| 1 |
MITRA S, TOMAR P C. Hybridoma technology; advancements, clinical significance, and future aspects [J]. Journal of Genetic Engineering and Biotechnology, 2021, 19: 1-12.
|
| 2 |
HOOGENBOOM H R, DE BRUıNE A P, HUFTON S E, et al. Antibody phage display technology and its applications [J]. Immunotechnology, 1998, 4(1): 1-20.
|
| 3 |
STOLTENBURG R, REINEMANN C, STREHLITZ B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands [J]. Biomolecular Engineering, 2007, 24(4): 381-403.
|
| 4 |
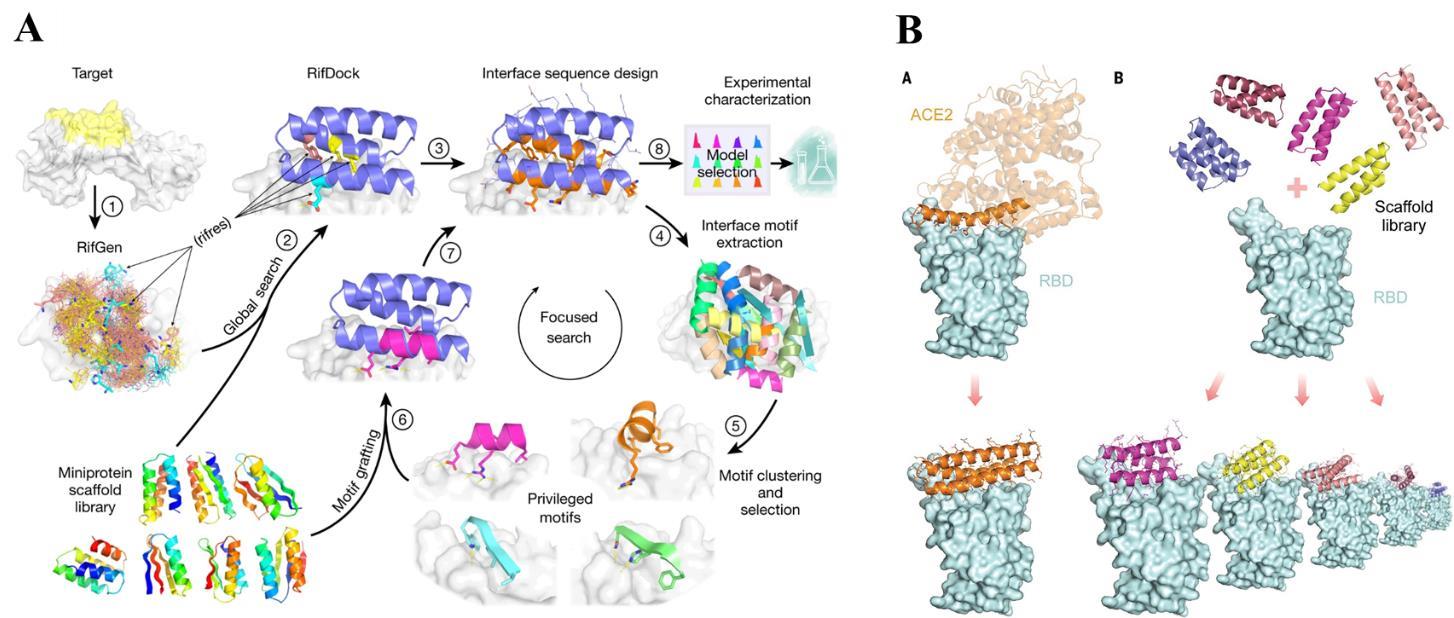
CAO L, GORESHNIK I, COVENTRY B, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors [J]. Science, 2020, 370(6515): 426-31.
|
| 5 |
CAO L, COVENTRY B, GORESHNIK I, et al. Design of protein-binding proteins from the target structure alone [J]. Nature, 2022, 605(7910): 551-60.
|
| 6 |
HIE B L, SHANKER V R, XU D, et al. Efficient evolution of human antibodies from general protein language models [J]. Nature Biotechnology, 2024, 42(2): 275-83.
|
| 7 |
HE H, HE B, GUAN L, et al. De novo generation of SARS-CoV-2 antibody CDRH3 with a pre-trained generative large language model [J]. Nature Communications, 2024, 15(1): 6867.
|
| 8 |
RUFFOLO J A, CHU L-S, MAHAJAN S P, et al. Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies [J]. Nature Communications, 2023, 14(1): 2389.
|
| 9 |
LI L, GUPTA E, SPAETH J, et al. Machine learning optimization of candidate antibody yields highly diverse sub-nanomolar affinity antibody libraries [J]. Nature Communications, 2023, 14(1): 3454.
|
| 10 |
JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold [J]. Nature, 2021, 596(7873): 583-9.
|
| 11 |
ABRAMSON J, ADLER J, DUNGER J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3 [J]. Nature, 2024: 1-3.
|
| 12 |
RIPOLL D R, CHAUDHURY S, WALLQVIST A. Using the antibody-antigen binding interface to train image-based deep neural networks for antibody-epitope classification [J]. Computational Biology, 2021, 17(3): e1008864.
|
| 13 |
CONTI S, LAU E Y, OVCHINNIKOV V. On the rapid calculation of binding affinities for antigen and antibody design and affinity maturation simulations [J]. Antibodies, 2022, 11(3): 51.
|
| 14 |
IWANO N, ADACHI T, AOKI K, et al. Generative aptamer discovery using RaptGen [J]. Nature Computational Science, 2022, 2(6): 378-86.
|