合成生物学 ›› 2025, Vol. 6 ›› Issue (1): 157-176.DOI: 10.12211/2096-8280.2024-066
DNA存储的关键技术:编码、纠错、随机访问与安全性
徐怀胜1, 石晓龙2, 刘晓光3, 徐苗苗4
- 1.广州商学院现代信息产业学院,广东 广州 511363
2.广州大学计算科技研究院,广东 广州 510006
3.广州体育学院运动健康学院,广东 广州 510620
4.广州中医药大学体育健康学院,广东 广州 510006
-
收稿日期:2024-08-26修回日期:2024-10-15出版日期:2025-02-28发布日期:2025-03-12 -
通讯作者:徐苗苗 -
作者简介:徐怀胜 (1996—),男,硕士,助教。研究方向为DNA信息存储。E-mail:hsxu@gcc.edu.cn徐苗苗 (1992—),女,师资博士后。研究方向为合成生物学及DNA存储。E-mail:miaomiaoxu@gzucm.edu.cn -
基金资助:国家自然科学基金(32300964)
Key technologies for DNA storage: encoding, error correction, random access, and security
XU Huaisheng1, SHI Xiaolong2, LIU Xiaoguang3, XU Miaomiao4
- 1.School of Modern Information Industry,Guangzhou College of Commerce,Guangzhou 511363,Guangdong,China
2.Institute of Computing Science and Technology,Guangzhou University,Guangzhou 510006,Guangdong,China
3.College of Sports and Health,Guangzhou Sport University,Guangzhou 510620,Guangdong,China
4.School of Physical Education and Health,Guangzhou University of Chinese Medicine,Guangzhou 510006,Guangdong,China
-
Received:2024-08-26Revised:2024-10-15Online:2025-02-28Published:2025-03-12 -
Contact:XU Miaomiao
摘要:
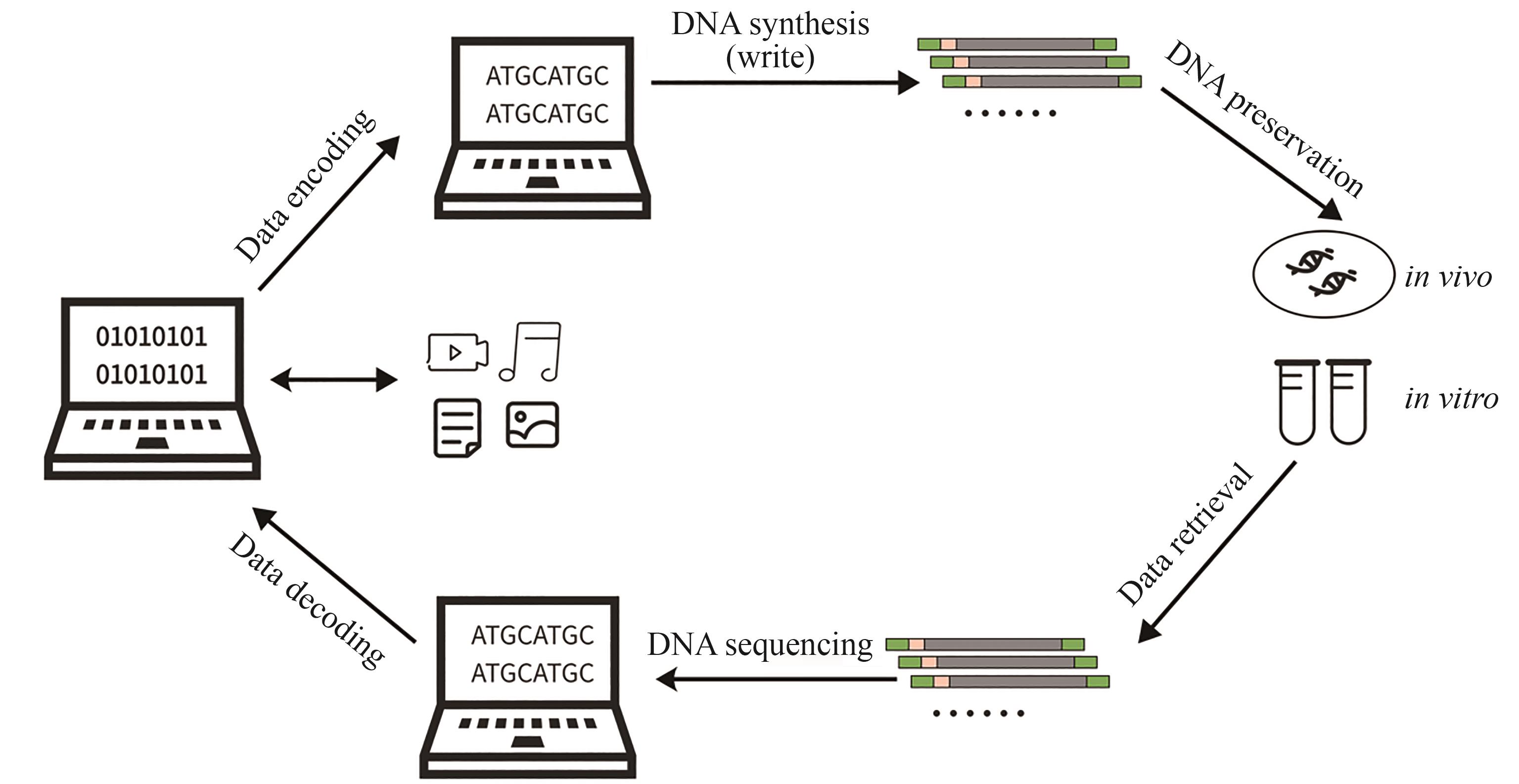
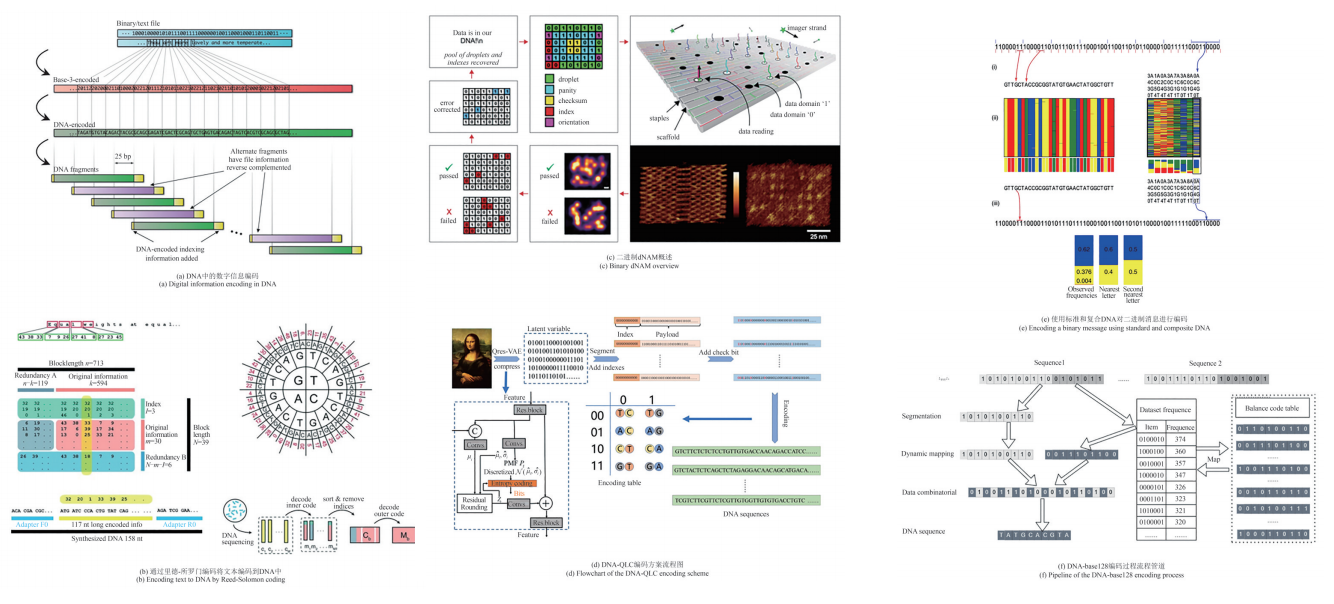
DNA信息存储是一种利用DNA分子作为数据载体的新型存储技术,通过合成特定序列的DNA来编码信息,并通过测序技术实现数据的读出。相比于传统的磁性、光学和电子存储介质,DNA存储在存储密度、数据保存时间和能源效率等方面具有显著优势,且不易受电磁干扰的影响。随着全球数据总量的猛增,DNA存储以其高效的存储能力、潜在的低维护成本和易于合成的化学特性,逐渐成为研究热点。本文首先介绍了DNA存储的基本流程,然后综述了DNA信息存储涉及的关键技术,尤其是编码策略、纠错技术、随机访问及DNA信息加密的研究进展。探讨了当前DNA存储技术的发展现状和主要挑战,如高成本、写入和读取速度慢等问题,并提出了可能的技术改进方向。并展望了DNA存储未来的发展前景,强调其在大数据时代的潜在应用和革命性影响,指出了实现商业化应用所需解决的关键技术瓶颈。
中图分类号:
引用本文
徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176.
XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security[J]. Synthetic Biology Journal, 2025, 6(1): 157-176.
| 作者 | 纠错码类型 | 码率 | 纠错类型 | 解码算法 | 参考文献 |
|---|---|---|---|---|---|
| Bornholt等 | XOR+重叠编码 | 约0.67 | 插入、删除和替换错误 | 聚类、对齐、多数投票 | [ |
| Sun等 | LDPC | 0.5,0.9 | 替换错误 | 非对称错误感知BP(ABP)解码算法 | [ |
| Antkowiak等 | RS纠错码 | 0.4 | 插入、缺失和替换错误 | 聚类、对齐、多数投票 | [ |
| Ding等 | 软判决译码方法+RS纠错码 | 0.83,0.92,0.945 | 插入、缺失和替换错误 | 软判决解码策略 | [ |
| Lu等 | LDPC+LLR | — | 插入、删除和替换错误 | LDPC码的和积算法 | [ |
| Fei等 | LDPC+类Turbo | 0.5 | 插入、删除和替换错误 | 基于LDPC的解码算法 | [ |
表1 DNA存储纠错技术研究对比
Table 1 Comparison of error correction technologies with DNA storage
| 作者 | 纠错码类型 | 码率 | 纠错类型 | 解码算法 | 参考文献 |
|---|---|---|---|---|---|
| Bornholt等 | XOR+重叠编码 | 约0.67 | 插入、删除和替换错误 | 聚类、对齐、多数投票 | [ |
| Sun等 | LDPC | 0.5,0.9 | 替换错误 | 非对称错误感知BP(ABP)解码算法 | [ |
| Antkowiak等 | RS纠错码 | 0.4 | 插入、缺失和替换错误 | 聚类、对齐、多数投票 | [ |
| Ding等 | 软判决译码方法+RS纠错码 | 0.83,0.92,0.945 | 插入、缺失和替换错误 | 软判决解码策略 | [ |
| Lu等 | LDPC+LLR | — | 插入、删除和替换错误 | LDPC码的和积算法 | [ |
| Fei等 | LDPC+类Turbo | 0.5 | 插入、删除和替换错误 | 基于LDPC的解码算法 | [ |
| 作者 | 数据大小 | 测序技术 | 覆盖率 | 随机访问 | 参考文献 |
|---|---|---|---|---|---|
| Organick等 | 200.2MB | Illumina/Nanopore | 5X/36X | ePCR | [ |
| Bögels等 | 25MB | Illumina | 30X | Thermoconfined PCR | [ |
| Hossein Tabatabaei Yazdi等 | 3KB | Nanopore | 200X | PCR | [ |
| Erlich等 | 2.15MB | Illumina | 250X | PCR | [ |
| Bornholt等 | 150KB | Illumina | 40X | PCR | [ |
| Lau等 | — | Nanopore | 30X | PCR | [ |
表2 使用PCR或变异PCR进行DNA存储随机访问的研究对比
Table 2 Comparison of studies using PCR or variant PCR for the random access of stored DNA
| 作者 | 数据大小 | 测序技术 | 覆盖率 | 随机访问 | 参考文献 |
|---|---|---|---|---|---|
| Organick等 | 200.2MB | Illumina/Nanopore | 5X/36X | ePCR | [ |
| Bögels等 | 25MB | Illumina | 30X | Thermoconfined PCR | [ |
| Hossein Tabatabaei Yazdi等 | 3KB | Nanopore | 200X | PCR | [ |
| Erlich等 | 2.15MB | Illumina | 250X | PCR | [ |
| Bornholt等 | 150KB | Illumina | 40X | PCR | [ |
| Lau等 | — | Nanopore | 30X | PCR | [ |
| 1 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. A DNA-based archival storage system[C/OL]//Proceedings of the Twenty-First International Conference on Architectural Support for Programming Languages and Operating Systems. New York, NY, USA: Association for Computing Machinery, 2016: 637-649. (2016-03-25)[2024-06-25]. . |
| 2 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337(6102): 1628. |
| 3 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes[J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555. |
| 4 | GOLDMAN N, BERTONE P, CHEN S Y, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494(7435): 77-80. |
| 5 | KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications[J]. Nature Methods, 2014, 11(5): 499-507. |
| 6 | LEE H H, KALHOR R, GOELA N, et al. Terminator-free template-independent enzymatic DNA synthesis for digital information storage[J]. Nature Communications, 2019, 10(1): 2383. |
| 7 | PALLUK S, ARLOW D H, DE ROND T, et al. De novo DNA synthesis using polymerase-nucleotide conjugates[J]. Nature Biotechnology, 2018, 36(7): 645-650. |
| 8 | ZHIRNOV V, ZADEGAN R M, SANDHU G S, et al. Nucleic acid memory[J]. Nature Materials, 2016, 15: 366-370. |
| 9 | HEINIS T, SOKOLOVSKII R, ALNASIR J J. Survey of information encoding techniques for DNA[J]. ACM Computing Surveys, 2024, 56(4): 1-30. |
| 10 | YU M, TANG X H, LI Z H, et al. High-throughput DNA synthesis for data storage[J]. Chemical Society Reviews, 2024, 53(9): 4463-4489. |
| 11 | TAN X, GE L Q, ZHANG T Z, et al. Preservation of DNA for data storage[J]. Russian Chemical Reviews, 2021, 90(2): 280-291. |
| 12 | LEE H, WIEGAND D J, GRISWOLD K, et al. Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage[J]. Nature Communications, 2020, 11(1): 5246. |
| 13 | KUBISTA M, ANDRADE J M, BENGTSSON M, et al. The real-time polymerase chain reaction[J]. Molecular Aspects of Medicine, 2006, 27(2-3): 95-125. |
| 14 | SHENDURE J, BALASUBRAMANIAN S, CHURCH G M, et al. DNA sequencing at 40: past, present and future[J]. Nature, 2017, 550(7676): 345-353. |
| 15 | MEISER L C, NGUYEN B H, CHEN Y J, et al. Synthetic DNA applications in information technology[J]. Nature Communications, 2022, 13(1): 352. |
| 16 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20(8): 456-466. |
| 17 | WELZEL M, SCHWARZ P M, LÖCHEL H F, et al. DNA-Aeon provides flexible arithmetic coding for constraint adherence and error correction in DNA storage[J]. Nature Communications, 2023, 14(1): 628. |
| 18 | LÖCHEL H F, WELZEL M, HATTAB G, et al. Fractal construction of constrained code words for DNA storage systems[J]. Nucleic Acids Research, 2022, 50(5): e30. |
| 19 | GAO X, LEPROUST E, ZHANG H, et al. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids[J]. Nucleic Acids Research, 2001, 29(22): 4744-4750. |
| 20 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36: 242-248. |
| 21 | BÖGELS B W A, NGUYEN B H, WARD D, et al. DNA storage in thermoresponsive microcapsules for repeated random multiplexed data access[J]. Nature Nanotechnology, 2023, 18(8): 912-921. |
| 22 | HAO M, QIAO H Y, GAO Y M, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale[J]. Communications Biology, 2020, 3(1): 416. |
| 23 | CHEN W G, HAN M Z, ZHOU J T, et al. An artificial chromosome for data storage[J]. National Science Review, 2021, 8(5): nwab028. |
| 24 | SUN F J, DONG Y M, NI M, et al. Mobile and self-sustained data storage in an extremophile genomic DNA[J]. Advanced Science, 2023, 10(10): e2206201. |
| 25 | DONG Y M, SUN F J, PING Z, et al. DNA storage: research landscape and future prospects[J]. National Science Review, 2020, 7(6): 1092-1107. |
| 26 | HOSSEIN TABATABAEI YAZDI S M, GABRYS R, MILENKOVIC O. Portable and error-free DNA-based data storage[J]. Scientific Reports, 2017, 7(1): 5011. |
| 27 | EL-SHAIKH A, WELZEL M, HEIDER D, et al. High-scale random access on DNA storage systems[J]. NAR Genomics and Bioinformatics, 2022, 4(1): lqab126. |
| 28 | CHOI Y, BAE H J, LEE A C, et al. DNA micro-disks for the management of DNA-based data storage with index and write-once-read-many (WORM) memory features[J]. Advanced Materials, 2020, 32(37): e2001249. |
| 29 | NEWMAN S, STEPHENSON A P, WILLSEY M, et al. High density DNA data storage library via dehydration with digital microfluidic retrieval[J]. Nature Communications, 2019, 10(1): 1706. |
| 30 | BANAL J L, SHEPHERD T R, BERLEANT J, et al. Random access DNA memory using Boolean search in an archival file storage system[J]. Nature Materials, 2021, 20(9): 1272-1280. |
| 31 | XU C T, MA B, GAO Z L, et al. Electrochemical DNA synthesis and sequencing on a single electrode with scalability for integrated data storage[J]. Science Advances, 2021, 7(46): eabk0100. |
| 32 | BENTLEY D R, BALASUBRAMANIAN S, SWERDLOW H P, et al. Accurate whole human genome sequencing using reversible terminator chemistry[J]. Nature, 2008, 456(7218): 53-59. |
| 33 | ROSS M G, RUSS C, COSTELLO M, et al. Characterizing and measuring bias in sequence data[J]. Genome Biology, 2013, 14(5): R51. |
| 34 | SCHWARTZ J J, LEE C, SHENDURE J. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules[J]. Nature Methods, 2012, 9(9): 913-915. |
| 35 | SHANNON C E. A mathematical theory of communication[J]. The Bell System Technical Journal, 1948, 27(3): 379-423. |
| 36 | REN Y B, ZHANG Y, LIU Y W, et al. DNA-based concatenated encoding system for high-reliability and high-density data storage[J]. Small Methods, 2022, 6(4): e2101335. |
| 37 | ERLICH Y, ZIELINSKI D. DNA fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355(6328): 950-954. |
| 38 | ANAVY L, VAKNIN I, ATAR O, et al. Data storage in DNA with fewer synthesis cycles using composite DNA letters[J]. Nature Biotechnology, 2019, 37(10): 1229-1236. |
| 39 | WANG K, CAO B, MA T, et al. Storing images in DNA via base128 encoding[J]. Journal of Chemical Information and Modeling, 2024, 64(5): 1719-1729. |
| 40 | DICKINSON G D, MORTUZA G M, CLAY W, et al. An alternative approach to nucleic acid memory[J]. Nature Communications, 2021, 12(1): 2371. |
| 41 | ZHENG Y F, CAO B, ZHANG X K, et al. DNA-QLC: an efficient and reliable image encoding scheme for DNA storage[J]. BMC Genomics, 2024, 25(1): 266. |
| 42 | ZHENG Y F, CAO B, WU J Q, et al. High net information density DNA data storage by the MOPE encoding algorithm[J]. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2023, 20(5): 2992-3000. |
| 43 | CHU H, WANG C, ZHANG Y. Improved constructions of secondary structure avoidance codes for DNA sequences[EB/OL]. arXiv, 2023: 2304.11403v1. (2023-04-22)[2024-06-25]. . |
| 44 | PARK S J, LEE Y W, NO J S. Iterative coding scheme satisfying GC balance and run-length constraints for DNA storage with robustness to error propagation[J]. Journal of Communications and Networks, 2022, 24(3): 283-291. |
| 45 | PING Z, CHEN S H, ZHOU G Y, et al. Towards practical and robust DNA-based data archiving using the Yin-Yang codec system[J]. Nature Computational Science, 2022, 2(4): 234-242. |
| 46 | LU X Z, KIM S H. Weakly mutually uncorrelated codes with maximum run length constraint for DNA storage[J]. Computers in Biology and Medicine, 2023, 165: 107439. |
| 47 | WANG Y X, NOOR-A-RAHIM M, ZHANG J Y, et al. High capacity DNA data storage with variable-length oligonucleotides using repeat accumulate code and hybrid mapping[J]. Journal of Biological Engineering, 2019, 13: 89. |
| 48 | WANG Y X, NOOR-A-RAHIM M D, GUNAWAN E, et al. Construction of bio-constrained code for DNA data storage[J]. IEEE Communications Letters, 2019, 23(6): 963-966. |
| 49 | XAVIER KOHLL A, ANTKOWIAK P L, CHEN W D, et al. Stabilizing synthetic DNA for long-term data storage with earth alkaline salts[J]. Chemical Communications, 2020, 56(25): 3613-3616. |
| 50 | HECKEL R, MIKUTIS G, GRASS R N. A characterization of the DNA data storage channel[J]. Scientific Reports, 2019, 9(1): 9663. |
| 51 | SUN Y, HAN G J, LIU C, et al. An asymmetric-error-aware LDPC decoding algorithm for DNA storage[J]. IEEE Communications Letters, 2023, 27(1): 32-36. |
| 52 | ANTKOWIAK P L, LIETARD J, DARESTANI M Z, et al. Low cost DNA data storage using photolithographic synthesis and advanced information reconstruction and error correction[J]. Nature Communications, 2020, 11(1): 5345. |
| 53 | DING L L, WU S G, HOU Z H, et al. Improving error-correcting capability in DNA digital storage via soft-decision decoding[J]. National Science Review, 2024, 11(2): nwad229. |
| 54 | LU X Z, JEONG J, KIM J W, et al. Error rate-based log-likelihood ratio processing for low-density parity-check codes in DNA storage[J]. IEEE Access, 2020, 8: 162892-162902. |
| 55 | FEI P, WANG Z Y. LDPC codes for portable DNA storage[C/OL]//2019 IEEE International Symposium on Information Theory (ISIT). July 7-12, 2019, Paris, France. IEEE, 2019: 76-80. (2019-09-26)[2024-06-25]. . |
| 56 | SUN J F, PHILPOTT M, LOI D, et al. Correcting PCR amplification errors in unique molecular identifiers to generate accurate numbers of sequencing molecules[J]. Nature Methods, 2024, 21(3): 401-405. |
| 57 | CHEN K K, ZHU J B, BOŠKOVIĆ F, et al. Nanopore-based DNA hard drives for rewritable and secure data storage[J]. Nano Letters, 2020, 20(5): 3754-3760. |
| 58 | MEISER L C, ANTKOWIAK P L, KOCH J, et al. Reading and writing digital data in DNA[J]. Nature Protocols, 2020, 15(1): 86-101. |
| 59 | CHANDAK S, TATWAWADI K, LAU B, et al. Improved read/write cost tradeoff in DNA-based data storage using LDPC codes[C/OL]//2019 57th Annual Allerton Conference on Communication, Control, and Computing (Allerton). September 24-27, 2019, Monticello, IL, USA. IEEE, 2019: 147-156. (2019-12-05)[2024-06-25]. . |
| 60 | CAO B, ZHANG X K, CUI S, et al. Adaptive coding for DNA storage with high storage density and low coverage[J]. NPJ Systems Biology and Applications, 2022, 8(1): 23. |
| 61 | ZHANG S F, WU J J, HUANG B B, et al. High-density information storage and random access scheme using synthetic DNA[J]. 3 Biotech, 2021, 11(7): 328. |
| 62 | RANU N, VILLANI A C, HACOHEN N, et al. Targeting individual cells by barcode in pooled sequence libraries[J]. Nucleic Acids Research, 2019, 47(1): e4. |
| 63 | MATANGE K, TUCK J M, KEUNG A J. DNA stability: a central design consideration for DNA data storage systems[J]. Nature Communications, 2021, 12(1): 1358. |
| 64 | BORNHOLT J, LOPEZ R, CARMEAN D M, et al. Toward a DNA-based archival storage system[J]. IEEE Micro, 2017, 37(3): 98-104. |
| 65 | LAU B, CHANDAK S, ROY S, et al. Magnetic DNA random access memory with nanopore readouts and exponentially-scaled combinatorial addressing[J]. Scientific Reports, 2023, 13(1): 8514. |
| 66 | HOSSEIN TABATABAEI YAZDI S M, YUAN Y B, MA J, et al. A rewritable, random-access DNA-based storage system[J]. Scientific Reports, 2015, 5: 14138. |
| 67 | LIMBACHIYA D, GUPTA M K, AGGARWAL V. Family of constrained codes for archival DNA data storage[J]. IEEE Communications Letters, 2018, 22(10): 1972-1975. |
| 68 | CAO B, II X, ZHANG X K, et al. Designing uncorrelated address constrain for DNA storage by DMVO algorithm[J]. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022, 19(2): 866-877. |
| 69 | WENGER A M, PELUSO P, ROWELL W J, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome[J]. Nature Biotechnology, 2019, 37(10): 1155-1162. |
| 70 | YIN Q, ZHENG Y F, WANG B, et al. Design of constraint coding sets for archive DNA storage[J]. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022, 19(6): 3384-3394. |
| 71 | YIN Q, CAO B, LI X, et al. An intelligent optimization algorithm for constructing a DNA storage code: NOL-HHO[J]. International Journal of Molecular Sciences, 2020, 21(6): 2191. |
| 72 | CAO B, ZHANG X K, WU J Q, et al. Minimum free energy coding for DNA storage[J]. IEEE Transactions on Nanobioscience, 2021, 20(2): 212-222. |
| 73 | RASOOL A, QU Q, JIANG Q S, et al. A strategy-based optimization algorithm to design codes for DNA data storage system[M/OL]// LAI Y X, WANG T, JIANG M, et al. Algorithms and architectures for parallel processing: lecture notes in computer science. Cham: Springer International Publishing, 2022: 284-299. (2022-02-23)[2024-06-28]. . |
| 74 | RASOOL A, QU Q, WANG Y, et al. Bio-constrained codes with neural network for density-based DNA data storage[J]. Mathematics, 2022, 10(5): 845. |
| 75 | CAO B, WANG B, ZHANG Q. GCNSA: DNA storage encoding with a graph convolutional network and self-attention[J]. iScience, 2023, 26(3): 106231. |
| 76 | WU J Q, ZHENG Y F, WANG B, et al. Enhancing physical and thermodynamic properties of DNA storage sets with end-constraint[J]. IEEE Transactions on Nanobioscience, 2022, 21(2): 184-193. |
| 77 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria[J]. Nature, 2017, 547(7663): 345-349. |
| 78 | ZHANG J Y, HOU C Y, LIU C C. CRISPR-powered quantitative keyword search engine in DNA data storage[J]. Nature Communications, 2024, 15(1): 2376. |
| 79 | BENCUROVA E, AKASH A, DOBSON R C J, et al. DNA storage-from natural biology to synthetic biology[J]. Computational and Structural Biotechnology Journal, 2023, 21: 1227-1235. |
| 80 | FAN C Y, DENG Q, ZHU T F. Bioorthogonal information storage in L-DNA with a high-fidelity mirror-image Pfu DNA polymerase[J]. Nature Biotechnology, 2021, 39(12): 1548-1555. |
| 81 | ZHANG J L, CHEN M Y, LIN G H, et al. Advanced DNA amplification for efficient data storage[J]. ACS Applied Materials & Interfaces, 2024, 16(37): 48870-48879. |
| 82 | ANDERSON R, MOORE T. The economics of information security[J]. Science, 2006, 314(5799): 610-613. |
| 83 | BRUNET T D P. Aims and methods of biosteganography[J]. Journal of Biotechnology, 2016, 226: 56-64. |
| 84 | DE SILVA P Y, GANEGODA G U. New trends of digital data storage in DNA[J]. BioMed Research International, 2016, 2016: 8072463. |
| 85 | CUI M Y, ZHANG Y X. Advancing DNA steganography with incorporation of randomness[J]. Chembiochem, 2020, 21(17): 2503-2511. |
| 86 | CLELLAND C T, RISCA V, BANCROFT C. Hiding messages in DNA microdots[J]. Nature, 1999, 399(6736): 533-534. |
| 87 | GEHANI A, LABEAN T, REIF J. DNA-based cryptography[M/OL]//JONOSKA N, PĂUN G, ROZENBERG G. Aspects of molecular computing: essays dedicated to Tom Head, on the Occasion of His 70th Birthday. Berlin, Heidelberg: Springer, 2003, 2950: 167-188 [2024-06-25]. . |
| 88 | LAI X J, LU M X, QIN L, et al. Asymmetric encryption and signature method with DNA technology[J]. Science China Information Sciences, 2010, 53(3): 506-514. |
| 89 | LU M X, LAI X J, XIAO G Z, et al. Symmetric-key cryptosystem with DNA technology[J]. Science in China Series F: Information Sciences, 2007, 50(3): 324-333. |
| 90 | GRASS R N, HECKEL R, DESSIMOZ C, et al. Genomic encryption of digital data stored in synthetic DNA[J]. Angewandte Chemie International Edition, 2020, 59(22): 8476-8480. |
| 91 | YAO X Y, XIE R Z, ZAN X Z, et al. A novel image encryption scheme for DNA storage systems based on DNA hybridization and gene mutation[J]. Interdisciplinary Sciences, Computational Life Sciences, 2023, 15(3): 419-432. |
| 92 | MARRAS A E, ZHOU L F, SU H J, et al. Programmable motion of DNA origami mechanisms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(3): 713-718. |
| 93 | ROTHEMUND P W K. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440(7082): 297-302. |
| 94 | VENEZIANO R, RATANALERT S, ZHANG K M, et al. Designer nanoscale DNA assemblies programmed from the top down[J]. Science, 2016, 352(6293): 1534. |
| 95 | BENSON E, MOHAMMED A, GARDELL J, et al. DNA rendering of polyhedral meshes at the nanoscale[J]. Nature, 2015, 523(7561): 441-444. |
| 96 | DOUGLAS S M, DIETZ H, LIEDL T, et al. Self-assembly of DNA into nanoscale three-dimensional shapes[J]. Nature, 2009, 459(7245): 414-418. |
| 97 | ZHANG Y N, CHAO J, LIU H J, et al. Transfer of two-dimensional oligonucleotide patterns onto stereocontrolled plasmonic nanostructures through DNA-origami-based nanoimprinting lithography[J]. Angewandte Chemie International Edition, 2016, 55(28): 8036-8040. |
| 98 | WANG D F, VIETZ C, SCHRÖDER T, et al. A DNA walker as a fluorescence signal amplifier[J]. Nano Letters, 2017, 17(9): 5368-5374. |
| 99 | KNUDSEN J B, LIU L, BANK KODAL A L, et al. Routing of individual polymers in designed patterns[J]. Nature Nanotechnology, 2015, 10(10): 892-898. |
| 100 | LANGECKER M, ARNAUT V, MARTIN T G, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures[J]. Science, 2012, 338(6109): 932-936. |
| 101 | VOIGT N V, TØRRING T, ROTARU A, et al. Single-molecule chemical reactions on DNA origami[J]. Nature Nanotechnology, 2010, 5(3): 200-203. |
| 102 | YANG J, MA J J, LIU S, et al. A molecular cryptography model based on structures of DNA self-assembly[J]. Chinese Science Bulletin, 2014, 59(11): 1192-1198. |
| 103 | ZHANG Y N, WANG F, CHAO J, et al. DNA origami cryptography for secure communication[J]. Nature Communications, 2019, 10(1): 5469. |
| 104 | FAN S S, WANG D F, CHENG J, et al. Information coding in a reconfigurable DNA origami domino array[J]. Angewandte Chemie, 2020, 132(31): 13091-13097. |
| 105 | ZHU J B, ERMANN N, CHEN K K, et al. Image encoding using multi-level DNA barcodes with nanopore readout[J]. Small, 2021, 17(28): e2100711. |
| 106 | SIDDARAMAPPA V, RAMESH K B. DNA-based XOR operation (DNAX) for data security using DNA as a storage medium[M/OL]//KRISHNA A, SRIKANTAIAH K, NAVEENA C. Integrated intelligent computing, communication and security: studies in computational intelligence. Singapore: Springer Singapore, 2019, 771: 343-351. (2028-09-15)[2024-03-01]. . |
| 107 | ZHU E Q, LUO X H, LIU C J, et al. An operational DNA strand displacement encryption approach[J]. Nanomaterials, 2022, 12(5): 877. |
| 108 | TENG Y, YANG S, LIU L Y, et al. Nanoscale storage encryption: data storage in synthetic DNA using a cryptosystem with a neural network[J]. Science China Life Sciences, 2022, 65(8): 1673-1676. |
| 109 | FONTANA R E JR, DECAD G M. Moore’s law realities for recording systems and memory storage components: HDD, tape, NAND, and optical[J]. AIP Advances, 2017, 8(5): 056506. |
| 110 | HUGHES R A, ELLINGTON A D. Synthetic DNA synthesis and assembly: putting the synthetic in synthetic biology[J]. Cold Spring Harbor Perspectives in Biology, 2017, 9(1): a023812. |
| 111 | BLAWAT M, GAEDKE K, HÜTTER I, et al. Forward error correction for DNA data storage[J]. Procedia Computer Science, 2016, 80: 1011-1022. |
| 112 | ZHOU Y, BI K, GE Q Y, et al. Advances and challenges in random access techniques for in vitro DNA data storage[J]. ACS Applied Materials & Interfaces, 2024, 16(33): 43102-43113. |
| 113 | LUO Y, CAO Z, LIU Y F, et al. The emerging landscape of microfluidic applications in DNA data storage[J]. Lab on a Chip, 2023, 23(8): 1981-2004. |
| 114 | LI X Y, CHEN M X, WU H M. Multiple errors correction for position-limited DNA sequences with GC balance and no homopolymer for DNA-based data storage[J]. Briefings in Bioinformatics, 2023, 24(1): bbac484. |
| 115 | PAN W J, BYRNE-STEELE M, WANG C L, et al. DNA polymerase preference determines PCR priming efficiency[J]. BMC Biotechnology, 2014, 14: 10. |
| 116 | AKASH A, BENCUROVA E, DANDEKAR T. How to make DNA data storage more applicable[J]. Trends in Biotechnology, 2024, 42(1): 17-30. |
| 117 | YANG S, BÖGELS B W A, WANG F, et al. DNA as a universal chemical substrate for computing and data storage[J]. Nature Reviews Chemistry, 2024, 8(3): 179-194. |
| 118 | ZHANG D Y, SEELIG G. Dynamic DNA nanotechnology using strand-displacement reactions[J]. Nature Chemistry, 2011, 3(2): 103-113. |
| 119 | CHERRY K M, QIAN L L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks[J]. Nature, 2018, 559(7714): 370-376. |
| 120 | CHEN C Z, WEN J D, WEN Z B, et al. DNA strand displacement based computational systems and their applications[J]. Frontiers in Genetics, 2023, 14: 1120791. |
| 121 | ZHANG Y Y, LI F, LI M, et al. Encoding carbon nanotubes with tubular nucleic acids for information storage[J]. Journal of the American Chemical Society, 2019, 141(44): 17861-17866. |
| 122 | COUDY D, COLOTTE M, LUIS A, et al. Long term conservation of DNA at ambient temperature. Implications for DNA data storage[J]. PLoS One, 2021, 16(11): e0259868. |
| 123 | BEE C, CHEN Y J, QUEEN M, et al. Molecular-level similarity search brings computing to DNA data storage[J]. Nature Communications, 2021, 12(1): 4764. |
| 124 | QIN Y, ZHU F, XI B, et al. Robust multi-read reconstruction from noisy clusters using deep neural network for DNA storage[J]. Computational and Structural Biotechnology Journal, 2024, 23: 1076-1087. |
| 125 | WANG S, MAO X, WANG F, et al. Data storage using DNA[J]. Advanced Materials, 2024, 36(6): e2307499. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [6] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [7] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [8] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [9] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [10] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [11] | 张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||