cn谭丹 (1986—),女,副教授,博士,硕士生导师。研究方向为基于嗜盐微生物的合成生物学与定量生物学、新活性天然产物的生物合成及关键酶挖掘与研究等。E-mail:tandan@mail.xjtu.edu.cn
第一联系人:汪昆昆、陈泓宇为同等贡献作者
A preliminary study on the regulation and analysis of indole mediated bidirectional chemotaxis behavior of Escherichia coli
WANG Kunkun1, CHEN Hongyu2, ZHANG Andong1, LU Xiaoyun1, TAN Dan1
- 1.Key Laboratory of Biomedical Information Engineering of Ministry of Education,School of Life Science and Technology,Xi'an Jiaotong University,Xi'an 710049,China
2.School of Life Sciences,Tsinghua University,Beijing 100084,China
-
Received:2025-04-29Revised:2025-07-31Online:2025-08-01 -
Contact:TAN Dan
摘要:
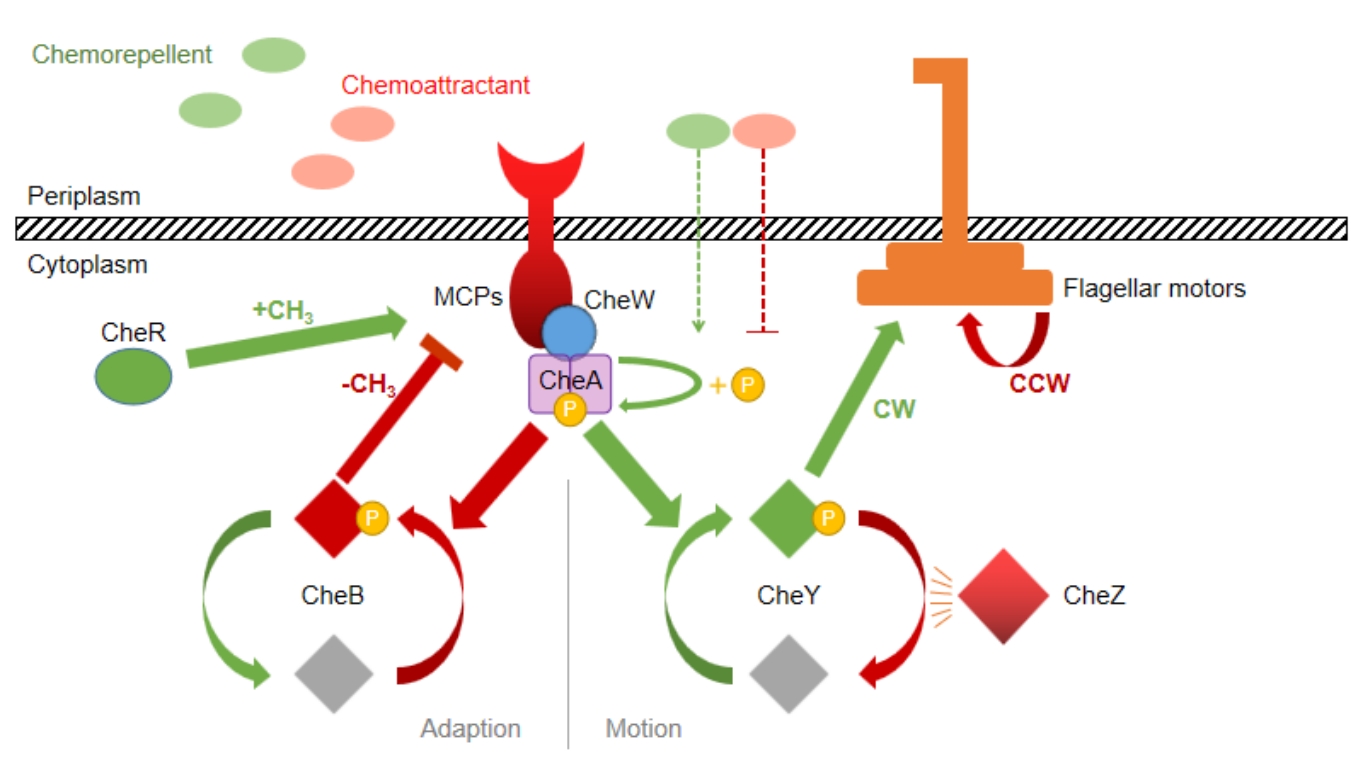
细菌趋化性在肠道菌群定植和癌症诊疗等领域发挥着重要作用,也是定量生物学与合成生物学的研究方向之一。大肠杆菌中吲哚诱导的趋化反应主要由两种效应相反的甲基化趋化受体Tsr与Tar介导,两者的拮抗效应可导致双向趋化反应的发生。本论文从数学模型的建立与仿真以及实验调控与观测两方面对该双向趋化反应进行研究。本论文基于信号转导动力学与马尔可夫随机游走模型,分别从个体水平和群体水平构建了大肠杆菌在双受体介导下趋化性的数学模型,并对不同受体比例下大肠杆菌的趋化响应行为进行了模拟和仿真分析。同时,利用CRISPRi技术对Tar受体进行了不同程度的可诱导敲低(分别敲低0%,40%和70%),并借助于Transwell细菌迁移实验和自主设计优化的微流控观测平台,对吲哚介导下不同Tar表达量菌株的趋化性行为进行了观测与分析。本文通过两种甲基化趋化受体Tsr/Tar相对比例的调节实现了细菌趋化行为的调控,同时结合数学建模初步揭示了相互拮抗的两种趋化性受体比例对细菌个体和群体趋化运动的影响,为扩展趋化行为的定量研究和精确调控手段,以及未来细菌趋化性在肠道菌群人工调节等多领域的应用奠定基础。
中图分类号:
引用本文
汪昆昆, 陈泓宇, 张安栋, 卢晓云, 谭丹. 大肠杆菌中吲哚介导的双向趋化行为的调控与分析[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-036.
WANG Kunkun, CHEN Hongyu, ZHANG Andong, LU Xiaoyun, TAN Dan. A preliminary study on the regulation and analysis of indole mediated bidirectional chemotaxis behavior of Escherichia coli[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-036.
| Name | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F- lacZΔ lacΔ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK+,mk+) phoA supE44 Y-thi-1 gyrA96 relA1 | TransGen Biotech |
| E. coli RP437 | F- thr-1 araC14 leuB6(Am) fhuA31 lacY1 tsx-78λ- eda-50 hisG4(Oc) rfbC1 rpsL136(strR) xylA5 metF159(Am) mtl-1 thiE1 | [ |
| E. coli RP437ΔTar | E. coli RP437 mutant with tar gene knockdown | This study |
| Plasmids | ||
| pUC19 (Bsa I free) | AmpR, pBR322 origin, no Bsa I restriction enzyme site | Tsingke |
| pUC-eGFP | pUC19-derived plasmid carrying eGFP, for cell visualization in Transwell and microfluidics experiment | This study |
| pdCas9 | CmR, backbone plasmid for CRISPRi, carrying dCas9 gene (tetracycline-induced) with sgRNA scaffold (arabinose-induced), and mRFP expression cassette | [ |
| pdCas9-Spacer 1~6 | pdCas9-derived plasmid inserted with Spacer 1~6 for tar gene knockdown | This study |
| Oligomers | Tsingke | |
| Spacer-F1 | TTCAGCCATACTTTTCATACTCCC | |
| Spacer-R1 | GAAGCGGAATATATCCCTAGGTAT | |
| Spacer-extend-F1 | GCAAGGCGATTAAGTTGGGTAA | |
| Spacer-extend-R1 | TGAGTTAGCTCACTCATTAGGCAC | |
| Spacer-extend-F2 | CGTAAGGAGAAAATACCGCATCAG | |
| Spacer-extend-R2 | ACAGGAAACAGCTATGACCAGAATT | |
| GFP-F-EcoR I | CCCAATCCGGAATTCCGCAATTAATGTGAGTTAGCTCAC | |
| GFP-R-EcoR I | CCCGGACACGAATTCGATCCGGATATAGTTCCTCCTTTC | |
| GFP-R-Spe I | CCCGGACACACTAGTGATCCGGATATAGTTCCTCCT | |
| Spacer-1 | CCAGGGAATGCAAAATGCAA | |
| Spacer-2 | CCAGCACGGCGGCAAAGTGG | |
| Spacer-3 | CGCACGCGCGGCTTCAACCG | |
| Spacer-4 | TGCTCACTGGCAGGACGGGA | |
| Spacer-5 | CGATAGCGCCAGGAAAACAT | |
| Spacer-6 | TTCAGTACGGGAGGAAAGAT |
表1 本研究所涉及的菌株、质粒与DNA序列信息
Table 1 Strains, Plasmids and Oligomers used in this study
| Name | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F- lacZΔ lacΔ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK+,mk+) phoA supE44 Y-thi-1 gyrA96 relA1 | TransGen Biotech |
| E. coli RP437 | F- thr-1 araC14 leuB6(Am) fhuA31 lacY1 tsx-78λ- eda-50 hisG4(Oc) rfbC1 rpsL136(strR) xylA5 metF159(Am) mtl-1 thiE1 | [ |
| E. coli RP437ΔTar | E. coli RP437 mutant with tar gene knockdown | This study |
| Plasmids | ||
| pUC19 (Bsa I free) | AmpR, pBR322 origin, no Bsa I restriction enzyme site | Tsingke |
| pUC-eGFP | pUC19-derived plasmid carrying eGFP, for cell visualization in Transwell and microfluidics experiment | This study |
| pdCas9 | CmR, backbone plasmid for CRISPRi, carrying dCas9 gene (tetracycline-induced) with sgRNA scaffold (arabinose-induced), and mRFP expression cassette | [ |
| pdCas9-Spacer 1~6 | pdCas9-derived plasmid inserted with Spacer 1~6 for tar gene knockdown | This study |
| Oligomers | Tsingke | |
| Spacer-F1 | TTCAGCCATACTTTTCATACTCCC | |
| Spacer-R1 | GAAGCGGAATATATCCCTAGGTAT | |
| Spacer-extend-F1 | GCAAGGCGATTAAGTTGGGTAA | |
| Spacer-extend-R1 | TGAGTTAGCTCACTCATTAGGCAC | |
| Spacer-extend-F2 | CGTAAGGAGAAAATACCGCATCAG | |
| Spacer-extend-R2 | ACAGGAAACAGCTATGACCAGAATT | |
| GFP-F-EcoR I | CCCAATCCGGAATTCCGCAATTAATGTGAGTTAGCTCAC | |
| GFP-R-EcoR I | CCCGGACACGAATTCGATCCGGATATAGTTCCTCCTTTC | |
| GFP-R-Spe I | CCCGGACACACTAGTGATCCGGATATAGTTCCTCCT | |
| Spacer-1 | CCAGGGAATGCAAAATGCAA | |
| Spacer-2 | CCAGCACGGCGGCAAAGTGG | |
| Spacer-3 | CGCACGCGCGGCTTCAACCG | |
| Spacer-4 | TGCTCACTGGCAGGACGGGA | |
| Spacer-5 | CGATAGCGCCAGGAAAACAT | |
| Spacer-6 | TTCAGTACGGGAGGAAAGAT |
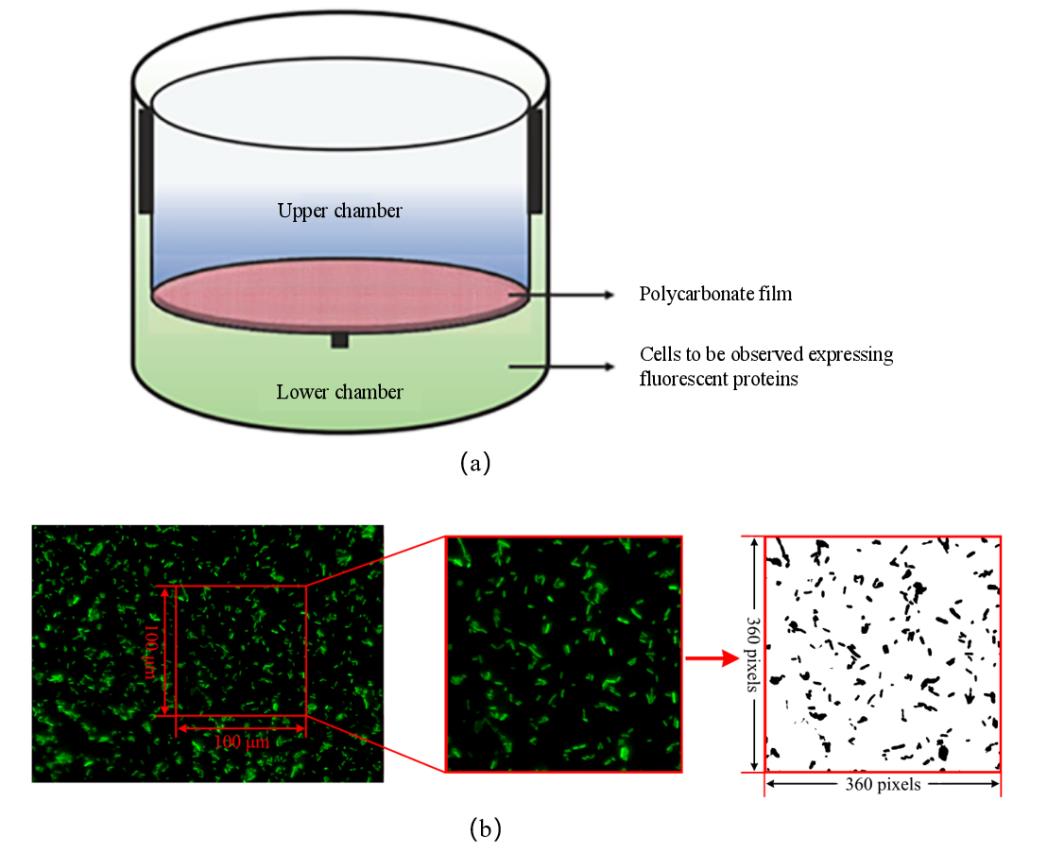
图2 Transwell迁移实验装置和图像处理流程(a)Transwell细菌迁移装置示意图 [24];(b)Transwell荧光成像图像处理流程
Fig. 2 Transwell migration experiment and image processing procedure(a)The Transwell bacterial migration device [24];(b)Transwell fluorescence image processing procedure
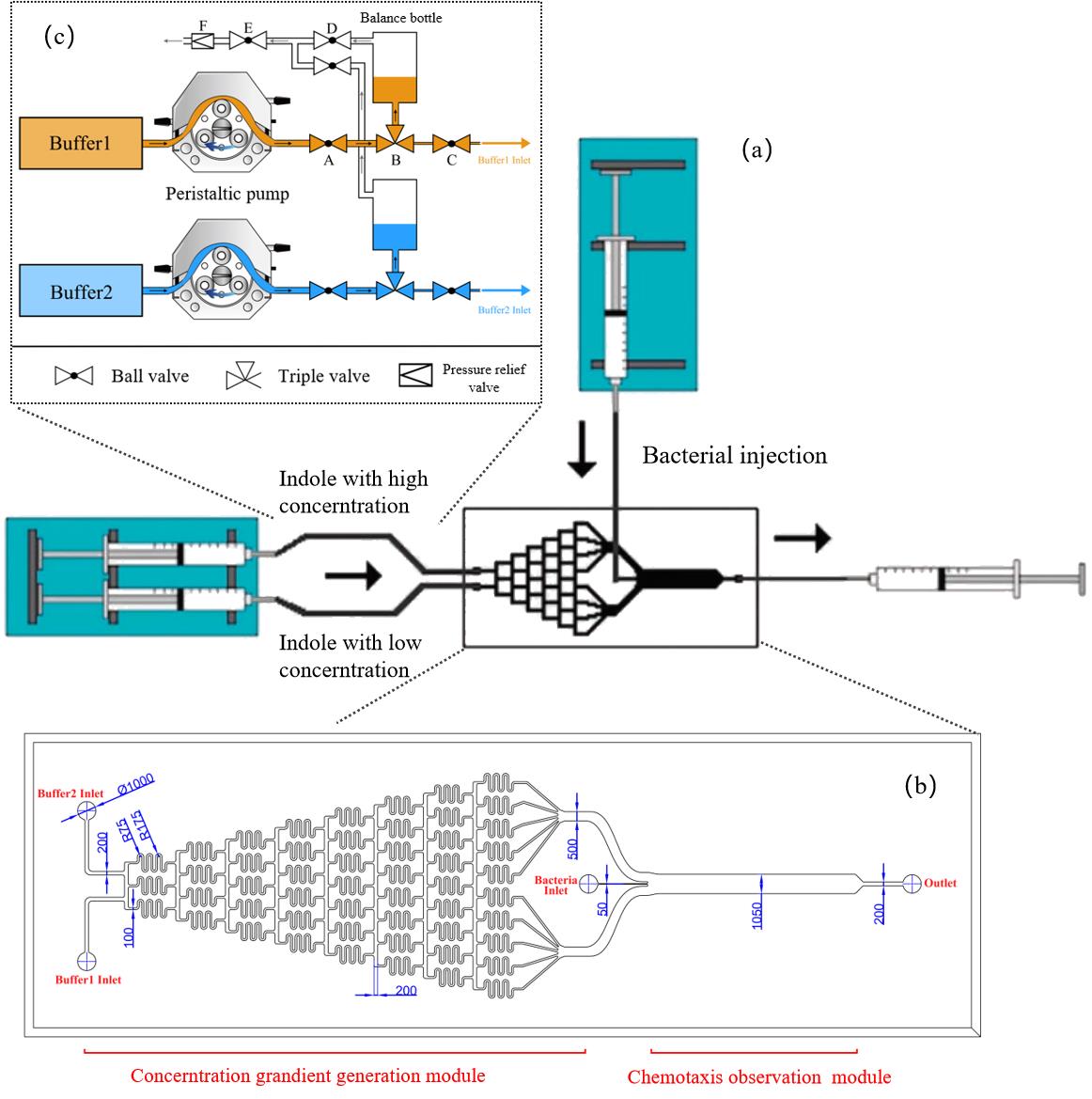
图3 微流控设计和优化示意图(a)μFlow装置示意图[27];(b)微流控芯片设计图;标注单位为 μm;(c)微流控进样系统示意图
Fig. 3 The design and optimization of microfluidics(a)μFlow design of microfluidics [27];(b)The detail of microfluidics;(c)The injection system of microfluidics
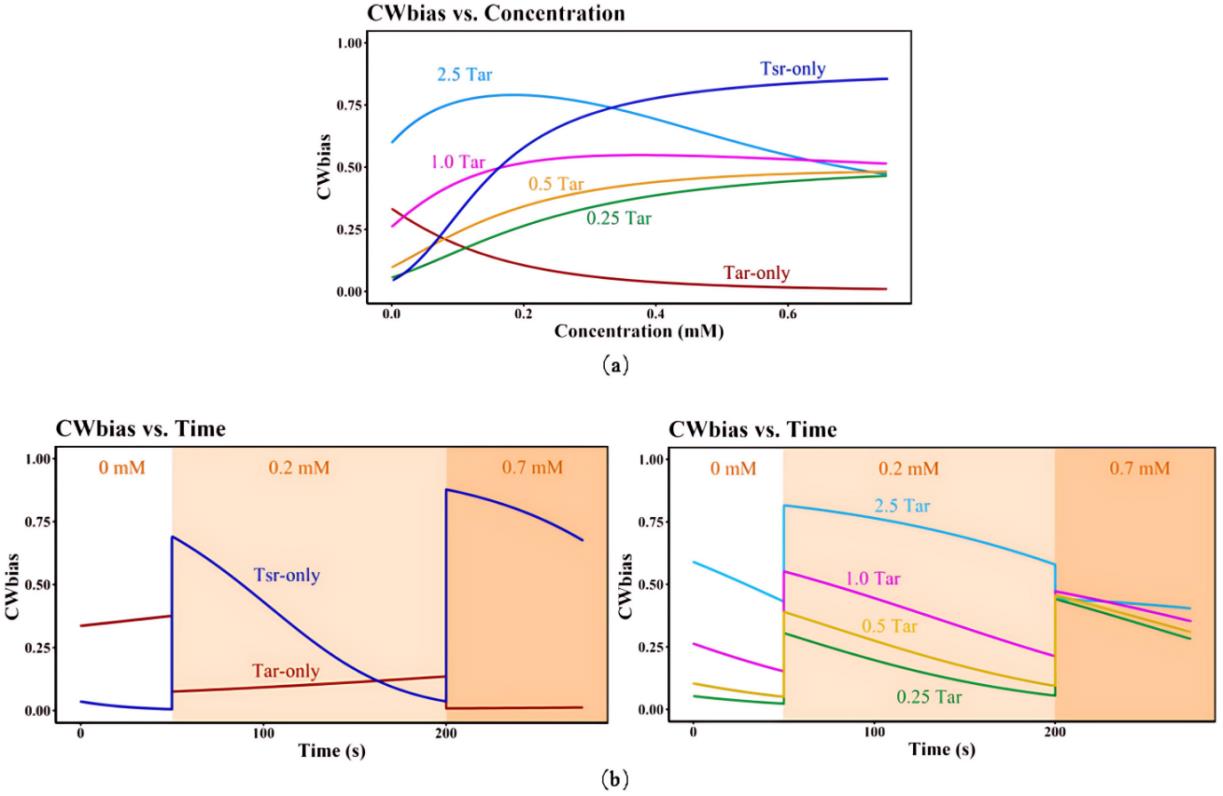
图4 信号转导动力学模型的模拟结果(a)不同吲哚浓度和不同Tsr/Tar受体比例下CWbias-c关系;(b)不同吲哚浓度下不同Tsr/Tar受体比例下CWbias-t变化关系:左图表示仅包含生理条件含量下的 Tar 受体与 Tsr 受体的CWbias-t变化关系,右图表示不同含量Tar(分别为2.5倍、1倍、0.5倍、0.25倍生理条件含量的Tar)条件下CWbias-t变化关系。其中,Tar-only 与 Tsr-only 分别代表仅包含生理条件含量下的 Tar 受体与 Tsr 受体。Tar前的倍数代表以生理条件含量作为 1Tar,分别对应增高/降低倍数。
Fig. 4 Simulation results of signal transduction kinetic model(a)CWbias-c relationship at different Tsr/Tar receptor ratios and different indole concentrations;(b)CWbias-t relationship at different Tsr/Tar receptor ratios and different indole concentrations;Tar-only and Tsr-only mean physiological concentration of Tar receptors and Tsr receptors. The number before Tar represent the corresponding increase/decrease fold when the physiological concentration of Tar served as 1Tar.
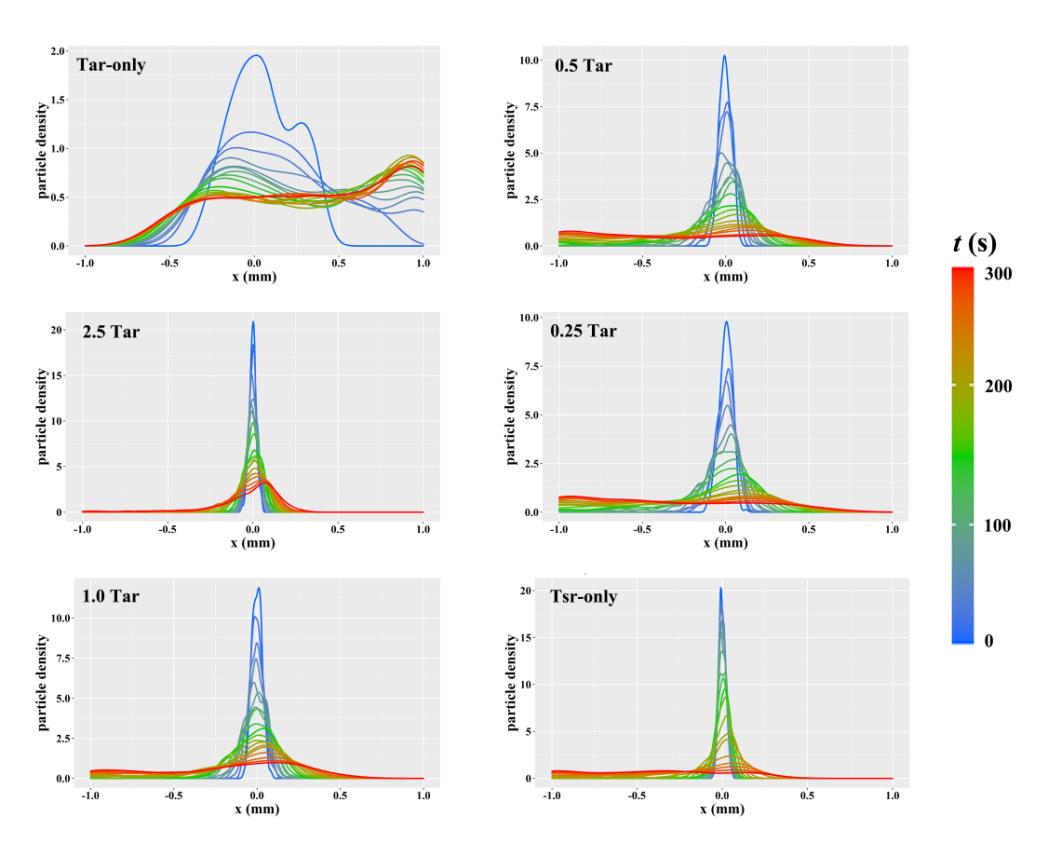
图5 随机游走模型模拟运动中粒子的密度分布曲线其中,Tar-only 与 Tsr-only 分别代表仅包含生理条件含量下的 Tar 受体与 Tsr 受体。2.5 Tar、1 Tar、0.5Tar、0.25Tar 分别表示2.5倍、1倍、0.5倍、0.25倍生理条件含量的Tar
Fig. 5 Density distribution curves of particles in simulated motion of random walk modelTar-only and Tsr-only mean physiological concentration of Tar receptors and Tsr receptors. The number before Tar represent the corresponding increase/decrease fold when the physiological concentration of Tar served as 1Tar.
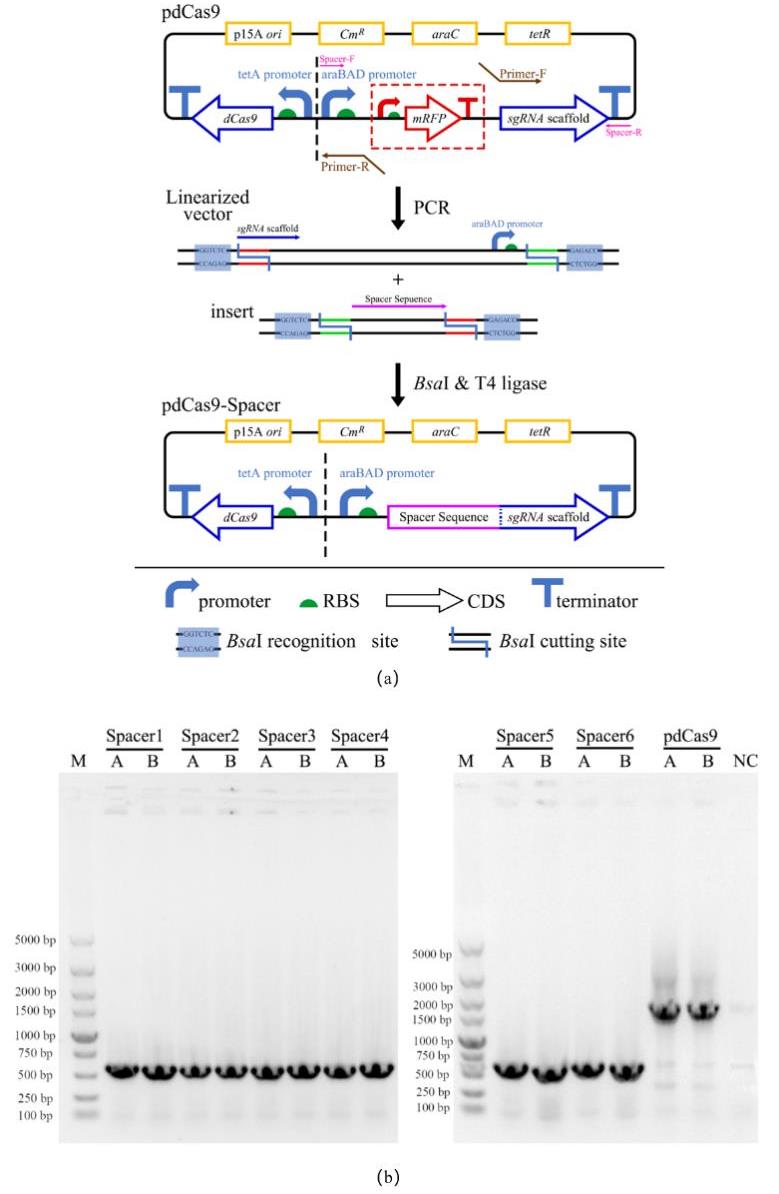
图6 CRISPRi工作质粒的设计、构建和验证(a)CRISPRi敲低质粒的设计与构建:Spacer序列与pdCas9线性化载体进行Golden gate 连接;(b)pdCas9-Spacer导入大肠杆菌E. coli DH5α的菌落PCR验证结果。其中左图为含pdCas9-Spacer 1~4的重组菌菌落PCR验证结果,其中右图为含pdCas9-Spacer 5~6的重组菌菌落PCR验证结果,阳性结果条带应为579 bp;M:DL5000 DNA Marker;A、B:各筛选平板上的两个平行单菌落;pdCas9孔道为只含有pdCas9空质粒的菌落PCR结果,条带应为1642 bp; NC:阴性对照,即无质粒的野生型E. coli DH5α
Fig. 6 Design, construction and verification of CRISPRi plasmids(a) Design of CRISPRi plasmid and its construction via Golden Gate of pdCas9 linear vector and spacer sequence;(b)Results of colony PCR after transformation;M:DL5000 DNA Marker;Spacer 1~6:Colony PCR results of recombinant strains with pdCas9-Spacer 1~6 plasmids;A、B:Two parallel single colonies on each screening plate;NC:negative control, that is, E. coli DH5α wild type without any plasmid
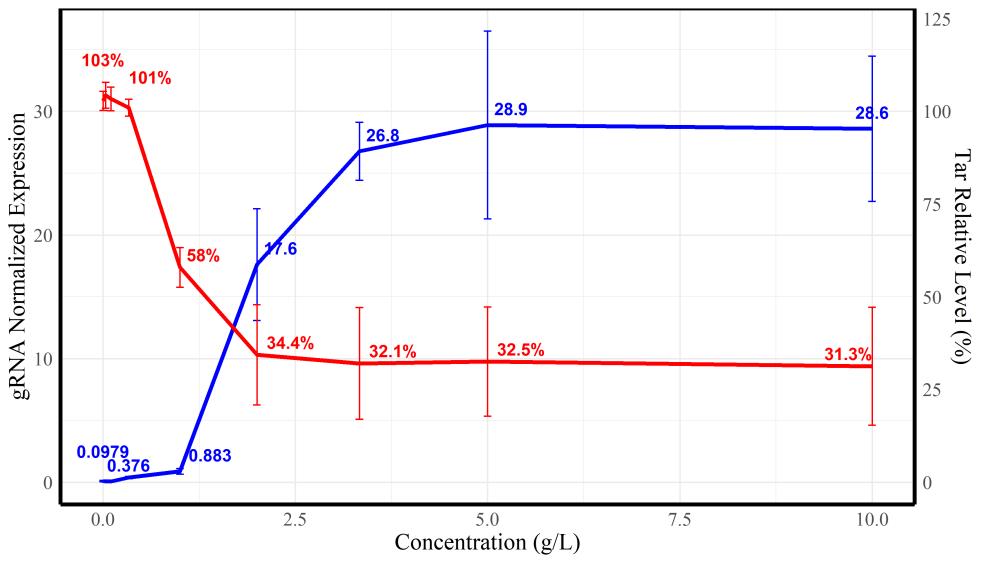
图7 不同阿拉伯糖诱导浓度下sgRNA转录水平(蓝色曲线)与tar相对转录水平(红色曲线)
Fig. 7 Relative transcriptional levels of gRNA (blue)and tar gene (red)under the induction of different arabinose concentrations
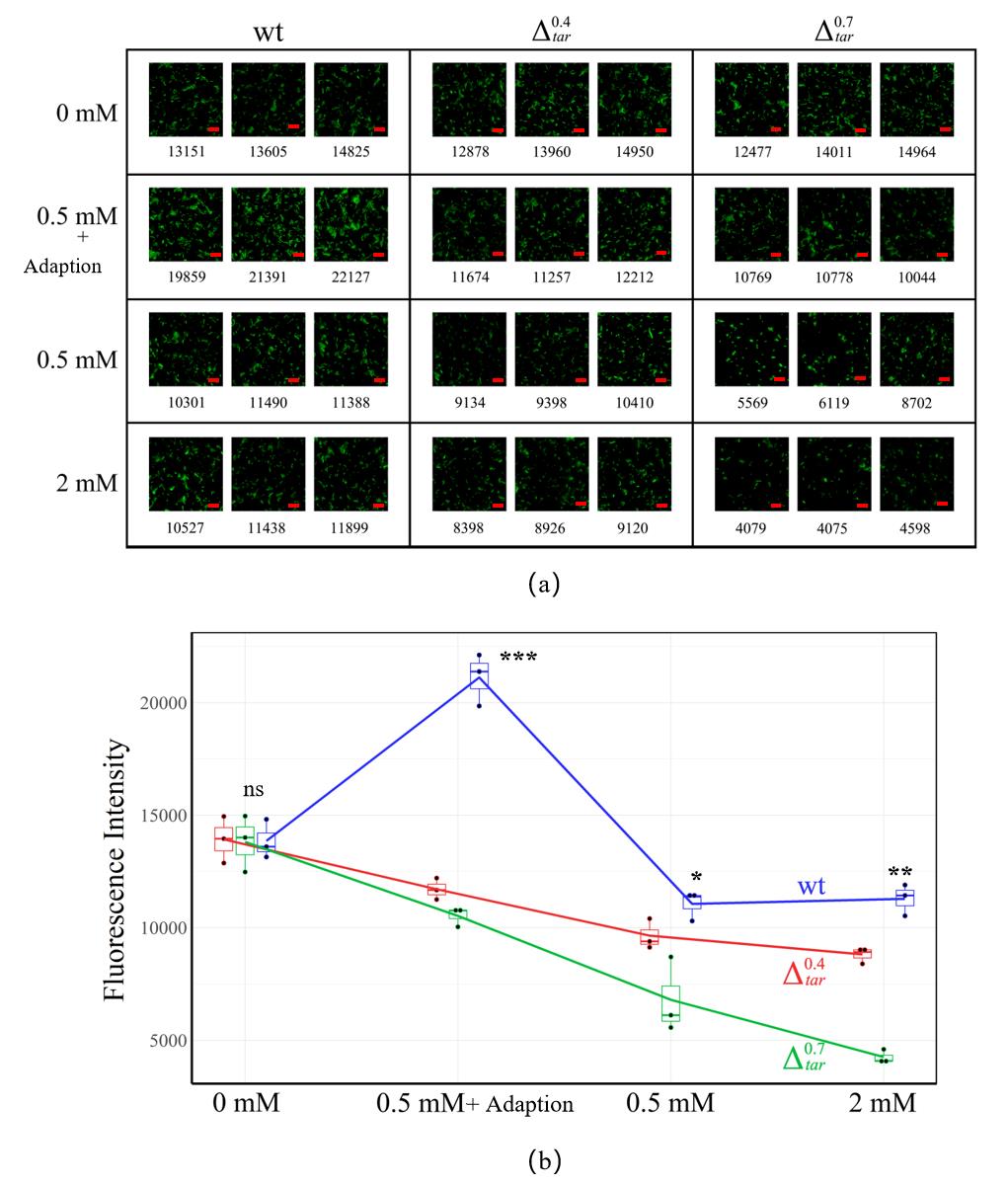
图8 利用Transwellx细菌迁移实验观测吲哚介导的趋化性结果(a)不同Tar表达量突变株在不同吲哚浓度条件下的Transwell成像结果。吲哚处理浓度分别为0、0.5mM、0.5mM+预适应、2 mM,每组含细菌分布较均匀的100 μm×100 μm的三个平行视野,红色标尺为2 μm。下方数字表示经过处理统计后的视野中的绿色荧光强度,并用于衡量细菌相对密度。(b)Transwell实验中不同菌株对吲哚浓度的响应的定量趋势统计。“ns”表示p>0.05,无显著差异;“*”表示p<0.05,“**”表示p<0.01,“***”表示p<0.001,以上三种代表存在显著性差异。
Fig. 8 Results of Transwell migration experiments(a)Transwell imaging results of tar knockdown mutants under different indole concentrations, which is 0 mM、0.5mM、0.5mM+adoption and 2 mM, respectively. Each group contains three parallel view fields with dimension of 100 μm × 100 μm. Number below each image means the fluorescence intensity in the view field after statistical processing. Bar is 2 μm;(b)Quantitative trend of the response of different tar knockdown mutants to indole concentration in Transwell experiments
| 菌株类型 | tar相对转录水平 | tsr相对转录水平 | tsr/tar |
|---|---|---|---|
| E. coli RP437野生型 | 7.80±0.10 | 14.66±0.03 | 1.88±0.06 |
| Tar 受体敲低菌株 | 4.68±0.08 | 14.81±0.02 | 3.16±0.05 |
| Tar 受体敲低菌株 | 2.53±0.02 | 14.31±0.11 | 5.66±0.03 |
表2 Transwell迁移实验的细菌中tar和tsr的相对转录水平(RT-qPCR测定)
Table 2 The relative transcriptional level of tar and tsr in E. coli used in Transwell experiment tested by RT-qPCR
| 菌株类型 | tar相对转录水平 | tsr相对转录水平 | tsr/tar |
|---|---|---|---|
| E. coli RP437野生型 | 7.80±0.10 | 14.66±0.03 | 1.88±0.06 |
| Tar 受体敲低菌株 | 4.68±0.08 | 14.81±0.02 | 3.16±0.05 |
| Tar 受体敲低菌株 | 2.53±0.02 | 14.31±0.11 | 5.66±0.03 |
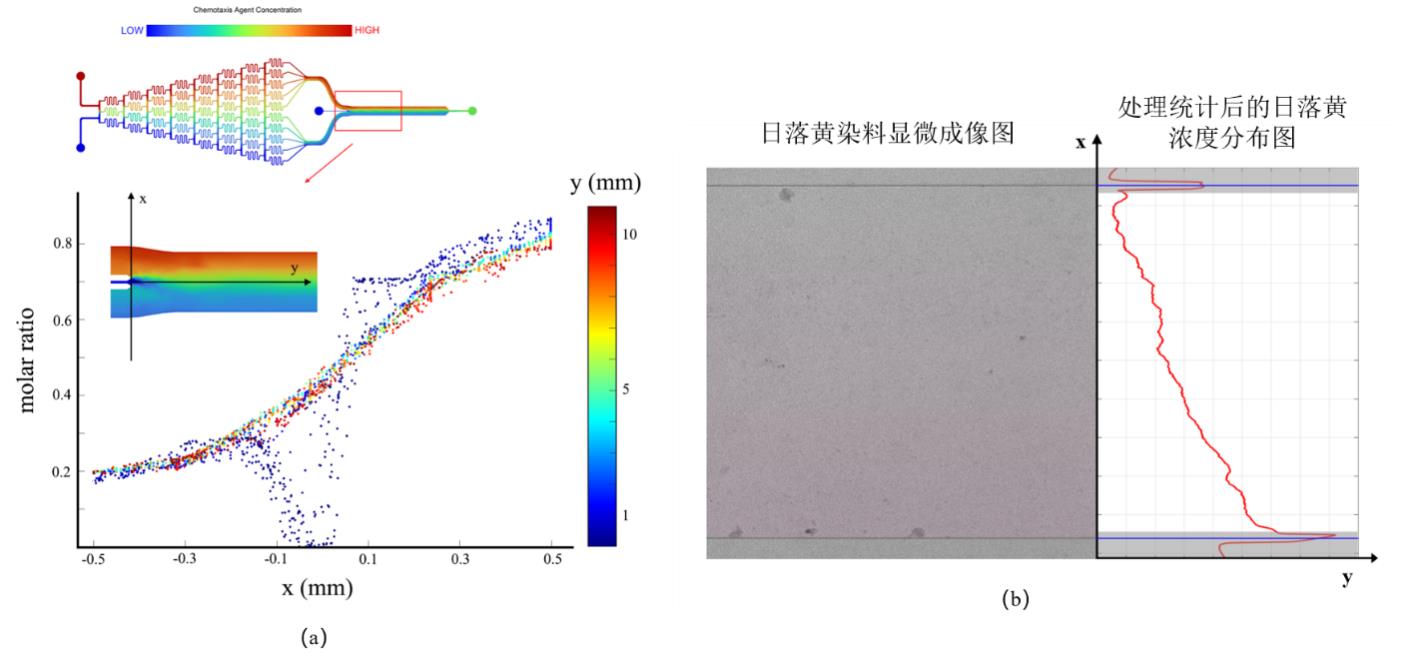
图9 微流控装置浓度场的仿真验证和实验验证结果(a)仿真验证微流控浓度场在各方向上分布;(b)以日落黄作为染料对微流控装置浓度场分布的实验验证
Fig. 9 Simulation and experimental verification results of concentration field in microfluidic devices(a)Simulation results of the distribution of concentration field in all directions; (b) Experimental verification of concentration field distribution in microfluidic device using sunset yellow as a dye
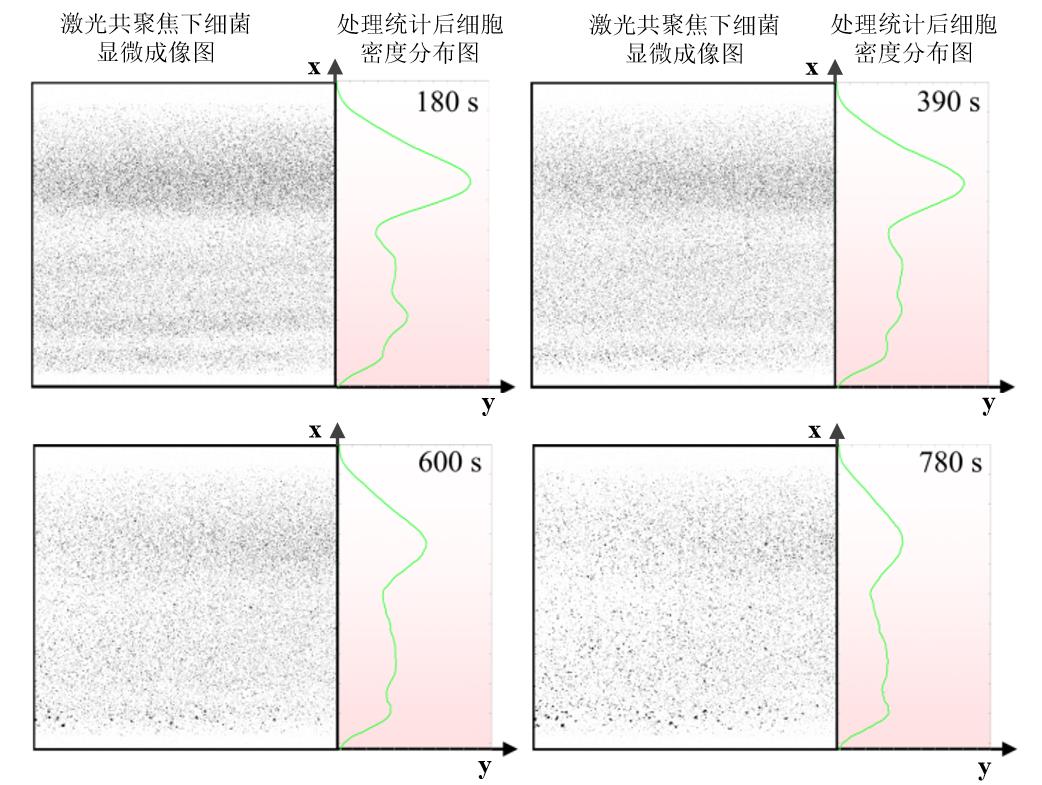
图10 微流控流道中大肠杆菌野生菌的密度分布随时间变化情况(左侧表示激光共聚焦显微镜下细菌成像结果(统一将视野中的绿色细菌处理成黑色以便于统计分析),右侧为统计分析后的细菌密度情况,且右侧背景的颜色深浅表示吲哚浓度高低,颜色越深代表此处对应的吲哚浓度越大。)
Fig. 10 Density distribution over time of Escherichia coli wild-type in microfluidic channel(The left part of each image represents the bacterial imaging results under confocal microscopy, and the right part represents the bacterial density after statistical analysis. The color of the background in right part indicates the concentration of indole, and the darker color means the higher indole concentration.)
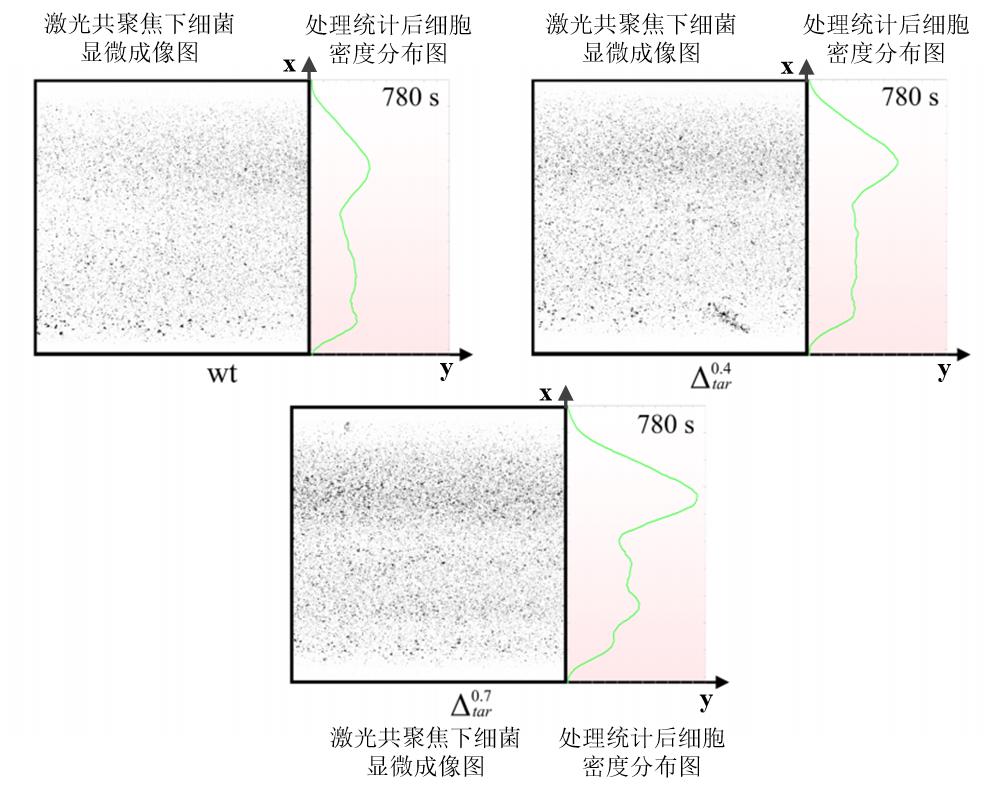
图11 微流控流道中不同Tar受体表达量菌株在同一时间的密度分布情况左侧表示激光共聚焦显微镜下细菌成像结果(统一将视野中的绿色细菌处理成黑色以便于统计分析),右侧为统计分析后的细菌密度情况,且右侧背景的颜色深浅表示吲哚浓度高低,颜色越深代表此处对应的吲哚浓度越大。
Fig. 11 Density distribution of strains with different Tar receptor expression levels in microfluidic channels at the same timeThe left part of each image represents the bacterial imaging results under confocal microscopy, and the right part represents the bacterial density after statistical analysis. The color of the background in right part indicates the concentration of indole, and the darker color means the higher indole concentration.
| [1] | ADLER J. Chemotaxis in bacteria [J]. Science, 1966, 153(3737): 708-16. |
| [2] | IMAE Y, OOSAWA K, MIZUNO T, et al. Phenol: A complex chemoeffector in bacterial chemotaxis [J]. J Bacteriol, 1987, 169(1): 371-9. |
| [3] | AIHARA E, CLOSSON C, MATTHIS A L, et al. Motility and chemotaxis mediate the preferential colonization of gastric injury sites by helicobacter pylori [J]. PLoS Pathog, 2014, 10(7): e1004275. |
| [4] | RAMOS H C, RUMBO M, SIRARD J C. Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa [J]. Trends Microbiol, 2004, 12(11): 509-17. |
| [5] | WADHAMS G H, ARMITAGE J P. Making sense of it all: Bacterial chemotaxis [J]. Nat Rev Mol Cell Biol, 2004, 5(12): 1024-37. |
| [6] | HOCH J A. Two-component and phosphorelay signal transduction [J]. Curr Opin Microbiol, 2000, 3(2): 165-70. |
| [7] | ADLER J. Chemotaxis in bacteria [J]. Annu Rev Biochem, 1975, 44: 341-56. |
| [8] | MACNAB R M. Examination of bacterial flagellation by dark-field microscopy [J]. J Clin Microbiol, 1976, 4(3): 258-65. |
| [9] | GREBE T W, STOCK J. Bacterial chemotaxis: The five sensors of a bacterium [J]. Curr Biol, 1998, 8(5): R154-7. |
| [10] | ZHULIN I B. The superfamily of chemotaxis transducers: From physiology to genomics and back [J]. Adv Microb Physiol, 2001, 45: 157-98. |
| [11] | NINFA E G, STOCK A, MOWBRAY S, et al. Reconstitution of the bacterial chemotaxis signal transduction system from purified components [J]. J Biol Chem, 1991, 266(15): 9764-70. |
| [12] | BOURRET R B, BORKOVICH K A, SIMON M I. Signal transduction pathways involving protein phosphorylation in prokaryotes [J]. Annu Rev Biochem, 1991, 60: 401-41. |
| [13] | MCEVOY M M, BREN A, EISENBACH M, et al. Identification of the binding interfaces on chey for two of its targets, the phosphatase CheZ and the flagellar switch protein flim [J]. J Mol Biol, 1999, 289(5): 1423-33. |
| [14] | DJORDJEVIC S, STOCK A M. Crystal structure of the chemotaxis receptor methyltransferase cher suggests a conserved structural motif for binding S-adenosylmethionine [J]. Structure, 1997, 5(4): 545-58. |
| [15] | YANG J, CHAWLA R, RHEE K Y, et al. Biphasic chemotaxis of Escherichia coli to the microbiota metabolite indole [J]. Proc Natl Acad Sci U S A, 2020, 117(11): 6114-20. |
| [16] | NORRIS N, ALCOLOMBRI U, KEEGSTRA J M, et al. Bacterial chemotaxis to saccharides is governed by a trade-off between sensing and uptake [J]. Biophys J, 2022, 121(11): 2046-59. |
| [17] | JIANG L, OUYANG Q, TU Y. Quantitative modeling of Escherichia coli chemotactic motion in environments varying in space and time [J]. PLoS Comput Biol, 2010, 6(4): e1000735. |
| [18] | KELLER E F, SEGEL L A. Traveling bands of chemotactic bacteria: A theoretical analysis [J]. J Theor Biol, 1971, 30(2): 235-48. |
| [19] | ZHU B, LI H, ZHANG L, et al. A markov random field model-based approach for differentially expressed gene detection from single-cell RNA-seq data [J]. Brief Bioinform, 2022, 23(5). |
| [20] | PARKINSON J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis [J]. J Bacteriol, 1978, 135(1): 45-53. |
| [21] | BAI Y, HE C, CHU P, et al. Spatial modulation of individual behaviors enables an ordered structure of diverse phenotypes during bacterial group migration [J]. Elife, 2021, 10. |
| [22] | BRADLEY R W. An easy-to-use crispri plasmid tool for inducible knockdown in E. coli [J]. Biotechnol Rep (Amst), 2021, 32: e00680. |
| [23] | ROCHA D, CASTRO T L P, AGUIAR E, et al. Gene expression analysis in bacteria by RT-qPCR [J]. Methods Mol Biol, 2020, 2065: 119-37. |
| [24] | JANI S. Visualizing chemoattraction of planktonic cells to a biofilm [J]. Methods Mol Biol, 2018, 1729: 61-9. |
| [25] | JUSTUS C R, MARIE M A, SANDERLIN E J, et al. Transwell in vitro cell migration and invasion assays [J]. Methods Mol Biol, 2023, 2644: 349-59. |
| [26] | ZANG X Q, LI Z Y, ZHANG X Y, et al. Advance in bacteria chemotaxis on microfluidic devices [J]. Chinese Journal of Analytical Chemistry, 2017, 45(11): 1734-44. |
| [27] | ENGLERT D L, MANSON M D, JAYARAMAN A. Investigation of bacterial chemotaxis in flow-based microfluidic devices [J]. Nat Protoc, 2010, 5(5): 864-72. |
| [28] | HINKAMP P E. A table of fikentscher K values versus relative viscosities for a concentration of 1·0 [J]. Polymer, 1967, 8: 381-4. |
| [29] | ANDERS C, NIEWOEHNER O, DUERST A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease [J]. Nature, 2014, 513(7519): 569-73. |
| [30] | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an rna-guided platform for sequence-specific control of gene expression [J]. Cell, 2021, 184(3): 844. |
| [31] | NISHIMASU H, RAN F A, HSU P D, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA [J]. Cell, 2014, 156(5): 935-49. |
| [32] | DOMINGUEZ A A, LIM W A, QI L S. Beyond editing: Repurposing CRISPR-Cas9 for precision genome regulation and interrogation [J]. Nat Rev Mol Cell Biol, 2016, 17(1): 5-15. |
| [33] | BYUNGJIN LEE H-H J, KANG KYOUNG-KU, LEE CHANG-SOO, LEE SANG-HO. Improvement of a diffusion-based microfluidic chemotaxis assay through stable formation of a chemical gradient [J]. Chemical Engineering Science, 2019, 202: 130-7. |
| [34] | LI JEON N, BASKARAN H, DERTINGER S K, et al. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device [J]. Nat Biotechnol, 2002, 20(8): 826-30. |
| [35] | DARNTON N C, TURNER L, ROJEVSKY S, et al. On torque and tumbling in swimming escherichia coli [J]. J Bacteriol, 2007, 189(5): 1756-64. |
| [36] | PING L. Cell orientation of swimming bacteria: From theoretical simulation to experimental evaluation [J]. Sci China Life Sci, 2012, 55(3): 202-9. |
| [37] | YANG Y, SOURJIK V. Opposite responses by different chemoreceptors set a tunable preference point in escherichia coli ph taxis [J]. Mol Microbiol, 2012, 86(6): 1482-9. |
| [38] | HU B, TU Y. Precision sensing by two opposing gradient sensors: How does Escherichia coli find its preferred ph level? [J]. Biophys J, 2013, 105(1): 276-85. |
| [39] | KHAN S, SPUDICH J L, MCCRAY J A, et al. Chemotactic signal integration in bacteria [J]. Proc Natl Acad Sci U S A, 1995, 92(21): 9757-61. |
| [40] | SALMAN H, LIBCHABER A. A concentration-dependent switch in the bacterial response to temperature [J]. Nat Cell Biol, 2007, 9(9): 1098-100. |
| [41] | PAULICK A, JAKOVLJEVIC V, ZHANG S, et al. Mechanism of bidirectional thermotaxis in Escherichia coli [J]. Elife, 2017, 6. |
| [42] | LAGANENKA L, LEE J W, MALFERTHEINER L, et al. Chemotaxis and autoinducer-2 signalling mediate colonization and contribute to co-existence of Escherichia coli strains in the murine gut [J]. Nat Microbiol, 2023, 8(2): 204-17. |
| [43] | SONG J, ZHANG Y, ZHANG C, et al. A microfluidic device for studying chemotaxis mechanism of bacterial cancer targeting [J]. Sci Rep, 2018, 8(1): 6394. |
| [1] | 王翠珍, 陈窕, 王健博. 酶催化杂Diels-Alder反应[J]. 合成生物学, 2024, 5(1): 107-125. |
| [2] | 郭肖杰, 剪兴金, 王立言, 张翀, 邢新会. 合成生物学表型测试生物反应器及其装备化研究进展[J]. 合成生物学, 2024, 5(1): 16-37. |
| [3] | 秦伟彤, 杨广宇. 微液滴高通量筛选方法的研究与应用进展[J]. 合成生物学, 2023, 4(5): 966-979. |
| [4] | 涂然, 李世新, 李昊霓, 王猛. 液滴微流控技术在微生物工程菌株选育中的应用进展[J]. 合成生物学, 2023, 4(1): 165-184. |
| [5] | 赵晓宇, 张浩, 李雪飞, 胡政. 进化视角下的定量生物学规律与人工生命合成[J]. 合成生物学, 2022, 3(1): 6-21. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||