合成生物学 ›› 2025, Vol. 6 ›› Issue (1): 45-64.DOI: 10.12211/2096-8280.2023-096
合成基因线路的工程化设计研究进展与展望
高歌1,2, 边旗1,2, 王宝俊1,2
- 1.浙江大学化学工程与生物工程学院,浙江 杭州 310058
2.浙江大学杭州国际科创中心,浙江 杭州 311200
-
收稿日期:2023-12-01修回日期:2024-04-10出版日期:2025-01-31发布日期:2025-03-12 -
通讯作者:王宝俊 -
作者简介:高歌 (1994—),女,博士,助理研究员。研究方向为合成生物学基因线路设计、肿瘤细菌疗法。 E-mail:gaoge@zju.edu.cn边旗 (1993—),女,博士,助理研究员。研究方向为合成生物学使能技术开发、代谢工程。 E-mail:bianqi@zju.edu.cn王宝俊 (1982—),男,浙江大学求是讲席教授,教育部长江学者讲席教授。研究方向为合成生物学和生物工程,长期从事合成生物使能技术、基因线路设计研究及其在生物传感、智能治疗和生物制造等领域的创新应用。 E-mail:baojun.wang@zju.edu.cn -
基金资助:国家重点研发计划“生物与信息融合(BT与IT融合)”重点专项(2023YFF1204500);浙江省“尖兵”“领雁”研发攻关计划项目(2024C03011);国家自然科学基金重点国际合作研究项目(32320103001);国家自然科学基金面上项目(32271475);中央高校基本科研业务费专项资金(226-2022-00214)
Synthetic genetic circuit engineering: principles, advances and prospects
GAO Ge1,2, BIAN Qi1,2, WANG Baojun1,2
- 1.College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310058,Zhejiang,China
2.ZJU-Hangzhou Global Scientific and Technological Innovation Center,Zhejiang University,Hangzhou 311200,Zhejiang,China
-
Received:2023-12-01Revised:2024-04-10Online:2025-01-31Published:2025-03-12 -
Contact:WANG Baojun
摘要:
合成基因线路利用合成生物学的技术和方法,将生物元件进行重新设计与构建,使人工设计的生物分子线路在活细胞中行使特定生物功能,在生物制造、医疗健康以及环境监测等领域具有巨大的潜力。但其工程化设计仍受到各种因素的制约,包括正交元器件数量有限、大规模线路组装困难、线路行为预测性低等。根据研究者们开发的各种调控元件工具箱和组装方法,本文逐点阐述了工程化设计基因线路所需遵循的几个核心原则:正交化、标准化、模块化与自动化。文章从DNA复制、转录和翻译层面介绍了正交基因元件库的构建和改造方法;全面总结了基因元件的标准化定量表征方法与标准元件设计方法;并介绍了本团队与其他团队在模块化基因线路设计方面的相关进展;分别从软件、硬件和人工智能角度展示如何实现基因线路的自动化设计。最后,本文探讨了基因线路设计的未来发展趋势,指出需要进一步融合人工智能和自动化等信息技术来加速基因线路“设计-构建-测试-学习”循环的迭代,提高线路设计的功能可预测性和复杂性,高效设计出符合目标需求的人造生命体。
中图分类号:
引用本文
高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64.
GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects[J]. Synthetic Biology Journal, 2025, 6(1): 45-64.
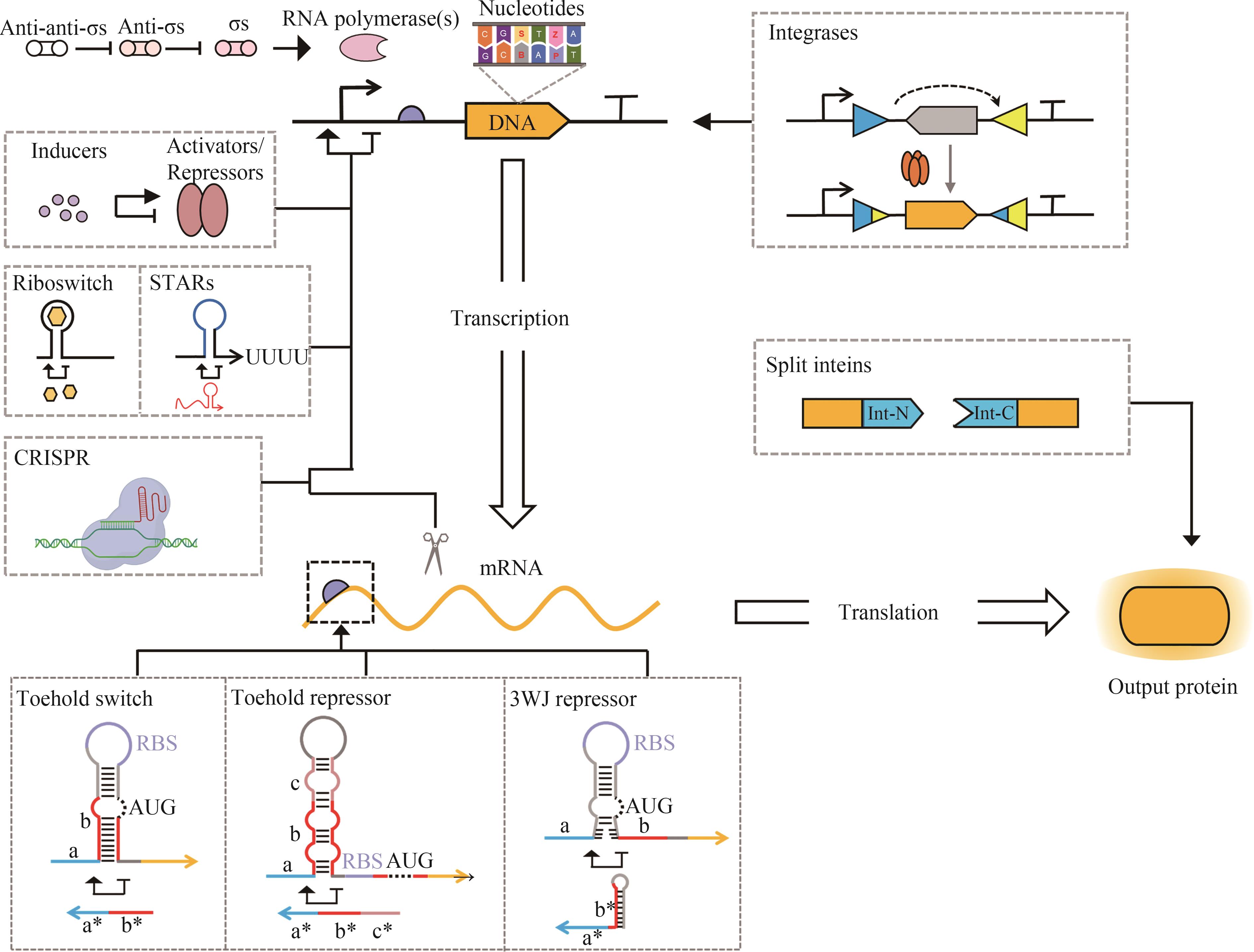
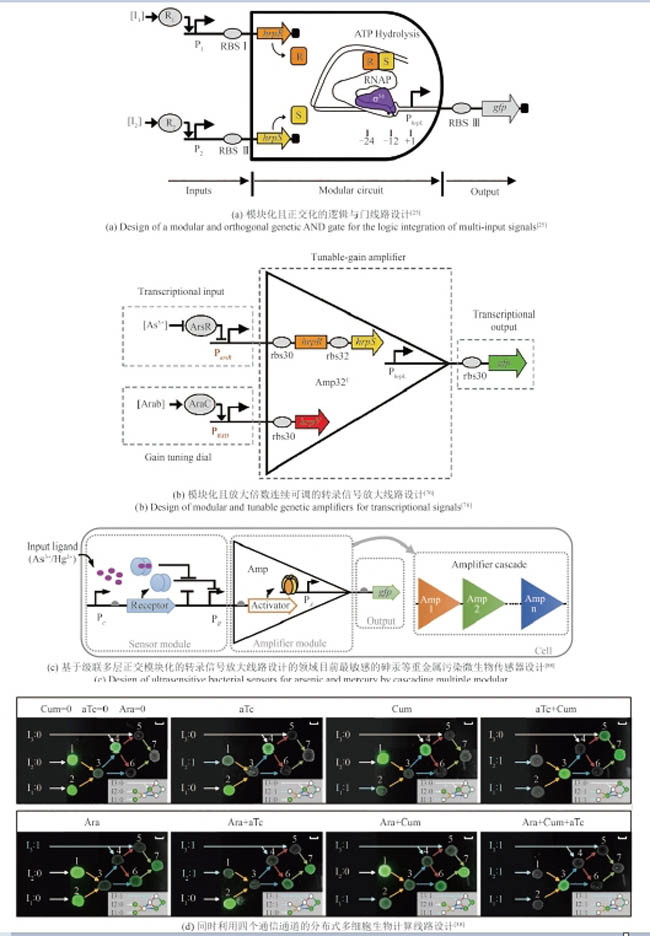
图1 合成基因线路中已经验证的用于基因表达控制的正交元件与调控工具(合成基因元件可以在遗传信息表达的不同过程中发挥调控作用,包括DNA存储与复制[14-20]、转录[21-45]、翻译[46-47]以及翻译后调控[48-50])
Fig. 1 Validated orthogonal parts and tools for precise gene expression control in genetic circuit design(Synthetic genetic parts can regulate various steps of gene expression, including DNA storage and replication[14-20], transcription[21-45], translation[46-47], and post-translational regulation[48-50].)
| 元件名称 | 获得正交元件的方法 | 元件数量 | 正交元件数量 | 参考文献 |
|---|---|---|---|---|
| T7 RNAP突变体 | 毒性降低的T7 RNAP突变体 | 4 | 4 | [ |
| 活性变高的T7 RNAP突变体 | 6 | 6 | [ | |
| ECF-σ因子 | 可替换的ECF-σ因子用于同源启动子的激活 | 52 | 20 | [ |
| LacI突变体 | N端序列突变的LacI与突变的LacO操纵子 | 5 | 5 | [ |
| Cl 突变体 | 基于噬菌粒的定向进化 | 12 | 6 | [ |
| TetR同系物 | 元件挖掘并鉴定TetR家族类似抑制子 | 20 | 17 | [ |
| 可诱导表达系统 | 金属离子(由金属离子诱导的调控因子和相应的启动子) | 5 | 5 | [ |
| 小分子(插入基因组的小分子生物传感器) | 12 | 12 | [ | |
| 代谢物(代谢的多样性) | 14 | 12 | [ | |
| 群体感应 | 对信号、遗传串扰优化后的群感调控因子和启动子 | 4 | 2 | [ |
| 突变pLux启动子序列 | 12 | 2 | [ | |
| 对不同来源群体感应系统进行同源和非同源表征 | 6 | 3 | [ | |
| 群感信号配体的筛选 | 10 | 6 | [ | |
| STARs | 目标RNA与小转录激活RNA | 100 | 6 | [ |
| CRISPRi | 高度非重复的超长sgRNA阵列 | 22 | 13 | [ |
| CRISPRa | 修饰的sgRNA与sigma 54激活因子 | 5 | 5 | [ |
| 核糖调控 | Toehold switches | 144 | 26 | [ |
| Toehold repressors | 95 | 15 | [ | |
| 断裂内含肽 | 元件挖掘并测试不同的断裂内含肽交叉活性 | 34 | 15 | [ |
| 遗传密码子 | 筛选技术:tREX | 71 | 23 | [ |
表1 典型正交基因元件库的设计与表征
Table 1 Design and characterization of the libraries of orthogonal genetic parts
| 元件名称 | 获得正交元件的方法 | 元件数量 | 正交元件数量 | 参考文献 |
|---|---|---|---|---|
| T7 RNAP突变体 | 毒性降低的T7 RNAP突变体 | 4 | 4 | [ |
| 活性变高的T7 RNAP突变体 | 6 | 6 | [ | |
| ECF-σ因子 | 可替换的ECF-σ因子用于同源启动子的激活 | 52 | 20 | [ |
| LacI突变体 | N端序列突变的LacI与突变的LacO操纵子 | 5 | 5 | [ |
| Cl 突变体 | 基于噬菌粒的定向进化 | 12 | 6 | [ |
| TetR同系物 | 元件挖掘并鉴定TetR家族类似抑制子 | 20 | 17 | [ |
| 可诱导表达系统 | 金属离子(由金属离子诱导的调控因子和相应的启动子) | 5 | 5 | [ |
| 小分子(插入基因组的小分子生物传感器) | 12 | 12 | [ | |
| 代谢物(代谢的多样性) | 14 | 12 | [ | |
| 群体感应 | 对信号、遗传串扰优化后的群感调控因子和启动子 | 4 | 2 | [ |
| 突变pLux启动子序列 | 12 | 2 | [ | |
| 对不同来源群体感应系统进行同源和非同源表征 | 6 | 3 | [ | |
| 群感信号配体的筛选 | 10 | 6 | [ | |
| STARs | 目标RNA与小转录激活RNA | 100 | 6 | [ |
| CRISPRi | 高度非重复的超长sgRNA阵列 | 22 | 13 | [ |
| CRISPRa | 修饰的sgRNA与sigma 54激活因子 | 5 | 5 | [ |
| 核糖调控 | Toehold switches | 144 | 26 | [ |
| Toehold repressors | 95 | 15 | [ | |
| 断裂内含肽 | 元件挖掘并测试不同的断裂内含肽交叉活性 | 34 | 15 | [ |
| 遗传密码子 | 筛选技术:tREX | 71 | 23 | [ |
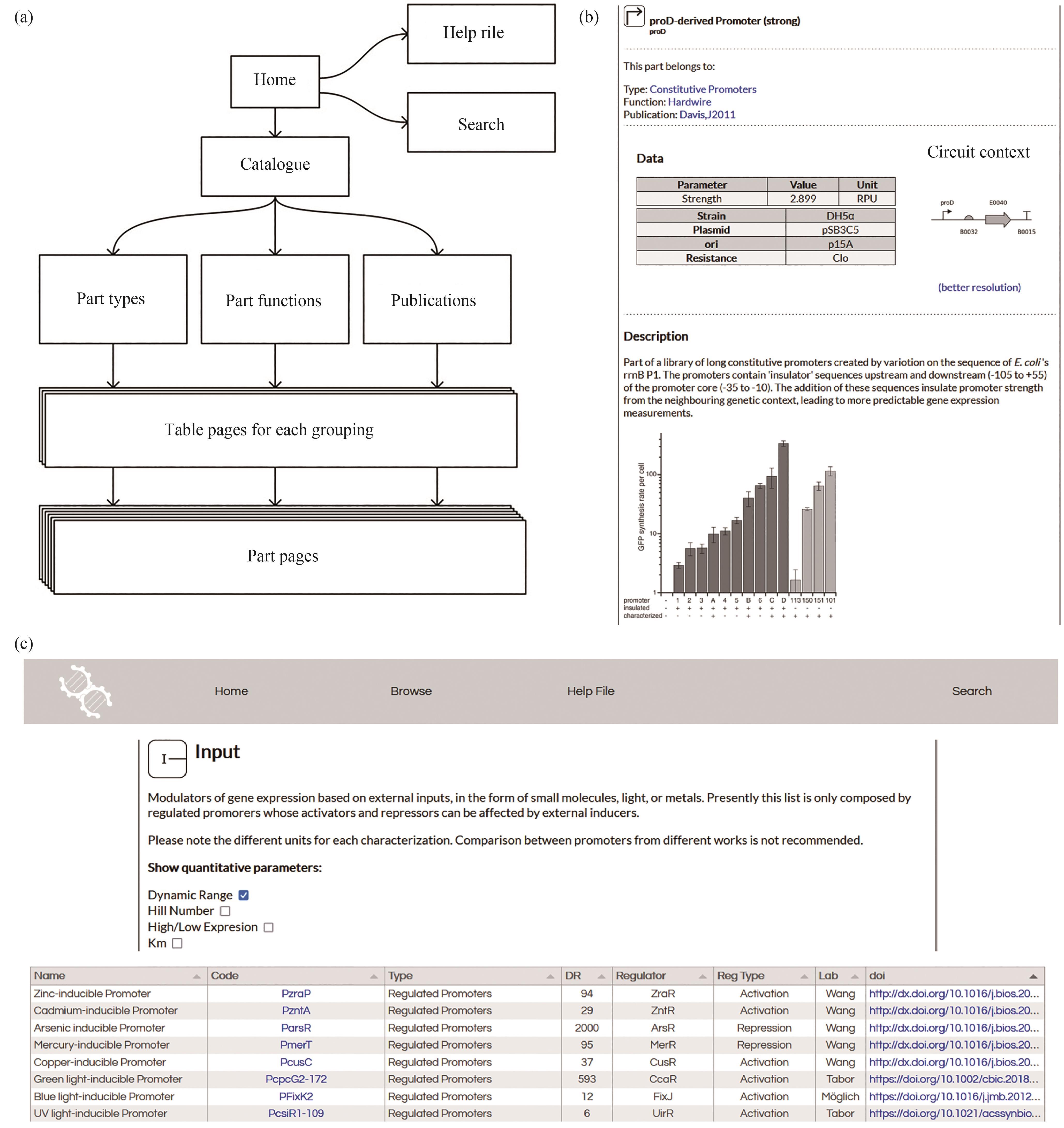
图2 基因元器件精选数据库BioPartsDB的网站结构与内容设计示意图[73](a) BioPartsDB数据库的设计架构图,箭头代表数据库里的预期用户流,箭头往下越来越多的堆叠面板代表该部分的页面数量越多,包含的信息越详细;(b)具体到某个基因元件数据的页面内容示例;(c)某种类型的基因元件列表页面内容示例,显示元件的简要说明和关键性能数据信息
Fig. 2 Design portfolio and the web architecture of the BioPartsDB platform[73](a) A simplified diagram showing the information flow of the database platform. Arrows indicate the intended user browsing along the platform's webpages. Increasingly stacked panels indicate the higher number of pages in each section and consequently the more detailed level of information. (b) A web page with in-depth description of the information, performance, and characterization conditions for a specific genetic part. (c) A table for parts with a brief description and data of their key performance.
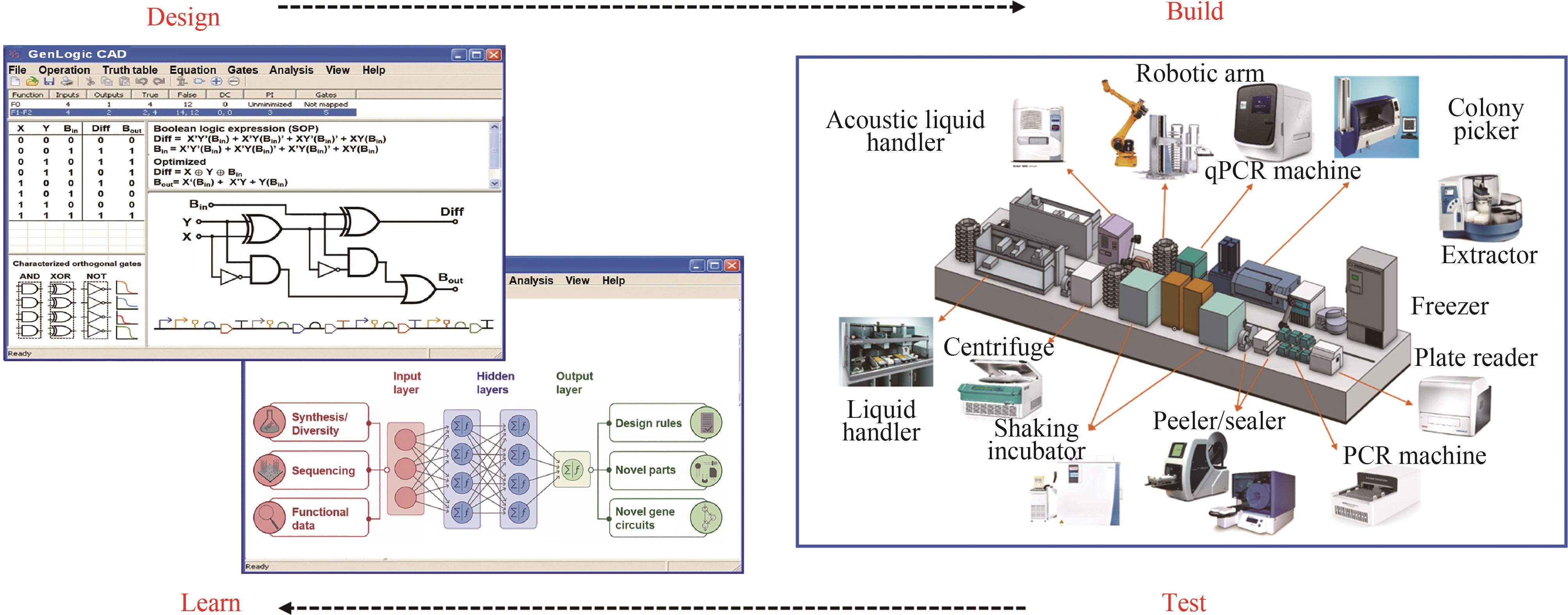
图4 合成基因线路的自动化“设计-构建-测试-学习”循环(“构建与测试”的生物铸造厂设施摘自文献[97],“学习”的模型改编自文献[98])
Fig. 4 An automated "design-build-test-learn" cycle for genetic circuit engineering.(Automated instrumentation in biofoundries is adapted from reference [97]. The neural network-based deep learning model is adapted from reference [98].)
| 1 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010—2020[J]. Nature Communications, 2020, 11(1): 5174. |
| 2 | WANG X Y, ZHOU N, WANG B J. Bacterial synthetic biology: tools for novel drug discovery[J]. Expert Opinion on Drug Discovery, 2023, 18(10): 1087-1097. |
| 3 | TAMSIR A, TABOR J J, VOIGT C A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'[J]. Nature, 2011, 469(7329): 212-215. |
| 4 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403(6767): 335-338. |
| 5 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 6 | ROQUET N, SOLEIMANY A P, FERRIS A C, et al. Synthetic recombinase-based state machines in living cells[J]. Science, 2016, 353(6297): aad8559. |
| 7 | RUBENS J R, SELVAGGIO G, LU T K. Synthetic mixed-signal computation in living cells[J]. Nature Communications, 2016, 7: 11658. |
| 8 | BRADLEY R W, BUCK M, WANG B J. Tools and principles for microbial gene circuit engineering[J]. Journal of Molecular Biology, 2016, 428(5 Pt B): 862-888. |
| 9 | GAO Y L, WANG L, WANG B J. Customizing cellular signal processing by synthetic multi-level regulatory circuits[J]. Nature Communications, 2023, 14(1): 8415. |
| 10 | XIANG Y Y, DALCHAU N, WANG B J. Scaling up genetic circuit design for cellular computing: advances and prospects[J]. Natural Computing, 2018, 17(4): 833-853. |
| 11 | ŞIMŞEK E, YAO Y, LEE D, et al. Toward predictive engineering of gene circuits[J]. Trends in Biotechnology, 2023, 41(6): 760-768. |
| 12 | PECCOUD J, BLAUVELT M F, CAI Y Z, et al. Targeted development of registries of biological parts[J]. PLoS One, 2008, 3(7): e2671. |
| 13 | COSTELLO A, BADRAN A H. Synthetic biological circuits within an orthogonal central dogma[J]. Trends in Biotechnology, 2021, 39(1): 59-71. |
| 14 | PARK M, PATEL N, KEUNG A J, et al. Engineering epigenetic regulation using synthetic read-write modules[J]. Cell, 2019, 176(1-2): 227-238.e20. |
| 15 | DHAMI K, MALYSHEV D A, ORDOUKHANIAN P, et al. Systematic exploration of a class of hydrophobic unnatural base pairs yields multiple new candidates for the expansion of the genetic alphabet[J]. Nucleic Acids Research, 2014, 42(16): 10235-10244. |
| 16 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363(6429): 884-887. |
| 17 | RAVIKUMAR A, ARZUMANYAN G A, OBADI M K A, et al. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds[J]. Cell, 2018, 175(7): 1946-1957.e13. |
| 18 | ZHONG Z W, RAVIKUMAR A, LIU C C. Tunable expression systems for orthogonal DNA replication[J]. ACS Synthetic Biology, 2018, 7(12): 2930-2934. |
| 19 | TIAN R Z, ZHAO R Z, GUO H Y, et al. Engineered bacterial orthogonal DNA replication system for continuous evolution[J]. Nature Chemical Biology, 2023, 19(12): 1504-1512. |
| 20 | YANG L, NIELSEN A A, FERNANDEZ-RODRIGUEZ J, et al. Permanent genetic memory with >1-byte capacity[J]. Nature Methods, 2014, 11(12): 1261-1266. |
| 21 | TEMME K, HILL R, SEGALL-SHAPIRO T H, et al. Modular control of multiple pathways using engineered orthogonal T7 polymerases[J]. Nucleic Acids Research, 2012, 40(17): 8773-8781. |
| 22 | MEYER A J, ELLEFSON J W, ELLINGTON A D. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry[J]. ACS Synthetic Biology, 2015, 4(10): 1070-1076. |
| 23 | RHODIUS V A, SEGALL-SHAPIRO T H, SHARON B D, et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters[J]. Molecular Systems Biology, 2013, 9: 702. |
| 24 | BERVOETS I, VAN BREMPT M, VAN NEROM K, et al. A sigma factor toolbox for orthogonal gene expression in Escherichia coli [J]. Nucleic Acids Research, 2018, 46(4): 2133-2144. |
| 25 | WANG B J, KITNEY R I, JOLY N, et al. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology[J]. Nature Communications, 2011, 2: 508. |
| 26 | ZHAN J, DING B, MA R, et al. Develop reusable and combinable designs for transcriptional logic gates[J]. Molecular Systems Biology, 2010, 6: 388. |
| 27 | BRÖDEL A K, JARAMILLO A, ISALAN M. Engineering orthogonal dual transcription factors for multi-input synthetic promoters[J]. Nature Communications, 2016, 7: 13858. |
| 28 | STANTON B C, NIELSEN A A, TAMSIR A, et al. Genomic mining of prokaryotic repressors for orthogonal logic gates[J]. Nature Chemical Biology, 2014, 10(2): 99-105. |
| 29 | WANG B J, BARAHONA M, BUCK M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals[J]. Biosensors & Bioelectronics, 2013, 40(1): 368-376. |
| 30 | LOPRESIDE A, WAN X Y, MICHELINI E, et al. Comprehensive profiling of diverse genetic reporters with application to whole-cell and cell-free biosensors[J]. Analytical Chemistry, 2019, 91(23): 15284-15292. |
| 31 | MEYER A J, SEGALL-SHAPIRO T H, GLASSEY E, et al. Escherichia coli "Marionette" strains with 12 highly optimized small-molecule sensors[J]. Nature Chemical Biology, 2019, 15(2): 196-204. |
| 32 | GROSECLOSE T M, RONDON R E, HERDE Z D, et al. Engineered systems of inducible anti-repressors for the next generation of biological programming[J]. Nature Communications, 2020, 11(1): 4440. |
| 33 | HANKO E K R, PAIVA A C, JONCZYK M, et al. A genome-wide approach for identification and characterisation of metabolite-inducible systems[J]. Nature Communications, 2020, 11(1): 1213. |
| 34 | SCOTT S R, HASTY J. Quorum sensing communication modules for microbial consortia[J]. ACS Synthetic Biology, 2016, 5(9): 969-977. |
| 35 | GRANT P K, DALCHAU N, BROWN J R, et al. Orthogonal intercellular signaling for programmed spatial behavior[J]. Molecular Systems Biology, 2016, 12(1): 849. |
| 36 | KYLILIS N, TUZA Z A, STAN G B, et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia[J]. Nature Communications, 2018, 9(1): 2677. |
| 37 | WU S B, QIAO J J, YANG A D, et al. Potential of orthogonal and cross-talk quorum sensing for dynamic regulation in cocultivation[J]. Chemical Engineering Journal, 2022, 445: 136720. |
| 38 | DU P, ZHAO H W, ZHANG H Q, et al. De novo design of an intercellular signaling toolbox for multi-channel cell-cell communication and biological computation[J]. Nature Communications, 2020, 11(1): 4226. |
| 39 | JONKERGOUW C, SAVOLA P, OSMEKHINA E, et al. Exploration of chemical diversity in intercellular quorum sensing signalling systems in prokaryotes[J]. Angewandte Chemie International Edition, 2024, 63(2): e202314469. |
| 40 | ESPAH BORUJENI A, MISHLER D M, WANG J Z, et al. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers[J]. Nucleic Acids Research, 2016, 44(1): 1-13. |
| 41 | CHAPPELL J, WESTBROOK A, VEROSLOFF M, et al. Computational design of small transcription activating RNAs for versatile and dynamic gene regulation[J]. Nature Communications, 2017, 8(1): 1051. |
| 42 | DIDOVYK A, BOREK B, HASTY J, et al. Orthogonal modular gene repression in Escherichia coli using engineered CRISPR/Cas9[J]. ACS Synthetic Biology, 2016, 5(1): 81-88. |
| 43 | NIELSEN A A K, VOIGT C A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks[J]. Molecular Systems Biology, 2014, 10(11): 763. |
| 44 | REIS A C, HALPER S M, VEZEAU G E, et al. Simultaneous repression of multiple bacterial genes using nonrepetitive extra-long sgRNA arrays[J]. Nature Biotechnology, 2019, 37(11): 1294-1301. |
| 45 | LIU Y, WAN X Y, WANG B J. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria[J]. Nature Communications, 2019, 10(1): 3693. |
| 46 | GREEN A A, SILVER P A, COLLINS J J, et al. Toehold switches: de-novo-designed regulators of gene expression[J]. Cell, 2014, 159(4): 925-939. |
| 47 | KIM J M, ZHOU Y, CARLSON P D, et al. De novo-designed translation-repressing riboregulators for multi-input cellular logic[J]. Nature Chemical Biology, 2019, 15(12): 1173-1182. |
| 48 | PINTO F, THORNTON E L, WANG B J. An expanded library of orthogonal split inteins enables modular multi-peptide assemblies[J]. Nature Communications, 2020, 11(1): 1529. |
| 49 | CERVETTINI D, TANG S, FRIED S D, et al. Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase-tRNA pairs[J]. Nature Biotechnology, 2020, 38(8): 989-999. |
| 50 | DUNKELMANN D L, WILLIS J C W, BEATTIE A T, et al. Engineered triply orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids[J]. Nature Chemistry, 2020, 12(6): 535-544. |
| 51 | MERRICK C A, ZHAO J, ROSSER S J. Serine integrases: advancing synthetic biology[J]. ACS Synthetic Biology, 2018, 7(2): 299-310. |
| 52 | PAGET M S. Bacterial sigma factors and anti-sigma factors: structure, function and distribution[J]. Biomolecules, 2015, 5(3): 1245-1265. |
| 53 | GUET C C, ELOWITZ M B, HSING W, et al. Combinatorial synthesis of genetic networks[J]. Science, 2002, 296(5572): 1466-1470. |
| 54 | HASTY J, DOLNIK M, ROTTSCHÄFER V, et al. Synthetic gene network for entraining and amplifying cellular oscillations[J]. Physical Review Letters, 2002, 88(14): 148101. |
| 55 | HOOSHANGI S, THIBERGE S, WEISS R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(10): 3581-3586. |
| 56 | WANG B J, BARAHONA M, BUCK M. Amplification of small molecule-inducible gene expression via tuning of intracellular receptor densities[J]. Nucleic Acids Research, 2015, 43(3): 1955-1964. |
| 57 | 李晓萌, 姜威, 梁泉峰, 等. 细菌群体感应系统在细胞间通讯中的应用及其合成生物学研究进展[J]. 合成生物学, 2020, 1(5): 540-555. |
| LI X M, JIANG W, LIANG Q F, et al. Application of bacterial quorum sensing system in intercellular communication and its progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(5): 540-555. | |
| 58 | 周爱林, 刘奕, 巴方, 等. 细菌群体感应元件构建和工程应用[J]. 合成生物学, 2021, 2(2): 234-246. |
| ZHOU A L, LIU Y, BA F, et al. Construction and engineering application of bacterial quorum sensing elements[J]. Synthetic Biology Journal, 2021, 2(2): 234-246. | |
| 59 | AMERUOSO A, GAMBILL L, LIU B Y, et al. Brave new 'RNA' world—advances in RNA tools and their application for understanding and engineering biological systems[J]. Current Opinion in Systems Biology, 2019, 14: 32-40. |
| 60 | ISAACS F J, DWYER D J, DING C M, et al. Engineered riboregulators enable post-transcriptional control of gene expression[J]. Nature Biotechnology, 2004, 22(7): 841-847. |
| 61 | 付宪, 林涛, 张帆, 等. 基因密码子拓展技术的方法原理和前沿应用研究进展[J]. 合成生物学, 2020, 1(1): 103-119. |
| FU X, LIN T, ZHANG F, et al. Progress in the study of genetic code expansion related methods, principles and applications[J]. Synthetic Biology Journal, 2020, 1(1): 103-119. | |
| 62 | CANTON B, LABNO A, ENDY D. Refinement and standardization of synthetic biological parts and devices[J]. Nature Biotechnology, 2008, 26(7): 787-793. |
| 63 | CASINI A, STORCH M, BALDWIN G S, et al. Bricks and blueprints: methods and standards for DNA assembly[J]. Nature Reviews Molecular Cell Biology, 2015, 16(9): 568-576. |
| 64 | GUO Y K, DONG J K, ZHOU T, et al. YeastFab: the design and construction of standard biological parts for metabolic engineering in Saccharomyces cerevisiae [J]. Nucleic Acids Research, 2015, 43(13): e88. |
| 65 | RAJKUMAR A S, VARELA J A, JUERGENS H, et al. Biological parts for Kluyveromyces marxianus synthetic biology[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 97. |
| 66 | WEBER E, ENGLER C, GRUETZNER R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PLoS One, 2011, 6(2): e16765. |
| 67 | LEE M E, DELOACHE W C, CERVANTES B, et al. A highly characterized yeast toolkit for modular, multipart assembly[J]. ACS Synthetic Biology, 2015, 4(9): 975-986. |
| 68 | WEI P L, FAN J, YU J W, et al. Quantitative characterization of filamentous fungal promoters on a single-cell resolution to discover cryptic natural products[J]. Science China Life Sciences, 2023, 66(4): 848-860. |
| 69 | REDDEN H, ALPER H S. The development and characterization of synthetic minimal yeast promoters[J]. Nature Communications, 2015, 6: 7810. |
| 70 | CURRAN K A, MORSE N J, MARKHAM K A, et al. Short synthetic terminators for improved heterologous gene expression in yeast[J]. ACS Synthetic Biology, 2015, 4(7): 824-832. |
| 71 | YIM S S, AN S J, KANG M, et al. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum [J]. Biotechnology and Bioengineering, 2013, 110(11): 2959-2969. |
| 72 | MCLAUGHLIN J A, MYERS C J, ZUNDEL Z, et al. SynBioHub: a standards-enabled design repository for synthetic biology[J]. ACS Synthetic Biology, 2018, 7(2): 682-688. |
| 73 | BUSON F, GAO Y L, WANG B J. Genetic parts and enabling tools for biocircuit design[J]. ACS Synthetic Biology, 2024, 13(3): 697-713. |
| 74 | HILLSON N, CADDICK M, CAI Y Z, et al. Building a global alliance of biofoundries[J]. Nature Communications, 2019, 10(1): 2040. |
| 75 | GRUNBERG T W, DEL VECCHIO D. Modular analysis and design of biological circuits[J]. Current Opinion in Biotechnology, 2020, 63: 41-47. |
| 76 | WANG B J, BARAHONA M, BUCK M. Engineering modular and tunable genetic amplifiers for scaling transcriptional signals in cascaded gene networks[J]. Nucleic Acids Research, 2014, 42(14): 9484-9492. |
| 77 | LI Y Q, JIANG Y, CHEN H, et al. Modular construction of mammalian gene circuits using TALE transcriptional repressors[J]. Nature Chemical Biology, 2015, 11(3): 207-213. |
| 78 | LEVINE M, CATTOGLIO C, TJIAN R. Looping back to leap forward: transcription enters a new era[J]. Cell, 2014, 157(1): 13-25. |
| 79 | HOU J R, ZENG W Q, ZONG Y Q, et al. Engineering the ultrasensitive transcription factors by fusing a modular oligomerization domain[J]. ACS Synthetic Biology, 2018, 7(5): 1188-1194. |
| 80 | PURNICK P E M, WEISS R. The second wave of synthetic biology: from modules to systems[J]. Nature Reviews Molecular Cell Biology, 2009, 10(6): 410-422. |
| 81 | CARDINALE S, ARKIN A P. Contextualizing context for synthetic biology: identifying causes of failure of synthetic biological systems[J]. Biotechnology Journal, 2012, 7(7): 856-866. |
| 82 | DEL VECCHIO D, NINFA A J, SONTAG E D. Modular cell biology: retroactivity and insulation[J]. Molecular Systems Biology, 2008, 4: 161. |
| 83 | LIU Q J, SCHUMACHER J, WAN X Y, et al. Orthogonality and burdens of heterologous AND gate gene circuits in E.coli [J]. ACS Synthetic Biology, 2018, 7(2): 553-564. |
| 84 | CERONI F, ALGAR R, STAN G B, et al. Quantifying cellular capacity identifies gene expression designs with reduced burden[J]. Nature Methods, 2015, 12(5): 415-418. |
| 85 | SEGALL-SHAPIRO T H, SONTAG E D, VOIGT C A. Engineered promoters enable constant gene expression at any copy number in bacteria[J]. Nature Biotechnology, 2018, 36(4): 352-358. |
| 86 | WAN X Y, PINTO F, YU L Y, et al. Synthetic protein-binding DNA sponge as a tool to tune gene expression and mitigate protein toxicity[J]. Nature Communications, 2020, 11(1): 5961. |
| 87 | MISHRA D, RIVERA P M, LIN A, et al. A load driver device for engineering modularity in biological networks[J]. Nature Biotechnology, 2014, 32(12): 1268-1275. |
| 88 | WAN X Y, VOLPETTI F, PETROVA E, et al. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals[J]. Nature Chemical Biology, 2019, 15(5): 540-548. |
| 89 | LIU Y, PINTO F, WAN X Y, et al. Reprogrammed tracrRNAs enable repurposing of RNAs as crRNAs and sequence-specific RNA biosensors[J]. Nature Communications, 2022, 13(1): 1937. |
| 90 | REGOT S, MACIA J, CONDE N, et al. Distributed biological computation with multicellular engineered networks[J]. Nature, 2011, 469(7329): 207-211. |
| 91 | AUSLÄNDER D, AUSLÄNDER S, PIERRAT X, et al. Programmable full-adder computations in communicating three-dimensional cell cultures[J]. Nature Methods, 2018, 15(1): 57-60. |
| 92 | DAVIS R M, MULLER R Y, HAYNES K A. Can the natural diversity of quorum-sensing advance synthetic biology?[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 30. |
| 93 | CURATOLO A I, ZHOU N, ZHAO Y, et al. Cooperative pattern formation in multi-component bacterial systems through reciprocal motility regulation[J]. Nature Physics, 2020, 16: 1152-1157. |
| 94 | 周楠, 夏婷颖, 黄建东. 合成生物学在探索生物图案形成基本原理中的应用与展望[J]. 合成生物学, 2020, 1(4): 470-480. |
| ZHOU N, XIA T Y, HUANG J D. Applications and prospects of synthetic biology in exploring the basic principles of biological pattern formation[J]. Synthetic Biology Journal, 2020, 1(4): 470-480. | |
| 95 | KHAN M S, KIM E, MCPHERSON A, et al. Adenovirus-vectored SARS-CoV-2 vaccine expressing S1-N fusion protein[J]. Antibody Therapeutics, 2022, 5(3): 177-191. |
| 96 | LI H S, ISRANI D V, GAGNON K A, et al. Multidimensional control of therapeutic human cell function with synthetic gene circuits[J]. Science, 2022, 378(6625): 1227-1234. |
| 97 | 唐婷, 付立豪, 郭二鹏, 等. 自动化合成生物技术与工程化设施平台[J]. 科学通报, 2021, 66(3): 300-309. |
| TANG T, FU L H, GUO E P, et al. Automation in synthetic biology using biological foundries[J]. Chinese Science Bulletin, 2021, 66(3): 300-309. | |
| 98 | CAMACHO D M, COLLINS K M, POWERS R K, et al. Next-generation machine learning for biological networks[J]. Cell, 2018, 173(7): 1581-1592. |
| 99 | NIELSEN A A K, DER B S, SHIN J, et al. Genetic circuit design automation[J]. Science, 2016, 352(6281): aac7341. |
| 100 | TAKETANI M, ZHANG J B, ZHANG S Y, et al. Genetic circuit design automation for the gut resident species Bacteroides thetaiotaomicron [J]. Nature Biotechnology, 2020, 38(8): 962-969. |
| 101 | CHEN Y, ZHANG S Y, YOUNG E M, et al. Genetic circuit design automation for yeast[J]. Nature Microbiology, 2020, 5(11): 1349-1360. |
| 102 | JONES T S, OLIVEIRA S M D, MYERS C J, et al. Genetic circuit design automation with Cello 2.0[J]. Nature Protocols, 2022, 17(4): 1097-1113. |
| 103 | MYERS C J, BARKER N, JONES K, et al. iBioSim: a tool for the analysis and design of genetic circuits[J]. Bioinformatics, 2009, 25(21): 2848-2849. |
| 104 | WATANABE L, NGUYEN T, ZHANG M, et al. iBioSim 3: a tool for model-based genetic circuit design[J]. ACS Synthetic Biology, 2019, 8(7): 1560-1563. |
| 105 | HILLSON N J, ROSENGARTEN R D, KEASLING J D. j5 DNA assembly design automation software[J]. ACS Synthetic Biology, 2012, 1(1): 14-21. |
| 106 | WILSON M L, HERTZBERG R, ADAM L, et al. A step-by-step introduction to rule-based design of synthetic genetic constructs using GenoCAD[J]. Methods in Enzymology, 2011, 498: 173-188. |
| 107 | HOLOWKO M B, FROW E K, REID J C, et al. Building a biofoundry[J]. Synthetic Biology, 2021, 6(1): ysaa026. |
| 108 | CHAO R, LIANG J, TASAN I, et al. Fully automated one-step synthesis of single-transcript TALEN pairs using a biological foundry[J]. ACS Synthetic Biology, 2017, 6(4): 678-685. |
| 109 | SI T, CHAO R, MIN Y H, et al. Automated multiplex genome-scale engineering in yeast[J]. Nature Communications, 2017, 8: 15187. |
| 110 | 崔金明, 张炳照, 马迎飞, 等. 合成生物学研究的工程化平台[J]. 中国科学院院刊, 2018, 33(11): 1249-1257. |
| CUI J M, ZHANG B Z, MA Y F, et al. Engineering platforms for synthetic biology research[J]. Bulletin of Chinese Academy of Sciences, 2018, 33(11): 1249-1257. | |
| 111 | ZHANG S Y, ZHU J, FAN S, et al. Directed evolution of a cyclodipeptide synthase with new activities via label-free mass spectrometric screening[J]. Chemical Science, 2022, 13(25): 7581-7586. |
| 112 | GUO E P, FU L H, FANG X T, et al. Robotic construction and screening of lanthipeptide variant libraries in Escherichia coli [J]. ACS Synthetic Biology, 2022, 11(12): 3900-3911. |
| 113 | ZHANG J Z, LI T, HONG Z L, et al. Biosynthesis of hybrid neutral lipids with archaeal and eukaryotic characteristics in engineered Saccharomyces cerevisiae [J]. Angewandte Chemie International Edition, 2023, 62(4): e202214344. |
| 114 | DENG H X, YU H, DENG Y W, et al. Pathway evolution through a bottlenecking-debottlenecking strategy and machine learning-aided flux balancing[J]. Advanced Science, 2024: e2306935. |
| 115 | 卢挥, 张芳丽, 黄磊. 合成生物学自动化装置iBioFoundry的构建与应用[J]. 合成生物学, 2023, 4(5): 877-891. |
| LU H, ZHANG F L, HUANG L. Establishment of iBioFoundry for synthetic biology applications[J]. Synthetic Biology Journal, 2023, 4(5): 877-891. | |
| 116 | HU R Y, FU L H, CHEN Y C, et al. Protein engineering via Bayesian optimization-guided evolutionary algorithm and robotic experiments[J]. Briefings in Bioinformatics, 2023, 24(1): bbac570. |
| 117 | WANG Y, WANG H C, WEI L, et al. Synthetic promoter design in Escherichia coli based on a deep generative network[J]. Nucleic Acids Research, 2020, 48(12): 6403-6412. |
| 118 | CONSTANT D A, GUTIERREZ J M, SASTRY A V, et al. Deep learning-based codon optimization with large-scale synonymous variant datasets enables generalized tunable protein expression[EB/OL]. bioRxiv, 2023, 2023.02.11.528149. (2023-02-12)[2023-12-01]. . |
| 119 | ALIPANAHI B, DELONG A, WEIRAUCH M T, et al. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning[J]. Nature Biotechnology, 2015, 33(8): 831-838. |
| 120 | WANG M, TAI C, E W N, et al. DeFine: deep convolutional neural networks accurately quantify intensities of transcription factor-DNA binding and facilitate evaluation of functional non-coding variants[J]. Nucleic Acids Research, 2018, 46(11): e69. |
| 121 | VALERI J A, COLLINS K M, RAMESH P, et al. Sequence-to-function deep learning frameworks for engineered riboregulators[J]. Nature Communications, 2020, 11(1): 5058. |
| 122 | SEAK L C U, LO O L I, SUEN W, et al. Next-generation biocomputing: mimicking artificial neural network with genetic circuits[EB/OL]. bioRxiv, 2021, 2021.03.12.435120. (2021-03-12)[2023-12-01]. . |
| [1] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [2] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [3] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [4] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| [5] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [6] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [7] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [8] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| [9] | 郭婷婷, 韩湘凝, 黄熙婷, 张婷婷, 孔健. 乳酸菌的合成生物学工具及在合成益肤因子中的应用[J]. 合成生物学, 2025, 6(2): 320-333. |
| [10] | 张萍, 张维娇, 胥睿睿, 李江华, 陈坚, 康振. 防晒化合物类菌孢素氨基酸的生物合成[J]. 合成生物学, 2025, 6(2): 306-319. |
| [11] | 黄姝涵, 马赫, 罗云孜. 生物合成红景天苷的研究进展[J]. 合成生物学, 2025, 6(2): 391-407. |
| [12] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [13] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [14] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [15] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||