合成生物学 ›› 2021, Vol. 2 ›› Issue (4): 598-611.DOI: 10.12211/2096-8280.2020-092
合成生物学研究中的微生物启动子工程策略
于慧敏, 郑煜堃, 杜岩, 王苗苗, 梁有向
- 清华大学化学工程系,教育部工业生物催化重点实验室,北京 100084
-
收稿日期:2020-12-30修回日期:2021-02-06出版日期:2021-08-31发布日期:2021-09-10 -
通讯作者:于慧敏 -
作者简介:于慧敏 (1973—),女,博士,教授,博士生导师。研究方向为工业生物催化、合成生物学与生物纳米技术等。E-mail:yuhm@tsinghua.edu.cn -
基金资助:国家重点研发计划(2018YFA0902200);国家自然科学基金面上项目(21776157)
Microbial promoter engineering strategies in synthetic biology
YU Huimin, ZHENG Yukun, DU Yan, WANG Miaomiao, LIANG Youxiang
- Key Laboratory of Industrial Biocatalysis,Ministry of Education,Department of Chemical Engineering,Tsinghua University,Beijing 100084,China
-
Received:2020-12-30Revised:2021-02-06Online:2021-08-31Published:2021-09-10 -
Contact:YU Huimin
摘要:
合成生物学研究对于我国绿色生物制造产业和可持续发展战略至关重要。启动子是合成生物学核心元件,是在转录水平上实现基因高效、精准表达调控的最关键因素之一。本文重点对原核微生物启动子工程研究的基本内容、研究进展及发展趋势进行了综述。首先概述了启动子序列基本特征及其受RNA聚合酶σ因子识别调控的一般规律;并以大肠杆菌乳糖操纵子为例简要介绍了诱导型启动子的负调控与正调控诱导机制。其次,分别从对靶基因自身内源启动子进行突变改造以及采用高效外源启动子进行替换改造这两个方面入手,阐述了启动子改造的常用策略。进一步对近年来公开报道的不同类型诱导型启动子进行了梳理,小结了代表性化学分子诱导剂以及物理信号诱导方式的种类及基本特征。简述了非模式和模式微生物组成型启动子的研究进展及研究侧重点。结合动态代谢调控技术及人工智能工具的突破性发展,提出具有动态调控功能的特殊启动子的发现与改造、全新性能启动子元件的人工智能设计与改造等将成为启动子工程研究的新方向与新前沿。最后分析了启动子工程领域存在的挑战性问题,展望了今后的研究重点,并结合合成生物学的发展,进一步强调了微生物启动子工程的重要作用。
中图分类号:
引用本文
于慧敏, 郑煜堃, 杜岩, 王苗苗, 梁有向. 合成生物学研究中的微生物启动子工程策略[J]. 合成生物学, 2021, 2(4): 598-611.
YU Huimin, ZHENG Yukun, DU Yan, WANG Miaomiao, LIANG Youxiang. Microbial promoter engineering strategies in synthetic biology[J]. Synthetic Biology Journal, 2021, 2(4): 598-611.
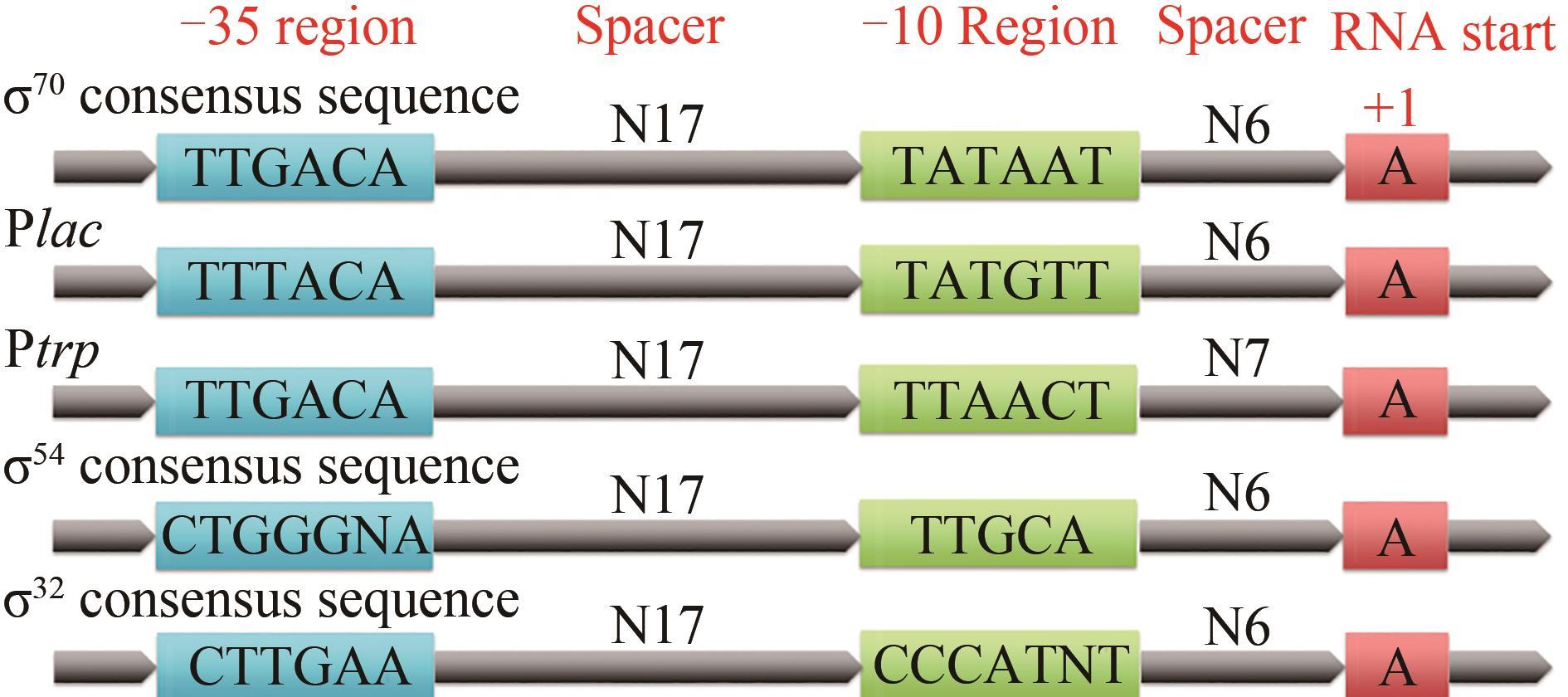
| σ 因子 | 微生物名称 | -35区 | -10区 | 参考 文献 |
|---|---|---|---|---|
| σ70 | 大肠杆菌 | TTGACA | TATAAT | [ |
| σ70 | 枯草芽孢杆菌 | TTGACA | TATAAT | [ |
| σA | 谷氨酸棒杆菌 | TTG(A/C)CA | TGN | [ |
| σA | 红色红球菌 | TTGNNN | (T/C)GN | [ |
| σA | 运动发酵单胞菌 | TTGNNN | TATNNN | [ |
| σ hrdB | 阿维链霉菌 | TTGACA | tAgATT | [ |
表1 几种模式/非模式微生物看家σ因子识别的启动子保守序列比较
Tab. 1 Some promoter consensus sequences for housekeeping σ factors of model/non-model microorganisms
| σ 因子 | 微生物名称 | -35区 | -10区 | 参考 文献 |
|---|---|---|---|---|
| σ70 | 大肠杆菌 | TTGACA | TATAAT | [ |
| σ70 | 枯草芽孢杆菌 | TTGACA | TATAAT | [ |
| σA | 谷氨酸棒杆菌 | TTG(A/C)CA | TGN | [ |
| σA | 红色红球菌 | TTGNNN | (T/C)GN | [ |
| σA | 运动发酵单胞菌 | TTGNNN | TATNNN | [ |
| σ hrdB | 阿维链霉菌 | TTGACA | tAgATT | [ |
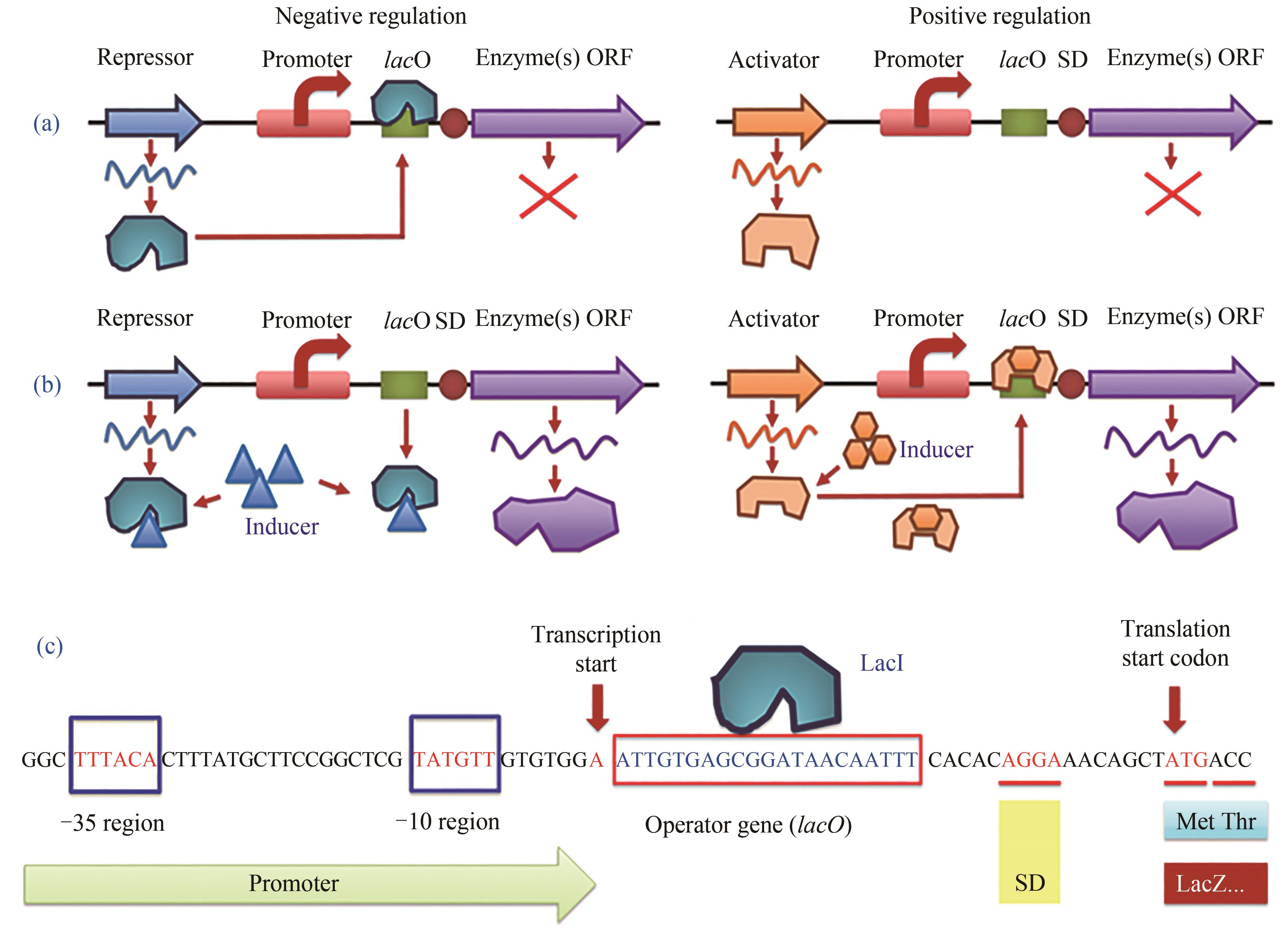
图2 诱导型启动子的负调控与正调控诱导机制以及大肠杆菌乳糖操纵子负调控的典型序列(a)无诱导剂条件下的基因调控;(b)诱导剂加入条件下的基因调控;(c) β-半乳糖苷酶(LacZ)启动子的乳糖操纵子负调控区域DNA序列。LacI—阻遏蛋白;lacO—操纵序列;SD序列—基因的核糖体结合位点(也称为RBS);Inducer—乳糖操纵子的lacI-lacO体系诱导剂,通常为异丙基-β-D-硫代吡喃半乳糖苷(IPTG)或乳糖(实际为1,6-别乳糖,乳糖的代谢物)
Fig. 2 Negative and positive regulation mechanism of inducible promoters and the partial sequence of lactose operon (lacO) of E. coli.(a, b) gene regulation mode under inducer-free or inducer-present condition, respectively. (c) partial DNA sequence of inducible promoter of β-galactosidase (LacZ) with negative regulation. LacI—inhibitor (repressor) protein; lacO—operator sequence for repressor binding; SD—Shine-Dalgarno sequence as ribosome biding site (RBS); Inducer—for lacI-lacO system, common inducer is isopropyl-β-D-thiogalactopyranoside (IPTG) or lactose (1,6-allolactose in fact, a metabolite of lactose), respectively
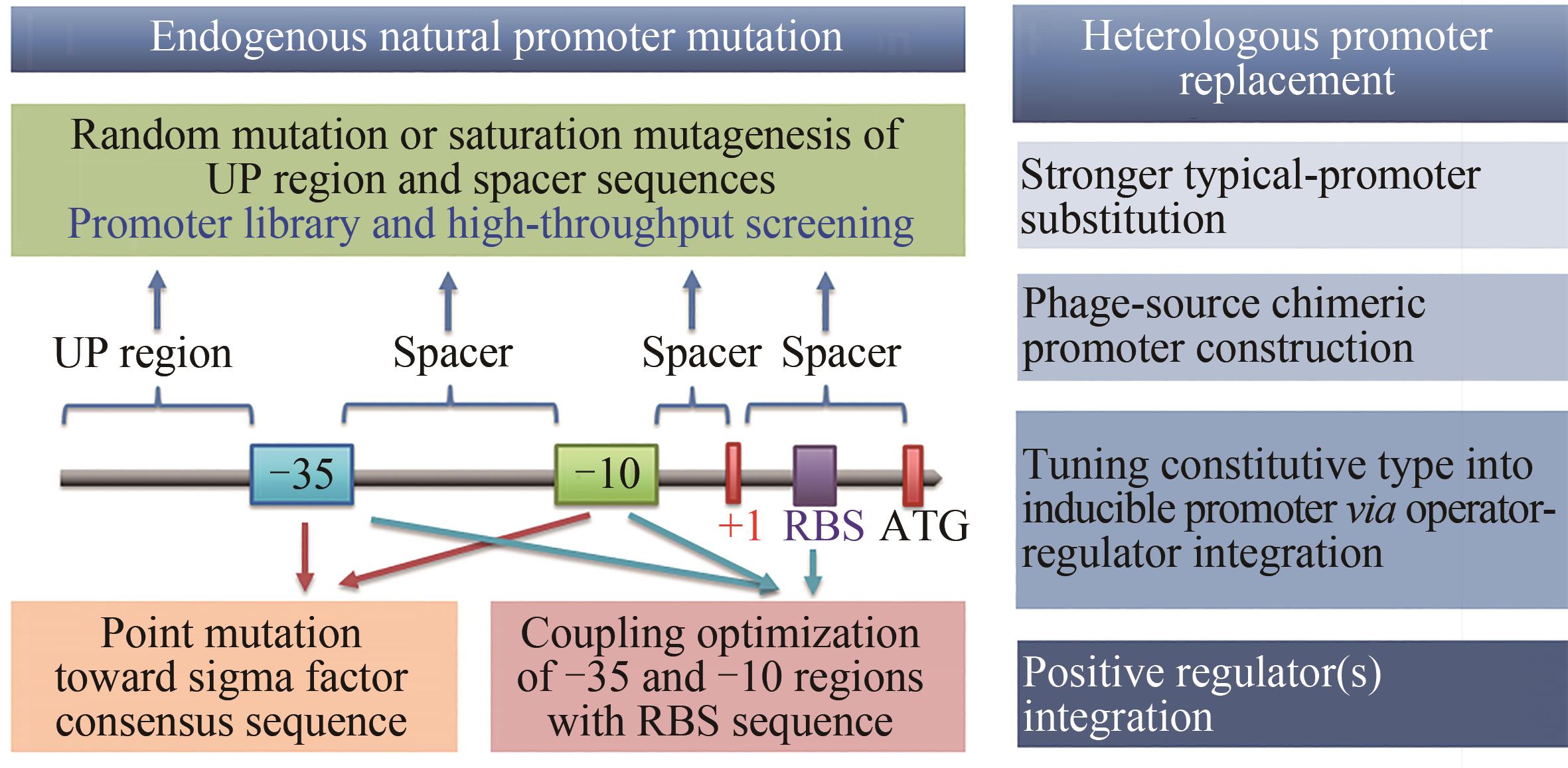
图3 启动子改造的常用有效策略-35和-10—启动子核心区;+1—转录起始位点;RBS—核糖体结合位点;ATG—翻译起始密码子
Fig. 3 Common efficient strategies for promoter engineering-35 and -10—core elements of promoter; +1— transcription initiation site; RBS— ribosome binding site; ATG— start codon of enzyme translation
| 诱导类型 | 诱导剂 | 相关微生物 | 启动子特征 | 参考 文献 |
|---|---|---|---|---|
| 化学信号 | IPTG/乳糖 | 大肠杆菌;谷氨酸棒杆菌;枯草芽孢杆菌等 | Plac、Ptac;广谱性好、诱导强度较高;诱导剂价格高;基于LacI阻遏蛋白(负调控蛋白)的负调控机制 | [ |
| IPTG | 大肠杆菌 | T7启动子,噬菌体来源;pET表达系统;常与溶源化宿主菌(即T7 RNA聚合酶基因插入在宿主菌基因组中)联用;诱导强度高、广谱性好;诱导剂价格高;基于阻遏蛋白的负调控机制 | [ | |
| L-鼠李糖/ L-阿拉伯糖 | 大肠杆菌;红球菌等 | rhaPBAD/araPBAD启动子;AraC正/负调控蛋白;pBAD表达系统;诱导剂价格较高;诱导强度较高;广谱性较好;其中rhaPBAD为基于正调控蛋白的正调控机制;araPBAD为基于阻遏蛋白的负调控机制 | [ | |
| 木糖 | 枯草芽孢杆菌;酵母菌等 | Pxyl;诱导剂价格高;基于阻遏蛋白的负调控机制 | [ | |
| 蔗糖 | 枯草芽孢杆菌等 | PsacB/PsacP;强度低,易泄露表达;基于不依赖于ρ因子的RNA抗终止子序列的负调控机制 | [ | |
| 四环素/脱水四环素 | 浑浊红球菌;丙酮丁醇梭菌等 | Ptet;诱导剂用量低;基于TetR阻遏蛋白的负调控机制 | [ | |
| 离子液体 | 木质素分解肠杆菌;大肠杆菌;酿酒酵母等 | eilAR表达盒;EilA表达离子液体自诱导内膜转运蛋白;EilR为负调控蛋白;负调控机制 | [ | |
| 尿酸 | 耐辐射奇球菌;大肠杆菌等 | 基于HucR阻遏蛋白的负调控机制;强度与pBAD/pET系统相当 | [ | |
| 巴豆酰胺/ 甲基丙烯酰胺 | 红球菌等 | 红球菌适用;基于正负调控蛋白协同作用的复合调控机制 | [ | |
| 联苯 | 红球菌RHA1;红串红球菌 | 基于BphS-BphT两组分调控元件的正调控机制 | [ | |
| ε-己内酰胺/十几种腈类 | 链霉菌;红球菌等 | PnitA-NitR杂合元件;强启动高表达;基于NitR正调控蛋白的正调控机制 | [ | |
| 尿素 | 红色红球菌 | Pnh,Pami,Pa2;基于正负调控蛋白协同作用的调控机制,具体调控机制尚未解析 | [ | |
| 铁离子 | 多能硫碱弧菌;大肠杆菌等 | P3AF启动子,携带19 bp大肠杆菌铁摄取调节子蛋白基因fur的核心区序列,铁离子严谨调控 | [ | |
| 碱性染料 | 大肠杆菌;恶臭假单胞菌;苜蓿中华根瘤菌;新月柄杆菌等 | 基于EilR阻遏蛋白-操纵序列的负调控机制;结晶紫等廉价、严谨、高效诱导;微生物广谱适用性;低成本 | [ | |
| 丙烯酸盐/葡萄糖酸/红霉素/柚皮素 | 大肠杆菌;类球红细菌;恶臭假单胞菌 | 属于TetR家族阻遏蛋白的AcuR/MphR/TtgR等调控蛋白;CdaR,转录激活蛋白;响应4种代谢物的生物传感器性能,分别为负调控和正调控机制;不同诱导信号的正交效果 | [ | |
| 甲醇 | 红串红球菌;毕赤酵母等 | 异柠檬酸裂解酶基因(iclRe)上游序列;基于RamB Re 阻遏蛋白的负调控机制 | [ | |
| 乙醇 | 运动发酵单胞菌等 | P0405、P0435和P0038启动子,调控机制尚未明确 | [ | |
| 对异丙苯甲酸酯/间苯二酚 | 链霉菌;放线菌 | 负调控机制;对异丙苯甲酸酯诱导,P21启动子-CymR调控蛋白;间苯二酚,PA3启动子-RolR调控蛋白 | [ | |
| 磷酸盐饥饿 | 大肠杆菌 | 磷酸盐饥饿诱导;基于PhoB激活的正调控机制 | [ | |
| 氮饥饿 | 三角褐指藻 | 铵转运蛋白AMT基因启动子;氮饥饿条件强诱导;机制尚未报道 | [ | |
| 低溶氧 | 大肠杆菌;谷氨酸棒杆菌;盐单胞菌等 | vgb启动子;nar启动子;响应低溶氧条件启动转录 | [ | |
| 低pH | 黑曲霉 | Pgas,低pH诱导(pH2.0/3.0);受未知功能双调控蛋白调控 | [ | |
| 物理信号 | 高温 | 大肠杆菌;枯草芽孢杆菌等 | pR/pL,受温敏型阻遏蛋白CI857调控;P2和P7等 | [ |
| 低温(-15 ℃) | 大肠杆菌 | PcspA耦合LacI负调控;杂合启动子,严谨低温调控,pCold载体效率与pET14相当 | [ | |
| 低温(-30 ℃) | 大肠杆菌 | 整合多个温敏转录开关与蛋白降解模块;从37 ℃到30 ℃严谨调控;复合调控机制 | [ | |
| 光 | 大肠杆菌;酵母菌等 | 10余种光敏调控蛋白Cph8/OmpR;YF1/FixJ;EL222等;白光诱导、蓝光诱导、黑暗诱导等 | [ |
表2 近年来报道的一些不同微生物来源、不同类型的诱导型启动子
Tab. 2 Some inducible promoters reported recently from different microorganisms with diverse mechanisms
| 诱导类型 | 诱导剂 | 相关微生物 | 启动子特征 | 参考 文献 |
|---|---|---|---|---|
| 化学信号 | IPTG/乳糖 | 大肠杆菌;谷氨酸棒杆菌;枯草芽孢杆菌等 | Plac、Ptac;广谱性好、诱导强度较高;诱导剂价格高;基于LacI阻遏蛋白(负调控蛋白)的负调控机制 | [ |
| IPTG | 大肠杆菌 | T7启动子,噬菌体来源;pET表达系统;常与溶源化宿主菌(即T7 RNA聚合酶基因插入在宿主菌基因组中)联用;诱导强度高、广谱性好;诱导剂价格高;基于阻遏蛋白的负调控机制 | [ | |
| L-鼠李糖/ L-阿拉伯糖 | 大肠杆菌;红球菌等 | rhaPBAD/araPBAD启动子;AraC正/负调控蛋白;pBAD表达系统;诱导剂价格较高;诱导强度较高;广谱性较好;其中rhaPBAD为基于正调控蛋白的正调控机制;araPBAD为基于阻遏蛋白的负调控机制 | [ | |
| 木糖 | 枯草芽孢杆菌;酵母菌等 | Pxyl;诱导剂价格高;基于阻遏蛋白的负调控机制 | [ | |
| 蔗糖 | 枯草芽孢杆菌等 | PsacB/PsacP;强度低,易泄露表达;基于不依赖于ρ因子的RNA抗终止子序列的负调控机制 | [ | |
| 四环素/脱水四环素 | 浑浊红球菌;丙酮丁醇梭菌等 | Ptet;诱导剂用量低;基于TetR阻遏蛋白的负调控机制 | [ | |
| 离子液体 | 木质素分解肠杆菌;大肠杆菌;酿酒酵母等 | eilAR表达盒;EilA表达离子液体自诱导内膜转运蛋白;EilR为负调控蛋白;负调控机制 | [ | |
| 尿酸 | 耐辐射奇球菌;大肠杆菌等 | 基于HucR阻遏蛋白的负调控机制;强度与pBAD/pET系统相当 | [ | |
| 巴豆酰胺/ 甲基丙烯酰胺 | 红球菌等 | 红球菌适用;基于正负调控蛋白协同作用的复合调控机制 | [ | |
| 联苯 | 红球菌RHA1;红串红球菌 | 基于BphS-BphT两组分调控元件的正调控机制 | [ | |
| ε-己内酰胺/十几种腈类 | 链霉菌;红球菌等 | PnitA-NitR杂合元件;强启动高表达;基于NitR正调控蛋白的正调控机制 | [ | |
| 尿素 | 红色红球菌 | Pnh,Pami,Pa2;基于正负调控蛋白协同作用的调控机制,具体调控机制尚未解析 | [ | |
| 铁离子 | 多能硫碱弧菌;大肠杆菌等 | P3AF启动子,携带19 bp大肠杆菌铁摄取调节子蛋白基因fur的核心区序列,铁离子严谨调控 | [ | |
| 碱性染料 | 大肠杆菌;恶臭假单胞菌;苜蓿中华根瘤菌;新月柄杆菌等 | 基于EilR阻遏蛋白-操纵序列的负调控机制;结晶紫等廉价、严谨、高效诱导;微生物广谱适用性;低成本 | [ | |
| 丙烯酸盐/葡萄糖酸/红霉素/柚皮素 | 大肠杆菌;类球红细菌;恶臭假单胞菌 | 属于TetR家族阻遏蛋白的AcuR/MphR/TtgR等调控蛋白;CdaR,转录激活蛋白;响应4种代谢物的生物传感器性能,分别为负调控和正调控机制;不同诱导信号的正交效果 | [ | |
| 甲醇 | 红串红球菌;毕赤酵母等 | 异柠檬酸裂解酶基因(iclRe)上游序列;基于RamB Re 阻遏蛋白的负调控机制 | [ | |
| 乙醇 | 运动发酵单胞菌等 | P0405、P0435和P0038启动子,调控机制尚未明确 | [ | |
| 对异丙苯甲酸酯/间苯二酚 | 链霉菌;放线菌 | 负调控机制;对异丙苯甲酸酯诱导,P21启动子-CymR调控蛋白;间苯二酚,PA3启动子-RolR调控蛋白 | [ | |
| 磷酸盐饥饿 | 大肠杆菌 | 磷酸盐饥饿诱导;基于PhoB激活的正调控机制 | [ | |
| 氮饥饿 | 三角褐指藻 | 铵转运蛋白AMT基因启动子;氮饥饿条件强诱导;机制尚未报道 | [ | |
| 低溶氧 | 大肠杆菌;谷氨酸棒杆菌;盐单胞菌等 | vgb启动子;nar启动子;响应低溶氧条件启动转录 | [ | |
| 低pH | 黑曲霉 | Pgas,低pH诱导(pH2.0/3.0);受未知功能双调控蛋白调控 | [ | |
| 物理信号 | 高温 | 大肠杆菌;枯草芽孢杆菌等 | pR/pL,受温敏型阻遏蛋白CI857调控;P2和P7等 | [ |
| 低温(-15 ℃) | 大肠杆菌 | PcspA耦合LacI负调控;杂合启动子,严谨低温调控,pCold载体效率与pET14相当 | [ | |
| 低温(-30 ℃) | 大肠杆菌 | 整合多个温敏转录开关与蛋白降解模块;从37 ℃到30 ℃严谨调控;复合调控机制 | [ | |
| 光 | 大肠杆菌;酵母菌等 | 10余种光敏调控蛋白Cph8/OmpR;YF1/FixJ;EL222等;白光诱导、蓝光诱导、黑暗诱导等 | [ |
| 87 | LIANG C N, ZHANG X X, WU J Y, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit [J]. Metabolic Engineering, 2020, 57: 239-246. |
| 88 | BREMPT M VAN, CLAUWAERT J, MEY F, et al. Predictive design of sigma factor-specific promoters [J]. Nature Communications, 2020, 11(1): 5822. |
| 89 | ZHAO M, ZHOU S H, WU L T, et al. Model-driven promoter strength prediction based on a fine-tuned synthetic promoter library in Escherichia coli [J]. bioRxiv, 2020, DOI:10.1101/2020.06.25.170365 . |
| 90 | KEASLING J D. Synthetic biology for synthetic chemistry [J]. ACS Chemical Biology, 2008, 3(1): 64-76. |
| 91 | 于慧敏,王勇. 全局转录机器工程——工业生物技术新方法[J]. 生物产业技术, 2009(4): 42-46. |
| YU H M, WANG Y. Global transcriptional machinery engineering: new method in industrial biotechnology [J]. Biotechnology & Business, 2009(4): 42-46. | |
| 92 | JIN L Q, JIN W R, MA Z C, et al. Promoter engineering strategies for the overproduction of valuable metabolites in microbes [J]. Applied Microbiology and Biotechnology, 2019, 103(21/22): 8725-8736. |
| 93 | HANNIG G, MAKRIDES S C. Strategies for optimizing heterologous protein expression in Escherichia coli [J]. Trends in Biotechnology, 1998, 16: 54-60. |
| 94 | KEASLING J D. Gene-expression tools for the metabolic engineering of bacteria [J]. Trends in Biotechnology, 1999, 17(11): 452-460. |
| 95 | HENRY K K, ROSS W, MYERS K S, et al. A majority of Rhodobacter sphaeroides promoters lack a crucial RNA polymerase recognition feature, enabling coordinated transcription activation [J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(47): 29658-29668. |
| 96 | TANG R Q, WAGNER J M, ALPER H S, et al. Design, evolution, and characterization of a xylose biosensor in Escherichia coli using the XylR/xylO system with an expanded operating range [J]. ACS Synthetic Biology, 2020, 9(10): 2714-2722. |
| 97 | LI N, ZENG W Z, XU S, et al. Obtaining a series of native gradient promoter-5′-UTR sequences in Corynebacterium glutamicum ATCC 13032 [J]. Microbial Cell Factories, 2020, 19(1): 120. |
| 98 | 陈江楠,陈潇宁,刘心怡,等. 基于工程化盐单胞菌的下一代工业生物技术[J]. 合成生物学, 2020, 1(5): 516-527. |
| 1 | CANTON B, LABNO A, ENDY D. Refinement and standardization of synthetic biological parts and devices [J]. Nature Biotechnology, 2008, 26(7): 787-793. |
| 2 | ARKIN A. Setting the standard in synthetic biology [J]. Nature Biotechnology, 2008, 26(7): 771-774. |
| 3 | BLAZECK J, ALPER H S. Promoter engineering: recent advances in controlling transcription at the most fundamental level [J]. Biotechnology Journal, 2013, 8(1): 46-58. |
| 4 | ROSS W, GOSINK K K, SALOMON J, et al. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase [J]. Science, 1993, 262(5138): 1407-1413. |
| 5 | ISHIHAMA A. Functional modulation of Escherichia coli RNA polymerase [J]. Annual Review of Microbiology, 2000, 54: 499-518. |
| 6 | NELSON D L, COX M M. Lehninger principles of biochemistry [M]. 7th ed. New York: W H Freeman and Company, 2017: 2711-2740. |
| 7 | WOSTEN M M. Eubacterial sigma-factors [J]. FEMS Microbiology Review, 1998, 22(3): 127-150. |
| 8 | PAGET M S, HELMANN J D. The Sigma70 family of sigma factors [J]. Genome Biology, 2003, 4(1): 203. |
| 9 | GRUBER T M, GROSS C A. Multiple sigma subunits and the partitioning of bacterial transcription space [J]. Annual Review of Microbiology, 2003, 57: 441-66. |
| 10 | BURGESS R R, ANTHONY L. How sigma docks to RNA polymerase and what sigma does [J]. Current Opinion in Microbiology, 2001, 4(2): 126-131. |
| 11 | YOUNG E, ALPER H. Synthetic biology: tools to design, build, and optimize cellular processes [J]. Journal of Biomedicine & Biotechnology, 2010, 2010: 130781. |
| 12 | YUKAWA H, INUI M. Corynebacterium glutamicum. Microbiology monographs [M]. Berlin Heidelberg: Springer, 2013, 23: 51-88. |
| 98 | CHEN J N, CHEN X N, LIU X Y, et al. Engineering Halomonas spp. for next generation industrial biotechnology (NGIB) [J]. Synthetic Biology Journal, 2020, 1(5): 516-527. |
| 99 | 武耀康,刘延峰,李江华, 等. 动态调控元件及其在微生物代谢工程中的应用[J]. 化工学报, 2018, 69(1): 272-281. |
| WU Y K, LIU Y F, LI J H, et al. Dynamic regulation elements and their applications in microbial metabolic engineering [J]. CIESC Journal, 2018, 69(1): 272-281. | |
| 100 | 钱秀娟, 陈琳, 章文明, 等. 人工多细胞体系设计与构建研究进展[J]. 合成生物学, 2020, 1(3): 267-284. |
| QIAN X J, CHEN L, ZHANG W M, et al. Recent research progress in the design and construction of synthetic microbial consortia [J]. Synthetic Biology Journal, 2020, 1(3): 267-284. | |
| 13 | ZHAO H, ZENG A P. Synthetic biology-metabolic engineering [M]. Berlin Cham: Springer, 2016, 162:21-44. |
| 14 | JIAO S, YU H M, SHEN Z Y. Core elements characterization of Rhodococcus promoters and development of a gradient intensity mini-pool for efficient gene expression [J]. New Biotechnology, 2018, 44: 41-49. |
| 15 | VERA J M, GHOSH I N, ZHANG Y P, et al. Genome-scale transcription-translation mapping reveals features of Zymomonas mobilis transcription units and promoters [J]. mSystems, 2020, 5(4): e00250-20. |
| 16 | ZHUO Y, ZHANG W Q, CHEN D F, et al. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(25): 11250-11254. |
| 17 | COX R S, SURETTE M G, ELOWITZ M B. Programming gene expression with combinatorial promoters [J]. Molecular System Biology, 2007, 3: 145. |
| 18 | BLAZECK J, ALPER H. Systems metabolic engineering: genome-scale models and beyond [J]. Biotechnology Journal, 2010, 5(7): 647-659. |
| 19 | WANG X Y, CHEN J, QUINN P. Reprogramming microbial metabolic pathways [M]. Dordrecht: Springer Science+Business Media, 2012: 181-224. |
| 20 | PHAN T T, NGUYEN H D, SCHUMANN W. Development of a strong intracellular expression system for Bacillus subtilis by optimizing promoter elements [J]. Journal of Biotechnology, 2012, 157(1): 167-172. |
| 21 | JIAO S, LI X, YU H M, et al. In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters [J]. Biotechnology and Bioengineering, 2017, 114: 832-842. |
| 22 | SONG Y F, NIKOLOFF J M, FU G, et al. Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications [J]. PLoS One, 2016, 11(7): e0158447. |
| 23 | SHANG X L, CHAI X, LU X M, et al. Native promoters of Corynebacterium glutamicum and its application in L-lysine production [J]. Biotechnology Letters, 2018, 40(2): 383-391. |
| 24 | CHOI Y J, MOREL L, LE FRANÇOIS T, et al. Novel, versatile, and tightly regulated expression system for Escherichia coli strains [J]. Applied and Environmental Microbiology, 2010, 76(15): 5058-5066. |
| 25 | LECOINTE F, COSTE G, SOMMER S, et al. Vectors for regulated gene expression in the radioresistant bacterium Deinococcus radiodurans [J]. Gene, 2004, 336(1): 25-35. |
| 26 | YANG M M, ZHANG W W, JI S Y, et al. Generation of an artificial double promoter for protein expression in Bacillus subtilis through a promoter trap system [J]. PLoS One, 2013, 8(2): e56321. |
| 27 | CASTILLO-HAIR S M, FUJITA M, IGOSHIN O A, et al. An engineered B. subtilis inducible promoter system with over 10 000-fold dynamic range [J]. ACS Synthetic Biology, 2019, 8: 1673-1678. |
| 28 | HERAI S, HASHIMOTO Y, HIGASHIBATA H, et al. Hyper-inducible expression system for streptomycetes [J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(39): 14031-14035. |
| 29 | GUIZIOU S, SAUVEPLANE V, CHANG H J, et al. A part toolbox to tune genetic expression in Bacillus subtilis [J]. Nucleic Acids Research, 2016, 44(15): 7495-7508. |
| 30 | ZHAO M, WANG S L, TAO X Y, et al. Engineering diverse eubacteria promoters for robust gene expression in Streptomyces lividans [J]. Journal of Biotechnology, 2019, 289: 93-102. |
| 31 | LI T T, LI T, JI W Y, et al. Engineering of core promoter regions enables the construction of constitutive and inducible promoters in Halomonas sp [J]. Biotechnology Journal, 2016, 11(2): 219-227. |
| 32 | SHEN R, YIN J, YE J W, et al. Promoter engineering for enhanced P(3HB-co-4HB) production by Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2018, 7(8):1897-1906. |
| 33 | JIN L Y, NAWAB S, XIA M L, et al. Context-dependency of synthetic minimal promoters in driving gene expression: a case study [J]. Microbial Biotechnology, 2019, 12(6): 1476-1486. |
| 34 | JI C H, KIM J P, KANG H S. Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters [J]. ACS Synthetic Biology, 2018, 7(8):1946-1955. |
| 35 | ZHOU S H, DING R P, CHEN J, et al. Obtaining a panel of cascade promoter-5′-UTR complexes in Escherichia coli [J]. ACS Synthetic Biology, 2017, 6(6): 1065-1075. |
| 36 | KÖBBING S, BLANK L M, WIERCKX N. Characterization of context-dependent effects on synthetic promoters [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 551. |
| 37 | GILMAN J, SINGLETON C, TENNANT R K, et al. Rapid, heuristic discovery and design of promoter collections in non-model microbes for industrial applications [J]. ACS Synthetic Biology, 2019, 8(5): 1175-1186. |
| 38 | YANG Y F, SHEN W, HUANG J, et al. Prediction and characterization of promoters and ribosomal binding sites of Zymomonas mobilis in system biology era [J]. Biotechnology for Biofuels, 2019, 12: 52. |
| 39 | WANG W Y, LI Y W B, WANG Y Q, et al. Bacteriophage T7 transcription system: an enabling tool in synthetic biology [J]. Biotechnology Advances, 2018, 36(8): 2129-2137. |
| 40 | LUTZ R, BUJARD H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements [J]. Nucleic Acids Research, 1997, 25(6): 1203-1210. |
| 41 | CHENG F Y, GONG Q Y, YU H M, et al. High-titer biosynthesis of hyaluronic acid by recombinant Corynebacterium glutamicum [J]. Biotechnology Journal, 2016, 11(4): 574-584. |
| 42 | CHENG F Y, YU H M, STEPHANOPOULOS G. Engineering Corynebacterium glutamicum for high-titer biosynthesis of hyaluronic acid [J]. Metabolic Engineering, 2019, 55: 276-289. |
| 43 | 贺根和, 刘强, 李晓红, 等. 大肠杆菌鼠李糖调节子[J]. 生命科学, 2008, 20(3): 477-482. |
| HE G H, LIU Q, LI X H, et al. Rhamnose regulon of Escherichia coli [J]. Chinese Bulletin of Life Sciences, 2008, 20(3): 477-482. | |
| 44 | MARSCHALL L, SAGMEISTER P, HERWIG C. Tunable recombinant protein expression in E. coli: promoter systems and genetic constraints [J]. Applied Microbiology and Biotechnology, 2017, 101(2): 501-512. |
| 45 | KIM L, MOGK A, SCHUMANN W. A xylose-inducible Bacillus subtilis integration vector and its application [J]. Gene, 1996, 181(1/2): 71-76. |
| 46 | TORTOSA P, DECLERCK N, DUTARTRE H, et al. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY[J]. Molecular Microbiology, 2001, 41(6): 1381-1393. |
| 47 | SKERRA A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli [J]. Gene, 1994, 151(1/2): 131-135. |
| 48 | DELORENZO D M, HENSON W R, MOON T S. Development of chemical and metabolite sensors for Rhodococcus opacus PD630 [J]. ACS Synthetic Biology, 2017, 6(10): 1973-1978. |
| 49 | DONG H J, TAO W W, ZHANG Y P, et al. Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: a useful tool for strain engineering [J]. Metabolic Engineering, 2012, 14(1): 59-67. |
| 50 | RUEGG T L, KIM E M, SIMMONS B A, et al. An auto-inducible mechanism for ionic liquid resistance in microbial biofuel production [J]. Nature Communications, 2014, 5: 3490. |
| 51 | LIANG C N, XIONG D D, ZHANG Y, et al. Development of a novel uric-acid-responsive regulatory system in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2015, 99(5): 2267-2275. |
| 52 | MIZUNASHI W, NISHIYAMA M, HORINOUCHI S, et al. Overexpression of high-molecular-mass nitrile hydratase from Rhodococcus rhodochrous J1 in recombinant Rhodococcus cells [J]. Applied Microbiology and Biotechnology, 1998, 49(5): 568-572. |
| 53 | TAKEDA H, HARA N, SAKAI Met al. Biphenyl-inducible promoters in a polychlorinated biphenyl-degrading bacterium, Rhodococcus sp. RHA1 [J]. Bioscience, Biotechnology and Biochemistry, 2004, 68(6): 1249-1258. |
| 54 | CHHIBA-GOVINDJEE V P, WESTHUYZEN C W VAN DER, BODE M L, et al. Bacterial nitrilases and their regulation [J]. Applied Microbiology and Biotechnology, 2019, 103(12): 4679-4692. |
| 55 | JIAO S, LI F L, YU H M, et al. Advances in acrylamide bioproduction catalyzed with Rhodococcus cells harboring nitrile hydratase [J]. Applied Microbiology and Biotechnology, 2020, 104(3): 1001-1012. |
| 56 | SHARSHARA M M, SAMAK N A, HAO X M, et al. Enhanced growth-driven stepwise inducible expression system development in haloalkaliphilic desulfurizing Thioalkalivibrio versutus [J]. Bioresource Technology, 2019, 288: 121486. |
| 57 | GUAN L Y, LIU Q, LI C, et al. Development of a Fur-dependent and tightly regulated expression system in Escherichia coli for toxic protein synthesis [J]. BMC Biotechnology, 2013, 13: 25-33. |
| 58 | RUEGG T L, PEREIRA J H, CHEN J C, et al. Jungle express is a versatile repressor system for tight transcriptional control [J]. Nature Communications, 2018, 9(1): 3617. |
| 59 | ROGERS J K, GUZMAN C D, TAYLOR N D, et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites [J]. Nucleic Acids Research, 2015, 43(15): 7648-7660. |
| 60 | KAGAWA Y, MITANI Y, YUN H Y, et al. Identification of a methanol-inducible promoter from Rhodococcus erythropolis PR4 and its use as an expression vector [J]. Journal of Bioscience and Bioengineering, 2012, 113(5): 596-603. |
| 61 | YANG Y F, RONG Z Y, SONG H Y, et al. Identification and characterization of ethanol-inducible promoters of Zymomonas mobilis based on omics data and dual reporter-gene system [J]. Biotechnology and Applied Biochemistry, 2020, 67(1):158-165. |
| 62 | HORBAL L, FEDORENKO V, LUZHETSKYY A. Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other actinobacteria [J]. Applied Microbiology and Biotechnology, 2014, 98(20): 8641-8655. |
| 63 | ULUŞEKER C, TORRES-BACETE J, GARCÍA J L, et al. Quantifying dynamic mechanisms of auto-regulation in Escherichia coli with synthetic promoter in response to varying external phosphate levels [J]. Scientific Reports, 2019, 9(1): 2076. |
| 64 | ADLER-AGNON Z, LEU S, ZARKA A, et al. Novel promoters for constitutive and inducible expression of transgenes in the diatom Phaeodactylum tricornutum under varied nitrate availability [J]. Journal of Applied Phycology, 2018, 30(5): 2763-2772. |
| 65 | YU H M, SHI Y, ZHANG Y P, et al. Effect of Vitreoscilla hemoglobin biosynthesis in Escherichia coli on production of poly(β-hydroxybutyrate) and fermentative parameters [J]. FEMS Microbiology Letters, 2002, 214(2): 223-227. |
| 66 | 周大袁, 林佳辉, 李霜. 利用溶氧调控型启动子Pvgb构建产surfactin的重组枯草芽孢杆菌[J]. 生物加工过程, 2020, 18(6): 690-695. |
| ZHOU D Y, LIN J H, LI S. Constructing recombinant Bacillus subtilis producing surfactin using aeration-inducible promoter Pvgb [J]. Chinese Journal of Bioprocess Engineering, 2020, 18(6): 690-695. | |
| 67 | HWANG H J, LEE S Y, LEE P C. Engineering and application of synthetic nar promoter for fine-tuning the expression of metabolic pathway genes in Escherichia coli [J]. Biotechnol Biofuels, 2018, 11: 103. |
| 68 | YIN X, SHIN H D, LI J, et al. Pgas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger [J]. Applied and Environmental Microbiology, 2017, 83(6): e03222-16. |
| 69 | HARDER B J, BETTENBROCK K, KLAMT S. Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli [J]. Biotechnology and Bioengineering, 2018, 115(1): 156-164. |
| 70 | LI W, LI H X, JI S Y, et al. Characterization of two temperature-inducible promoters newly isolated from B. subtilis [J]. Biochemical and Biophysical Research Communications, 2007, 358(4): 1148-1153. |
| 71 | QING G, MA L C, KHORCHID A, et al. Cold-shock induced high-yield protein production in Escherichia coli [J]. Nature Biotechnology, 2004, 22(7): 877-882. |
| 72 | ZHENG Y, MENG F K, ZHU Z H, et al. A tight cold-inducible switch built by coupling thermosensitive transcriptional and proteolytic regulatory parts [J]. Nucleic Acids Research, 2019, 47(21): e137. |
| 73 | LIU Z D, ZHANG J Z, JIN J, et al. Programming bacteria with light-sensors and applications in synthetic biology [J]. Frontiers in Microbiology, 2018, 9: 2692. |
| 74 | ZHAO E M, ZHANG Y, MEHL J, et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production [J]. Nature, 2018, 555: 683-687. |
| 75 | LALWANI M A, IP S S, CARRASCO-LÓPEZ C, et al. Optogenetic control of the lac operon for bacterial chemical and protein production [J]. Nature Chemical Biology, 2021, 17(1): 71-79. |
| 76 | GILMAN J, LOVE J. Synthetic promoter design for new microbial chassis [J]. Biochemical Society Transactions, 2016, 44(3): 731-737. |
| 77 | BARRICK D, VILLANUEBA K, CHILDS J, et al. Quantitative analysis of ribosome binding sites in E.coli [J]. Nucleic Acids Research, 1994, 22(7): 1287-1295. |
| 78 | TRISRIVIRAT D, HUGHES J M X, HOEVEN R, et al. Promoter engineering for microbial bio-alkane gas production [J]. Synthetic Biology, 2020, 5(1): ysaa022. |
| 79 | SUN H, YANG J L, SONG H. Engineering mycobacteria artificial promoters and ribosomal binding sites for enhanced sterol production [J]. Biochemical Engineering Journal, 2020, 162: 107739. |
| 80 | LIU R, LIU L Q, LI X, et al. Engineering yeast artificial core promoter with designated base motifs [J]. Microbial Cell Factories, 2020, 19(1): 38. |
| 81 | ZHANG S H, LIU D Y, MAO Z T, et al. Model-based reconstruction of synthetic promoter library in Corynebacterium glutamicum [J]. Biotechnology Letters, 2018, 40(5): 819-827. |
| 82 | WANG Y, WANG H C, WEI L, et al. Synthetic promoter design in Escherichia coli based on a deep generative network [J]. Nucleic Acids Research, 2020, 48(12): 6403-6412. |
| 83 | ÖZTÜRK S, ERGÜN B G, ÇALIK P. Double promoter expression systems for recombinant protein production by industrial microorganisms [J]. Applied Microbiology and Biotechnology, 2017, 101(20): 7459-7475. |
| 84 | LANDBERG J, MUNDHADA H, NIELSEN A T. An autoinducible trp-T7 expression system for production of proteins and biochemicals in Escherichia coli [J]. Biotechnology and Bioengineering, 2020, 117(5): 1513-1524. |
| 85 | MOREB E A, YE Z X, EFROMSON J P, et al. Media robustness and scalability of phosphate regulated promoters useful for two-stage autoinduction in E. coli [J]. ACS Synthetic Biology, 2020, 9(6): 1483-1486. |
| 86 | IKEGAYA R, SHINTANI M, KIMBARA K, et al. Identification of a transcriptional regulator for oligotrophy-responsive promoter in Rhodococcus erythropolis N9T-4 [J]. Bioscience, Biotechnology, and Biochemistry, 2020, 84(4): 865-868. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||