合成生物学 ›› 2025, Vol. 6 ›› Issue (2): 334-356.DOI: 10.12211/2096-8280.2024-057
合成生物学助力萜类香精香料可持续生产
张梦瑶1,2,3, 蔡鹏1,2, 周雍进1,2
- 1.中国科学院大连化学物理研究所生物技术研究部,辽宁 大连 116023
2.大连市能源生物技术重点实验室,辽宁 大连 116023
3.中国科学院大学,北京 100049
-
收稿日期:2024-07-31修回日期:2024-09-18出版日期:2025-04-30发布日期:2025-05-20 -
通讯作者:周雍进 -
作者简介:张梦瑶 (1999—),女,博士研究生。研究方向为天然产物的生物合成。E-mail:myzhang@dicp.ac.cn周雍进 (1984—),男,博士,研究员。研究方向为基于甲醇生物转化与天然产物生物合成。E-mail:zhouyongjin@dicp.ac.cn -
基金资助:国家重点研发计划(2022YFC2105900);中国科学院特别研究助理资助项目
Synthetic biology drives the sustainable production of terpenoid fragrances and flavors
ZHANG Mengyao1,2,3, CAI Peng1,2, ZHOU Yongjin1,2
- 1.Division of Biotechnology,Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian 116023,Liaoning,China
2.Dalian Key Laboratory of Energy Biotechnology,Dalian 116023,Liaoning,China
3.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2024-07-31Revised:2024-09-18Online:2025-04-30Published:2025-05-20 -
Contact:ZHOU Yongjin
摘要:
香精香料是个人护理产品中的重要成分,其中,萜类化合物及其衍生物在天然香料市场中有着重要的地位。近年来,合成生物学的蓬勃发展为解决萜类香料产能瓶颈及开发更多元化的新型香料化合物带来了新机遇。本文探讨了合成生物学在萜类香料可持续生产中的应用和发展,介绍了数据驱动的合成生物学和生物技术创新如何赋能萜类香料生产,讨论比较了萜类合成的经典合成途径和替代合成途径,并探讨了萜类合酶挖掘与改造进展。在此基础上,着重介绍了单萜类、倍半萜类和降异戊二烯类香料的细胞工厂合成现状,包括元件改造、途径优化和萜类解毒等关键技术策略。最后,对当前专利布局和产业化竞争格局进行了总结分析,并对未来发展的挑战和机遇进行了展望,包括生物合成技术的挑战、新产物的发掘与设计,以及市场监管与安全性问题。
中图分类号:
引用本文
张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356.
ZHANG Mengyao, CAI Peng, ZHOU Yongjin. Synthetic biology drives the sustainable production of terpenoid fragrances and flavors[J]. Synthetic Biology Journal, 2025, 6(2): 334-356.
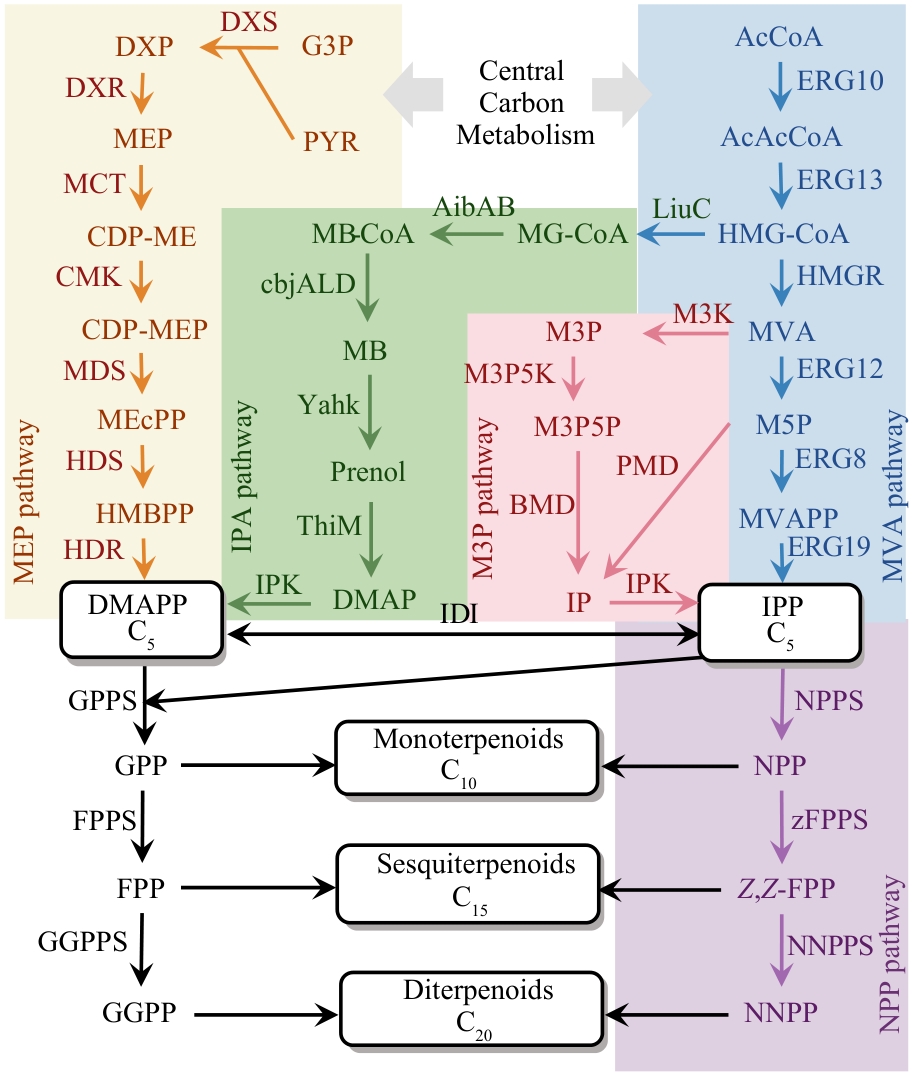
图2 经典的萜类合成途径与常见替代合成途径(G3P—D-甘油醛-3-磷酸; PYR—丙酮酸; DXP—1-脱氧-D- 木酮糖-5-磷酸酯; MEP—2-C-甲基-D-赤藓糖醇-4-磷酸; CDP-ME—4-焦磷酸胞苷-2-C-甲基-D-赤藓糖醇; CDP-MEP—4-焦磷酸胞苷-2-C-甲基-D-赤藓糖醇-2-磷酸; MEcPP—2-C-甲基-D-赤藓糖醇-2,4-环焦磷酸; HMBPP—4-羟基-3-甲基-2-丁烯基焦磷酸; DMAPP—二甲基烯丙基焦磷酸; GPP—香叶基焦磷酸; FPP—法尼基焦磷酸; GGPP—香叶基香叶基焦磷酸; MG-CoA—3-甲基戊烯二酰辅酶A; MB-CoA—3-甲基-2-丁烯酰辅酶A; MB—3-甲基-2-丁烯醛; DMAP—二甲基烯丙基磷酸; M3P—甲羟戊酸-3-磷酸; M3P5P—甲羟戊酸-3,5-焦磷酸; IP—异戊烯基磷酸; AcCoA—乙酰辅酶A; AcAcCoA—乙酰乙酰辅酶A; HMG-CoA—3-羟基-3-甲基戊二酰辅酶A; MVA—甲羟戊酸; M5P—甲羟戊酸-5-磷酸; MVAPP—甲羟戊酸焦磷酸; IPP—异戊烯基焦磷酸; NPP—橙花基焦磷酸; Z,Z-FPP—Z,Z-法尼基焦磷酸; NNPP—橙花橙花基焦磷酸; DXS—DXP合酶; DXR—DXP还原异构酶; MCT—MEP胞苷酰转移酶; CMK—CDP-ME激酶; MDS—ME-CPP合酶; HDS—HMB-PP合酶; HDR—HMB-PP还原酶; ERG10—乙酰辅酶A硫解酶; ERG13—3-羟基-3-甲基戊二酰-CoA合酶; HMGR—3-羟基-3-甲基戊二酰辅酶A还原酶; ERG12/MK—甲羟戊酸激酶; ERG8/PMVK—磷酸甲羟戊酸激酶; ERG19/MVD—甲羟戊酸焦磷酸脱羧酶; LiuC—烯酰辅酶A水合酶; AibAB—戊烯二酰辅酶A脱羧酶; cbjALD—酰基辅酶A还原酶; YahK—醇脱氢酶; ThiM—羟乙基噻唑激酶; IPK—异戊烯基磷酸激酶; IDI—异戊烯基焦磷酸异构酶; GPPS—香叶基焦磷酸合酶; FPPS—法尼基焦磷酸合酶; GGPPS—香叶基香叶基焦磷酸合酶; NPPS—橙花基焦磷酸合酶; zFPPS—Z,Z-法尼基焦磷酸合酶; NNPPS—橙花橙花基焦磷酸合酶; M3K—甲羟戊酸-3-激酶; M3P5K—甲羟戊酸-3-磷酸-5-激酶; BMD—甲羟戊酸焦磷酸脱羧酶; IPK—异戊烯基磷酸激酶;PMD—甲羟戊酸-5-磷酸脱羧酶)
Fig. 2 Classical terpene biosynthetic pathways and alternative chemical synthetic routes(G3P—D-glyceraldehyde 3-phosphate; PYR—pyruvate; DXP—1-deoxy-D-xylulose 5-phosphate; MEP—2-C-methyl-D-erythritol 4-phosphate; CDP-ME—4-diphophocytidyl-2-C-methyl-D-erythrito; CDP-MEP—4-diphophocytidyl-2-C-methyl-D-erythritol 2phosphate; MEcPP—2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBPP—4-hydroxy-3-methyl-butenyl diphosphate; DMAPP—dimethylallyl diphosphate; GPP—geranyl diphosphate; FPP—farnesyl diphosphate; GGPP—geranylgeranyl diphosphate; MG-CoA—3-methylglutaconyl-CoA; MB-CoA—3-methyl-2-butenoyl-CoA; MB—3-methyl-2-butenal; DMAP—dimethylallyl phosphate; M3P—mevalonate 3-phosphate; M3P5P—mevalonate 3,5-biphosphate; IP—isopentenyl phosphate; AcCoA—acetyl-CoA; AcAcCoA—acetoacetyl-CoA; HMGCoA—3-hydroxy-3-methylglutaryl-CoA; MVA—mevalonate; M5P—mevalonate 5-phosphate; MVAPP—mevalonate diphosphate; IPP—isopentenyl diphosphate; NPP—neryl diphosphate; Z,Z-FPP—Z,Z-farnesyl diphosphate; NNPP—nerylneryl diphosphate; DXS—DXP synthase; DXR—DXP reductoisomerase; MCT—MEP cytidylyltransferase; CMK—CDP-ME kinase; MDS—ME-CPP synthase; HDS—HMB-PP synthase; HDR—HMB-PP reductase; ERG10—ACCT acetyl-CoA C-acetyl transferase; ERG13—HMGS HMG-CoA synthase; HMGR—HMG-CoA reductase; ERG12—MK MVA kinase; ERG8—PMVK phosphomevalonate kinase; ERG19—MVD diphosphomevalonate decarboxylase; LiuC—enoyl-CoA hydratase; AibAB—glutaconyl-CoA decarboxylase; cbjALD—acyl-CoA reductase; YahK—alcohol dehydrogenase; ThiM—hydroxyethylthiazole kinase; IPK—isopentenyl phosphate kinase; IDI—isopentenyl-diphosphate isomerase; GPPS—geranyl pyrophosphate synthase; FPPS—farnesyl pyrophosphate synthase; GGPPS—geranylgeranyl diphosphate synthase; NPPS—nerol pyrophosphate synthase; zFPPS—Z,Z-Farnesyl diphosphate synthase; NNPPS—nerylneryl diphosphate synthase; M3K—mevalonate 3-kinase; M3P5K—mevalonate 3-phosphate 5-kinase; BMD—mevalonate biphosphate decarboxylase; IPK—isopentenyl phosphate kinase; PMD—mevalonate 5-phosphate decarboxylase.)
| 结构 | 化合物 | 气味特征 | 底盘菌株 | 培养条件 | 产量/(g/L) | 参考文献 |
|---|---|---|---|---|---|---|
| 链状单萜 | 香叶醇 Geraniol | 温和、甜香、花果香气 | 大肠杆菌 | 摇瓶发酵 | 2.1 | [ |
| 巴斯德毕赤酵母 | 24孔板发酵 | 1.2 | [ | |||
月桂烯 Myrcene | 甜香脂香气 | 大肠杆菌 | 摇瓶发酵 | 1.2 | [ | |
香茅醇 Citronellol | 甜润玫瑰花香 | 酿酒酵母 | 5 L生物反应器 | 8.3 | [ | |
芳樟醇 Linalool | 铃兰清香 | 菠萝潘托氏菌 | 5 mL试管 | S型5.6 R型3.7 | [ | |
| 单环单萜 | 柠檬烯 Limonene | 柠檬香气 | 大肠杆菌 | 3 L生物反应器 | 3.6 | [ |
α-松油醇 α-Terpineol | 丁香花气味 | 酿酒酵母 | 5 L生物反应器 | 21.8 | [ | |
左旋香芹酮 (-)-Carvone | 兰花香气、类清新薄荷气味 | 大肠杆菌 | 摇瓶发酵 柠檬烯为底物 | 44.3 | [ | |
| 双环单萜 | 龙脑 Borneol | 极强的樟脑和松木香气 | 酿酒酵母 | 摇瓶补料 | 0.75 | [ |
α-蒎烯 α-Pinene | 松木芳香,清新草本气味 | 大肠杆菌 | 摇瓶发酵 | 0.17 | [ | |
香桧烯 Sabinene | 湿泥土气味与木质香气 | 酿酒酵母 | 摇瓶发酵 | 0.15 | [ | |
| 链状倍半萜 | 橙花叔醇 Nerolidol | 清新柔和的木质香及花果香 | 酿酒酵母 | 摇瓶发酵 | 3.5 | [ |
β-法尼烯 β-Farnesol | 青草与柑橘混合的清新香气 | 酿酒酵母 | 200 t生物反应器 | 130 | [ | |
法尼醇 Farnesol | 温和的花香调 | 大肠杆菌 | 试管 | 0.53 | [ | |
| 单环倍半萜 | 红没药烯 Bisabolene | 果香与香脂香 | 酿酒酵母 | 5 L生物反应器 | 9.8 | [ |
α-葎草烯 α-Humulene | 丁香香型 | 热带念珠菌 | 30 L生物反应器 | 4.1 | [ | |
右旋大根香叶烯D (-)-Germacrene D | 辛辣的胡椒香气 | 酿酒酵母 | 5 L生物反应器 | 7.9 | [ | |
| 双环倍半萜 | 右旋瓦伦烯 (+)-Valencene | 愉悦的柑橘香 | 酿酒酵母 | 15 L生物反应器 | 16.6 | [ |
右旋诺卡酮 (+)-Nootkatone | 葡萄柚的香气 | 酿酒酵母 | 5 L生物反应器 | 0.80 | [ | |
檀香醇 Santalol | 柔和温暖的木质香型 | 酿酒酵母 | 5 L生物反应器 | 1.3 | [ | |
| 三环倍半萜 | α-檀香烯 α-Santalene | 温暖细腻的檀香木香气 | 巴斯德毕赤酵母 | 1 L生物反应器 | 21.5 | [ |
长叶烯 Longifolene | 鸢尾花与木质香 | 大肠杆菌 | 5 L生物反应器 | 0.38 | [ | |
广藿香醇 Patchoulol | 广藿芳香 | 酿酒酵母 | 5 L生物反应器 | 1.6 | [ | |
| 单环降异戊二烯 | α-紫罗酮 α-Ionone | 强烈的紫罗兰花与鸢尾根香 | 酿酒酵母 | 摇瓶补料 | 0.48 | [ |
β-紫罗酮 β-Ionone | 紫罗兰与玫瑰香,含木质香 | 解脂耶氏酵母 | 3 L生物反应器 | 0.98 | [ |
表1 经典的萜类香料细胞工厂汇总
Table 1 Summary of cell factories for producing terpene fragrances
| 结构 | 化合物 | 气味特征 | 底盘菌株 | 培养条件 | 产量/(g/L) | 参考文献 |
|---|---|---|---|---|---|---|
| 链状单萜 | 香叶醇 Geraniol | 温和、甜香、花果香气 | 大肠杆菌 | 摇瓶发酵 | 2.1 | [ |
| 巴斯德毕赤酵母 | 24孔板发酵 | 1.2 | [ | |||
月桂烯 Myrcene | 甜香脂香气 | 大肠杆菌 | 摇瓶发酵 | 1.2 | [ | |
香茅醇 Citronellol | 甜润玫瑰花香 | 酿酒酵母 | 5 L生物反应器 | 8.3 | [ | |
芳樟醇 Linalool | 铃兰清香 | 菠萝潘托氏菌 | 5 mL试管 | S型5.6 R型3.7 | [ | |
| 单环单萜 | 柠檬烯 Limonene | 柠檬香气 | 大肠杆菌 | 3 L生物反应器 | 3.6 | [ |
α-松油醇 α-Terpineol | 丁香花气味 | 酿酒酵母 | 5 L生物反应器 | 21.8 | [ | |
左旋香芹酮 (-)-Carvone | 兰花香气、类清新薄荷气味 | 大肠杆菌 | 摇瓶发酵 柠檬烯为底物 | 44.3 | [ | |
| 双环单萜 | 龙脑 Borneol | 极强的樟脑和松木香气 | 酿酒酵母 | 摇瓶补料 | 0.75 | [ |
α-蒎烯 α-Pinene | 松木芳香,清新草本气味 | 大肠杆菌 | 摇瓶发酵 | 0.17 | [ | |
香桧烯 Sabinene | 湿泥土气味与木质香气 | 酿酒酵母 | 摇瓶发酵 | 0.15 | [ | |
| 链状倍半萜 | 橙花叔醇 Nerolidol | 清新柔和的木质香及花果香 | 酿酒酵母 | 摇瓶发酵 | 3.5 | [ |
β-法尼烯 β-Farnesol | 青草与柑橘混合的清新香气 | 酿酒酵母 | 200 t生物反应器 | 130 | [ | |
法尼醇 Farnesol | 温和的花香调 | 大肠杆菌 | 试管 | 0.53 | [ | |
| 单环倍半萜 | 红没药烯 Bisabolene | 果香与香脂香 | 酿酒酵母 | 5 L生物反应器 | 9.8 | [ |
α-葎草烯 α-Humulene | 丁香香型 | 热带念珠菌 | 30 L生物反应器 | 4.1 | [ | |
右旋大根香叶烯D (-)-Germacrene D | 辛辣的胡椒香气 | 酿酒酵母 | 5 L生物反应器 | 7.9 | [ | |
| 双环倍半萜 | 右旋瓦伦烯 (+)-Valencene | 愉悦的柑橘香 | 酿酒酵母 | 15 L生物反应器 | 16.6 | [ |
右旋诺卡酮 (+)-Nootkatone | 葡萄柚的香气 | 酿酒酵母 | 5 L生物反应器 | 0.80 | [ | |
檀香醇 Santalol | 柔和温暖的木质香型 | 酿酒酵母 | 5 L生物反应器 | 1.3 | [ | |
| 三环倍半萜 | α-檀香烯 α-Santalene | 温暖细腻的檀香木香气 | 巴斯德毕赤酵母 | 1 L生物反应器 | 21.5 | [ |
长叶烯 Longifolene | 鸢尾花与木质香 | 大肠杆菌 | 5 L生物反应器 | 0.38 | [ | |
广藿香醇 Patchoulol | 广藿芳香 | 酿酒酵母 | 5 L生物反应器 | 1.6 | [ | |
| 单环降异戊二烯 | α-紫罗酮 α-Ionone | 强烈的紫罗兰花与鸢尾根香 | 酿酒酵母 | 摇瓶补料 | 0.48 | [ |
β-紫罗酮 β-Ionone | 紫罗兰与玫瑰香,含木质香 | 解脂耶氏酵母 | 3 L生物反应器 | 0.98 | [ |
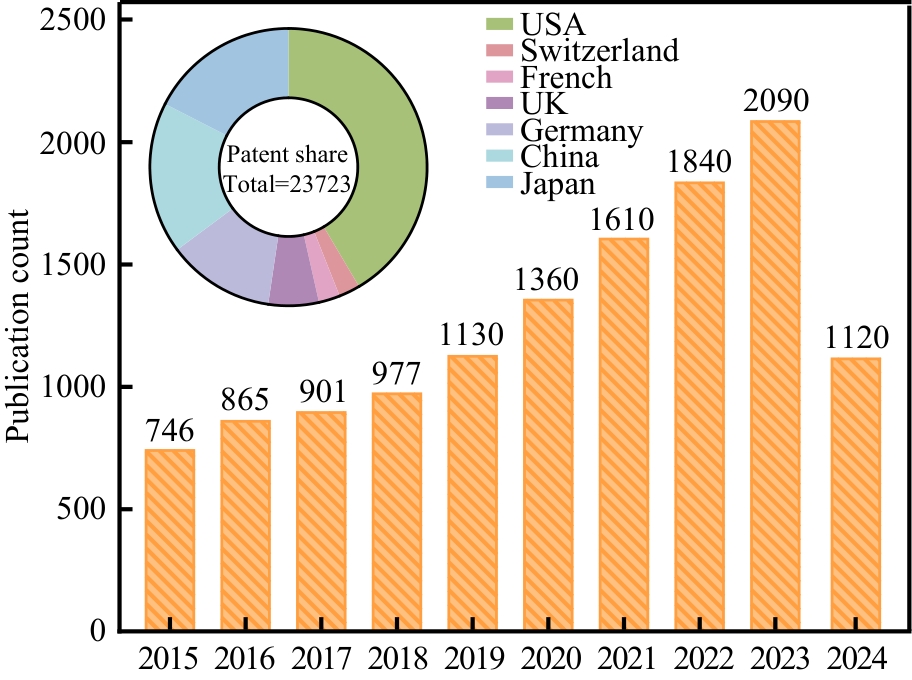
图7 全球十年内萜类香料生物合成相关文献与专利分布(The data were from Google Patents and Google Scholar using “terpene fragrances” and “synthetic biology” as the keywords, July 17th, 2024.)
Fig. 7 Global literatures and patents related to terpene fragrance biosynthesis that were publicized within the last decade
| 1 | BOM S, JORGE J, RIBEIRO H M, et al. A step forward on sustainability in the cosmetics industry: a review[J]. Journal of Cleaner Production, 2019, 225: 270-290. |
| 2 | EL-OTMANI N, ZEOUK I, HAMMANI O, et al. Analysis and quality control of bio-actives and herbal cosmetics: the case of traditional cooperatives from Fes-Meknes Region[J]. Tropical Journal of Natural Product Research, 2024, 8(5): 7181-7195. |
| 3 | MICHAILIDOU D F. The scent of change: sustainable fragrances through industrial biotechnology[J]. ChemBioChem, 2023, 24(19): e202300309. |
| 4 | WESTON-GREEN K, CLUNAS H, JIMENEZ NARANJO C. A review of the potential use of pinene and linalool as terpene-based medicines for brain health: discovering novel therapeutics in the flavours and fragrances of Cannabis [J]. Frontiers in Psychiatry, 2021, 12: 583211. |
| 5 | JIANG H, WANG X. Biosynthesis of monoterpenoid and sesquiterpenoid as natural flavors and fragrances[J]. Biotechnology Advances, 2023, 65: 108151. |
| 6 | ZHANG C B, LI M, ZHAO G R, et al. Harnessing yeast peroxisomes and cytosol acetyl-CoA for sesquiterpene α-humulene production[J]. Journal of Agricultural and Food Chemistry, 2020, 68(5): 1382-1389. |
| 7 | MASYITA A, MUSTIKA SARI R, ASTUTI A DWI, et al. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives[J]. Food Chemistry: X, 2022, 13: 100217. |
| 8 | CAO X, YU W, CHEN Y, et al. Engineering yeast for high-level production of diterpenoid sclareol[J]. Metabolic Engineering, 2023, 75: 19-28. |
| 9 | Linkedin. Terpenes Market Research Report: Company Snapshot 2031[EB/OL]. LinkedIn Pulse. [2024-03-01]. . |
| 10 | CAO C Y, CAO X, YU W, et al. Global metabolic rewiring of yeast enables overproduction of sesquiterpene (+)-valencene[J]. Journal of Agricultural and Food Chemistry, 2022, 70(23): 7180-7187. |
| 11 | VOIGT C A. Synthetic biology 2020—2030: six commercially-available products that are changing our world[J]. Nature Communications, 2020, 11(1): 6379. |
| 12 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age[J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 13 | VOLK M J, TRAN V G, TAN S I, et al. Metabolic engineering: methodologies and applications[J]. Chemical Reviews, 2023, 123(9): 5521-5570. |
| 14 | CIMERMANCIC P, MEDEMA M H, CLAESEN J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters[J]. Cell, 2014, 158(2): 412-421. |
| 15 | BLIN K, SHAW S, AUGUSTIJN H E, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation[J]. Nucleic Acids Research, 2023, 51(W1): W46-W50. |
| 16 | SKINNIDER M A, JOHNSTON C W, GUNABALASINGAM M, et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences[J]. Nature Communications, 2020, 11(1): 6058. |
| 17 | WANG X R, CHEN N X, CRUZ-MORALES P, et al. Elucidation of genes enhancing natural product biosynthesis through co-evolution analysis[J]. Nature Metabolism, 2024, 6(5): 933-946. |
| 18 | ARKIN A P, COTTINGHAM R W, HENRY C S, et al. KBase: the United States Department of Energy systems biology knowledgebase[J]. Nature Biotechnology, 2018, 36(7): 566-569. |
| 19 | SEAVER S M D, LIU F, ZHANG Q Z, et al. The ModelSEED Biochemistry Database for the integration of metabolic annotations and the reconstruction, comparison and analysis of metabolic models for plants, fungi and microbes[J]. Nucleic Acids Research, 2021, 49(D1): D575-D588. |
| 20 | AGREN R, LIU L M, SHOAIE S, et al. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum [J]. PLoS Computational Biology, 2013, 9(3): e1002980. |
| 21 | BURGARD A P, PHARKYA P, MARANAS C D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization[J]. Biotechnology and Bioengineering, 2003, 84(6): 647-657. |
| 22 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 23 | ABRAMSON J, ADLER J, DUNGER J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 630(8016): 493-500. |
| 24 | GREENING C, CABOTAJE P R, VALENTIN ALVARADO L E, et al. Minimal and hybrid hydrogenases are active from Archaea[J]. Cell, 2024, 187(13): 3357-3372. e19. |
| 25 | TRIVEDI V, RAMESH A, WHEELDON I. Analyzing CRISPR screens in non-conventional microbes[J]. Journal of Industrial Microbiology & Biotechnology, 2023, 50(1): kuad006. |
| 26 | LIU X Q, CUI Z Y, SU T Y, et al. Identification of genome integration sites for developing a CRISPR-based gene expression toolkit in Yarrowia lipolytica [J]. Microbial Biotechnology, 2022, 15(8): 2223-2234. |
| 27 | HOLKENBRINK C, DAM M I, KILDEGAARD K R, et al. EasyCloneYALI: CRISPR/Cas9-based synthetic toolbox for engineering of the yeast Yarrowia lipolytica [J]. Biotechnology Journal, 2018, 13(9): 1700543. |
| 28 | CUI Z Y, JIANG X, ZHENG H H, et al. Homology-independent genome integration enables rapid library construction for enzyme expression and pathway optimization in Yarrowia lipolytica [J]. Biotechnology and Bioengineering, 2019, 116(2): 354-363. |
| 29 | LIU Q, SHI X N, SONG L L, et al. CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris [J]. Microbial Cell Factories, 2019, 18(1): 144. |
| 30 | CAI P, DUAN X P, WU X Y, et al. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris [J]. Nucleic Acids Research, 2021, 49(13): 7791-7805. |
| 31 | GAO J C, JIANG L H, LIAN J Z. Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products[J]. Synthetic and Systems Biotechnology, 2021, 6(2): 110-119. |
| 32 | GAO J Q, GAO N, ZHAI X X, et al. Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha [J]. iScience, 2021, 24(3): 102168. |
| 33 | YU W, GAO J Q, ZHAI X X, et al. Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha [J]. Synthetic and Systems Biotechnology, 2021, 6(2): 63-68. |
| 34 | FATMA Z, TAN S I, BOOB A G, et al. A landing pad system for multicopy gene integration in Issatchenkia orientalis [J]. Metabolic Engineering, 2023, 78: 200-208. |
| 35 | KIM S K, HAN G H, SEONG W, et al. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production[J]. Metabolic Engineering, 2016, 38: 228-240. |
| 36 | LIAN J Z, HAMEDIRAD M, HU S M, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system[J]. Nature Communications, 2017, 8(1): 1688. |
| 37 | DE MUNTER S, VAN PARYS A, BRAL L, et al. Rapid and effective generation of nanobody based CARs using PCR and Gibson assembly[J]. International Journal of Molecular Sciences, 2020, 21(3): 883. |
| 38 | SORIDA M, BONASIO R. An efficient cloning method to expand vector and restriction site compatibility of Golden Gate Assembly[J]. Cell Reports Methods, 2023, 3(8): 100564. |
| 39 | MA Y, SU S X, FU Z H, et al. Convenient synthesis and delivery of a megabase-scale designer accessory chromosome empower biosynthetic capacity[J]. Cell Research, 2024, 34(4): 309-322. |
| 40 | CARBONELL P, RADIVOJEVIC T, GARCÍA MARTÍN H. Opportunities at the intersection of synthetic biology, machine learning, and automation[J]. ACS Synthetic Biology, 2019, 8(7): 1474-1477. |
| 41 | IGNEA C, RAADAM M H, MOTAWIA M S, et al. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate[J]. Nature Communications, 2019, 10(1): 3799. |
| 42 | LIPKO A, PĄCZKOWSKI C, PEREZ-FONS L, et al. Divergent contribution of the MVA and MEP pathways to the formation of polyprenols and dolichols in Arabidopsis [J]. Biochemical Journal, 2023, 480(8): 495-520. |
| 43 | PAN X M, DU W Y, ZHANG X W, et al. Discovery, structure, and mechanism of a class Ⅱ sesquiterpene cyclase[J]. Journal of the American Chemical Society, 2022, 144(48): 22067-22074. |
| 44 | ZHOU F, PICHERSKY E. More is better: the diversity of terpene metabolism in plants[J]. Current Opinion in Plant Biology, 2020, 55: 1-10. |
| 45 | ZENG L P, DEHESH K. The eukaryotic MEP-pathway genes are evolutionarily conserved and originated from Chlaymidia and cyanobacteria[J]. BMC Genomics, 2021, 22(1): 137. |
| 46 | ALLAMAND A, PIECHOWIAK T, LIÈVREMONT D, et al. The multifaceted MEP pathway: towards new therapeutic perspectives[J]. Molecules, 2023, 28(3): 1403. |
| 47 | BANERJEE A, WU Y, BANERJEE R, et al. Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway[J]. Journal of Biological Chemistry, 2013, 288(23): 16926-16936. |
| 48 | DIAO J J, SONG X Y, ZHANG L, et al. Tailoring cyanobacteria as a new platform for highly efficient synthesis of astaxanthin[J]. Metabolic Engineering, 2020, 61: 275-287. |
| 49 | VOLKE D C, ROHWER J, FISCHER R, et al. Investigation of the methylerythritol 4-phosphate pathway for microbial terpenoid production through metabolic control analysis[J]. Microbial Cell Factories, 2019, 18(1): 192. |
| 50 | DU F L, YU H L, XU J H, et al. Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli [J]. Bioresources and Bioprocessing, 2014, 1(1): 10. |
| 51 | MA Y W, MCCLURE D D, SOMERVILLE M V, et al. Metabolic engineering of the MEP pathway in Bacillus subtilis for increased biosynthesis of menaquinone-7[J]. ACS Synthetic Biology, 2019, 8(7): 1620-1630. |
| 52 | BRÖKER J N, MÜLLER B, VAN DEENEN N, et al. Upregulating the mevalonate pathway and repressing sterol synthesis in Saccharomyces cerevisiae enhances the production of triterpenes[J]. Applied Microbiology and Biotechnology, 2018, 102(16): 6923-6934. |
| 53 | LIU F, LIU S C, QI Y K, et al. Enhancing trans-nerolidol productivity in Yarrowia lipolytica by improving precursor supply and optimizing nerolidol synthase activity[J]. Journal of Agricultural and Food Chemistry, 2022, 70(48): 15157-15165. |
| 54 | MUKHERJEE M, BLAIR R H, WANG Z Q. Machine-learning guided elucidation of contribution of individual steps in the mevalonate pathway and construction of a yeast platform strain for terpenoid production[J]. Metabolic Engineering, 2022, 74: 139-149. |
| 55 | RINALDI M A, FERRAZ C A, SCRUTTON N S. Alternative metabolic pathways and strategies to high-titre terpenoid production in Escherichia coli [J]. Natural Product Reports, 2022, 39(1): 90-118. |
| 56 | CHEN H L, LIU C Q, LI M J, et al. Directed evolution of mevalonate kinase in Escherichia coli by random mutagenesis for improved lycopene[J]. RSC Advances, 2018, 8(27): 15021-15028. |
| 57 | CAO C Y, ZHANG H Y, CAO X, et al. Construction and optimization of nonclassical isoprenoid biosynthetic pathways in yeast peroxisomes for (+)-valencene production[J]. Journal of Agricultural and Food Chemistry, 2023, 71(29): 11124-11130. |
| 58 | CLOMBURG J M, QIAN S, TAN Z G, et al. The isoprenoid alcohol pathway, a synthetic route for isoprenoid biosynthesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(26): 12810-12815. |
| 59 | MA Y S, ZU Y X, HUANG S W, et al. Engineering a universal and efficient platform for terpenoid synthesis in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(1): e2207680120. |
| 60 | LIU S S, ZHANG M Y, REN Y Y, et al. Engineering Rhodosporidium toruloides for limonene production[J]. Biotechnology for Biofuels, 2021, 14(1): 243. |
| 61 | SCHILMILLER A L, SCHAUVINHOLD I, LARSON M, et al. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(26): 10865-10870. |
| 62 | 程晓雷, 刘天罡, 陶慧. 萜类化合物的非常规生物合成研究进展[J]. 合成生物学, 2024, 5(5): 1050-1071. |
| CHENG X L, LIU T G, TAO H. Recent research progress in non-canonical biosynthesis of terpenoids[J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. | |
| 63 | LUO P, HUANG J H, LÜ J M, et al. Biosynthesis of fungal terpenoids[J]. Natural Product Reports, 2024, 41(5): 748-783. |
| 64 | LI Z, ZHANG L L, XU K W, et al. Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase[J]. Nature Communications, 2023, 14(1): 4001. |
| 65 | ABE T, SHIRATORI H, KASHIWAZAKI K, et al. Structural-model-based genome mining can efficiently discover novel non-canonical terpene synthases hidden in genomes of diverse species[J]. Chemical Science, 2024, 15(27): 10402-10407. |
| 66 | DENG X M, YE Z L, DUAN J Y, et al. Complete pathway elucidation and heterologous reconstitution of (+)-nootkatone biosynthesis from Alpinia oxyphylla [J]. New Phytologist, 2024, 241(2): 779-792. |
| 67 | CHEN R, JIA Q D, MU X, et al. Systematic mining of fungal chimeric terpene synthases using an efficient precursor-providing yeast chassis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(29): e2023247118. |
| 68 | YANG D Y, ZHANG S S, MA K, et al. Discovery and characterization of a new family of diterpene cyclases in bacteria and fungi[J]. Angewandte Chemie, 2017, 129(17): 4827-4830. |
| 69 | DALETOS G, STEPHANOPOULOS G. Protein engineering strategies for microbial production of isoprenoids[J]. Metabolic Engineering Communications, 2020, 11: e00129. |
| 70 | YE Z L, HUANG Y L, SHI B, et al. Coupling cell growth and biochemical pathway induction in Saccharomyces cerevisiae for production of (+)-valencene and its chemical conversion to (+)-nootkatone[J]. Metabolic Engineering, 2022, 72: 107-115. |
| 71 | 祁延萍, 朱晋, 张凯, 等. 定向进化在蛋白质工程中的应用研究进展[J]. 合成生物学, 2022, 3(6): 1081-1108. |
| QI Y P, ZHU J, ZHANG K, et al. Recent development of directed evolution in protein engineering[J]. Synthetic Biology Journal, 2022, 3(6): 1081-1108. | |
| 72 | LAUCHLI D R, RABE D K S, KALBARCZYK K Z, et al. High-throughput screening for terpene-synthase-cyclization activity and directed evolution of a terpene synthase[J]. Angewandte Chemie International Edition, 2013, 52(21): 5571-5574. |
| 73 | WANG X, CHEN J M, ZHANG J, et al. Engineering Escherichia coli for production of geraniol by systematic synthetic biology approaches and laboratory-evolved fusion tags[J]. Metabolic Engineering, 2021, 66: 60-67. |
| 74 | YE C F, LI M X, GAO J C, et al. Metabolic engineering of Pichia pastoris for overproduction of cis-trans nepetalactol[J]. Metabolic Engineering, 2024, 84: 83-94. |
| 75 | WANG X, WANG J J, ZHANG X Y, et al. Efficient myrcene production using linalool dehydratase isomerase and rational biochemical process in Escherichia coli [J]. Journal of Biotechnology, 2023, 371: 33-40. |
| 76 | JIANG G Z, YAO M D, WANG Y, et al. A “push-pull-restrain” strategy to improve citronellol production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2021, 66: 51-59. |
| 77 | HOSHINO Y, MORIYA M, MATSUDAIRA A, et al. Stereospecific linalool production utilizing two-phase cultivation system in Pantoea ananatis [J]. Journal of Biotechnology, 2020, 324: 21-27. |
| 78 | ROLF J, JULSING M K, ROSENTHAL K, et al. A gram-scale limonene production process with engineered Escherichia coli [J]. Molecules, 2020, 25(8): 1881. |
| 79 | ZHANG C B, LI M, ZHAO G R, et al. α-Terpineol production from an engineered Saccharomyces cerevisiae cell factory[J]. Microbial Cell Factories, 2019, 18(1): 160. |
| 80 | YOSHIDA E, KOJIMA M, SUZUKI M, et al. Increased carvone production in Escherichia coli by balancing limonene conversion enzyme expression via targeted quantification concatamer proteome analysis[J]. Scientific Reports, 2021, 11(1): 22126. |
| 81 | ZHANG H Y, CAI P, GUO J, et. al . Engineering cellular dephosphorylation boosts (+)-borneol production in yeast[J]. Acta Pharmaceutica Sinica B. Unpublished. |
| 82 | NIU F X, HE X, WU Y Q, et al. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering[J]. Frontiers in Microbiology, 2018, 9: 1623. |
| 83 | JIA H J, CHEN T H, QU J Z, et al. Collaborative subcellular compartmentalization to improve GPP utilization and boost sabinene accumulation in Saccharomyces cerevisiae [J]. Biochemical Engineering Journal, 2020, 164: 107768. |
| 84 | LU Z Y, PENG B Y, EBERT B E, et al. Auxin-mediated protein depletion for metabolic engineering in terpene-producing yeast[J]. Nature Communications, 2021, 12(1): 1051. |
| 85 | MEADOWS A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production[J]. Nature, 2016, 537(7622): 694-697. |
| 86 | WANG C L, PARK J E, CHOI E S, et al. Farnesol production in Escherichia coli through the construction of a farnesol biosynthesis pathway-application of PgpB and YbjG phosphatases[J]. Biotechnology Journal, 2016, 11(10): 1291-1297. |
| 87 | ZHANG W X, GUO J Q, WANG Z, et al. Improved production of germacrene A, a direct precursor of β-elemene, in engineered Saccharomyces cerevisiae by expressing a cyanobacterial germacrene A synthase[J]. Microbial Cell Factories, 2021, 20(1): 7. |
| 88 | ZHANG L H, YANG H Q, XIA Y Y, et al. Engineering the oleaginous yeast Candida tropicalis for α-humulene overproduction[J]. Biotechnology for Biofuels and Bioproducts, 2022, 15(1): 59. |
| 89 | LIU J J, CHEN C, WAN X K, et al. Identification of the sesquiterpene synthase AcTPS1 and high production of (–)-germacrene D in metabolically engineered Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2022, 21(1): 89. |
| 90 | GUO J Y, ZHOU W, LI Y Y, et al. Combination of protein engineering and metabolic engineering to enhance (+)-nootkatone production in Saccharomyces cerevisiae [J]. Food Bioengineering, 2022, 1(2): 192-202. |
| 91 | ZHA W L, AN T Y, LI T, et al. Reconstruction of the biosynthetic pathway of santalols under control of the GAL regulatory system in yeast[J]. ACS Synthetic Biology, 2020, 9(2): 449-456. |
| 92 | ZUO Y M, XIAO F, GAO J C, et al. Establishing Komagataella phaffii as a cell factory for efficient production of sesquiterpenoid α-santalene[J]. Journal of Agricultural and Food Chemistry, 2022, 70(26): 8024-8031. |
| 93 | CAO Y J, ZHANG R B, LIU W, et al. Manipulation of the precursor supply for high-level production of longifolene by metabolically engineered Escherichia coli [J]. Scientific Reports, 2019, 9(1): 95. |
| 94 | LIU M, LIN Y C, GUO J J, et al. High-level production of sesquiterpene patchoulol in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2021, 10(1): 158-172. |
| 95 | ZHANG C Q, CHEN X X, LINDLEY N D, et al. A “plug-n-play” modular metabolic system for the production of apocarotenoids[J]. Biotechnology and Bioengineering, 2018, 115(1): 174-183. |
| 96 | LU Y P, YANG Q Y, LIN Z L, et al. A modular pathway engineering strategy for the high-level production of β-ionone in Yarrowia lipolytica [J]. Microbial Cell Factories, 2020, 19(1): 49. |
| 97 | MAGNARD J L, ROCCIA A, CAISSARD J C, et al. Biosynthesis of monoterpene scent compounds in roses[J]. Science, 2015, 349(6243): 81-83. |
| 98 | ZHU K, KONG J, ZHAO B X, et al. Metabolic engineering of microbes for monoterpenoid production[J]. Biotechnology Advances, 2021, 53: 107837. |
| 99 | JIANG G Z, YAO M D, WANG Y, et al. Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2017, 41: 57-66. |
| 100 | CAO X, LÜ Y B, CHEN J, et al. Metabolic engineering of oleaginous yeast Yarrowia lipolytica for limonene overproduction[J]. Biotechnology for Biofuels, 2016, 9: 214. |
| 101 | JONGEDIJK E, CANKAR K, RANZIJN J, et al. Capturing of the monoterpene olefin limonene produced in Saccharomyces cerevisiae [J]. Yeast, 2015, 32(1): 159-171. |
| 102 | ZHAO Y, LIANG F Y, XIE Y M, et al. Oxetane ring formation in taxol biosynthesis is catalyzed by a bifunctional cytochrome P450 enzyme[J]. Journal of the American Chemical Society, 2024, 146(1): 801-810. |
| 103 | YUAN W, LÜ S, CHEN L Y, et al. Production of sesterterpene ophiobolin by a bifunctional terpene synthase in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2019, 103(21-22): 8785-8797. |
| 104 | TANG N C, SU J C, SHMIDOV Y, et al. Synthetic intrinsically disordered protein fusion tags that enhance protein solubility[J]. Nature Communications, 2024, 15(1): 3727. |
| 105 | CHEAH L C, LIU L, STARK T, et al. Metabolic flux enhancement from the translational fusion of terpene synthases is linked to terpene synthase accumulation[J]. Metabolic Engineering, 2023, 77: 143-151. |
| 106 | GUO Q, SHI T Q, PENG Q Q, et al. Harnessing Yarrowia lipolytica peroxisomes as a subcellular factory for α-humulene overproduction[J]. Journal of Agricultural and Food Chemistry, 2021, 69(46): 13831-13837. |
| 107 | MILLER B R, KUNG Y. Structural features and domain movements controlling substrate binding and cofactor specificity in class Ⅱ HMG-CoA reductase[J]. Biochemistry, 2018, 57(5): 654-662. |
| 108 | KWAK S, KIM S R, XU H Q, et al. Enhanced isoprenoid production from xylose by engineered Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2017, 114(11): 2581-2591. |
| 109 | KIM T Y, PARK H, KIM S K, et al. Production of (-)-α-bisabolol in metabolically engineered Saccharomyces cerevisiae [J]. Journal of Biotechnology, 2021, 340: 13-21. |
| 110 | YUZBASHEVA E Y, AGRIMI G, YUZBASHEV T V, et al. The mitochondrial citrate carrier in Yarrowia lipolytica: its identification, characterization and functional significance for the production of citric acid[J]. Metabolic Engineering, 2019, 54: 264-274. |
| 111 | RODRIGUEZ S, DENBY C M, VAN VU T, et al. ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2016, 15: 48. |
| 112 | HELLGREN J, GODINA A, NIELSEN J, et al. Promiscuous phosphoketolase and metabolic rewiring enables novel non-oxidative glycolysis in yeast for high-yield production of acetyl-CoA derived products[J]. Metabolic Engineering, 2020, 62: 150-160. |
| 113 | YE M, GAO J Q, LI J J, et al. Promoter engineering enables precise metabolic regulation towards efficient β-elemene production in Ogataea polymorpha [J]. Synthetic and Systems Biotechnology, 2024, 9(2): 234-241. |
| 114 | CHEN Y, DAVIET L, SCHALK M, et al. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism[J]. Metabolic Engineering, 2013, 15: 48-54. |
| 115 | LI Q Y, FAN F Y, GAO X, et al. Balanced activation of IspG and IspH to eliminate MEP intermediate accumulation and improve isoprenoids production in Escherichia coli [J]. Metabolic Engineering, 2017, 44: 13-21. |
| 116 | WU T, LI S W, ZHANG B L, et al. Engineering Saccharomyces cerevisiae for the production of the valuable monoterpene ester geranyl acetate[J]. Microbial Cell Factories, 2018, 17(1): 85. |
| 117 | DUSSÉAUX S, WAJN W T, LIU Y X, et al. Transforming yeast peroxisomes into microfactories for the efficient production of high-value isoprenoids[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(50): 31789-31799. |
| 118 | ZHAO J Z, LI C, ZHANG Y, et al. Dynamic control of ERG20 expression combined with minimized endogenous downstream metabolism contributes to the improvement of geraniol production in Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2017, 16(1): 17. |
| 119 | ZHAO B X, ZHANG Y H, WANG Y P, et al. Biosynthesis of α-bisabolene from low-cost renewable feedstocks by peroxisome engineering and systems metabolic engineering of the yeast Yarrowia lipolytica [J]. Green Chemistry, 2023, 25(20): 8145-8159. |
| 120 | SON S H, KIM J E, PARK G R, et al. Metabolite trafficking enables membrane-impermeable-terpene secretion by yeast[J]. Nature Communications, 2022, 13(1): 2605. |
| 121 | QIN L, MA D S, LIN G Y, et al. Low temperature promotes the production and efflux of terpenoids in yeast[J]. Bioresource Technology, 2024, 395: 130376. |
| 122 | CHENG T, ZHANG K, GUO J, et al. Highly efficient biosynthesis of β-caryophyllene with a new sesquiterpene synthase from tobacco[J]. Biotechnology for Biofuels and Bioproducts, 2022, 15(1): 39. |
| 123 | SCANDIFFIO R, GEDDO F, COTTONE E, et al. Protective effects of (E)-β-caryophyllene (BCP) in Chronic inflammation [J]. Nutrients, 2020, 12(11): 3273. |
| 124 | TANG Q H, XU F, WEI X C, et al. Investigation of β-caryophyllene as terpene penetration enhancer: role of stratum corneum retention[J]. European Journal of Pharmaceutical Sciences, 2023, 183: 106401. |
| 125 | LIM H S, KIM S K, WOO S G, et al. (–)-α-Bisabolol production in engineered Escherichia coli expressing a novel (-)-α-bisabolol synthase from the globe artichoke Cynara cardunculus var. Scolymus [J]. Journal of Agricultural and Food Chemistry, 2021, 69(30): 8492-8503. |
| 126 | EDDIN L B, JHA N K, GOYAL S N, et al. Health benefits, pharmacological effects, molecular mechanisms, and therapeutic potential of α-bisabolol[J]. Nutrients, 2022, 14(7): 1370. |
| 127 | JIANG Y K, XIA L, GAO S, et al. Engineering Saccharomyces cerevisiae for enhanced (–)-α-bisabolol production[J]. Synthetic and Systems Biotechnology, 2023, 8(2): 187-195. |
| 128 | LIU S C, LIU Z J, WEI L J, et al. Pathway engineering and medium optimization for α-farnesene biosynthesis in oleaginous yeast Yarrowia lipolytica [J]. Journal of Biotechnology, 2020, 319: 74-81. |
| 129 | LI S, LUO S S, YIN X R, et al. Screening of ent-copalyl diphosphate synthase and metabolic engineering to achieve de novo biosynthesis of ent-copalol in Saccharomyces cerevisiae [J]. Synthetic and Systems Biotechnology, 2024, 9(4): 784-792. |
| 130 | ZHANG X Y, CHEN S T, LIN Y, et al. Metabolic engineering of Pichia pastoris for high-level production of lycopene[J]. ACS Synthetic Biology, 2023, 12(10): 2961-2972. |
| 131 | YE M, GAO J Q, ZHOU Y J. Global metabolic rewiring of the nonconventional yeast Ogataea polymorpha for biosynthesis of the sesquiterpenoid β-elemene[J]. Metabolic Engineering, 2023, 76: 225-231. |
| 132 | LUO G J, LIN Y, CHEN S T, et al. Overproduction of patchoulol in metabolically engineered Komagataella phaffii [J]. Journal of Agricultural and Food Chemistry, 2023, 71(4): 2049-2058. |
| 133 | WEI Y, MENG N, WANG Y C, et al. Transcription factor VvWRKY70 inhibits both norisoprenoid and flavonol biosynthesis in grape[J]. Plant Physiology, 2023, 193(3): 2055-2070. |
| 134 | MOSAFERI S, JELLEY R E, FEDRIZZI B, et al. Synthesis of d6-deuterated analogues of aroma molecules-β-damascenone, β-damascone and safranal[J]. Results in Chemistry, 2022, 4: 100264. |
| 135 | TOMASINO E, BOLMAN S. The potential effect of β-ionone and β-damascenone on sensory perception of pinot noir wine aroma[J]. Molecules, 2021, 26(5): 1288. |
| 136 | CHUDASAMA D. Importance of intellectual property rights[J]. Journal of Intellectual Property Rights Law, 2022, 4(2): 2582-9742. |
| 137 | 陈大明, 周光明, 刘晓, 等. 从全球专利分析看合成生物学技术发展趋势[J]. 合成生物学, 2020, 1(3): 372-384. |
| CHEN D M, ZHOU G M, LIU X, et al. Analysis of global patents for the trend of synthetic biology inventions[J]. Synthetic Biology Journal, 2020, 1(3): 372-384. | |
| 138 | WANG S H, SUN X, HAN Y Q, et al. Sustainable biosynthesis of squalene from waste cooking oil by the yeast Yarrowia lipolytica [J]. Metabolic Engineering Communications, 2024, 18: e00240. |
| 139 | PANG Y R, ZHAO Y K, LI S L, et al. Engineering the oleaginous yeast Yarrowia lipolytica to produce limonene from waste cooking oil[J]. Biotechnology for Biofuels, 2019, 12: 241. |
| 140 | ZHAO Y K, ZHU K, LI J, et al. High-efficiency production of bisabolene from waste cooking oil by metabolically engineered Yarrowia lipolytica [J]. Microbial Biotechnology, 2021, 14(6): 2497-2513. |
| [1] | 吴柯, 罗家豪, 李斐然. 机器学习驱动的基因组规模代谢模型构建与优化[J]. 合成生物学, 2025, 6(3): 566-584. |
| [2] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [3] | 章益蜻, 刘高雯. 合成生物学视角下的基因功能探索与酵母工程菌株文库构建[J]. 合成生物学, 2025, 6(3): 685-700. |
| [4] | 黄怡, 司同, 陆安静. 生物制造标准体系建设的现状、问题与建议[J]. 合成生物学, 2025, 6(3): 701-714. |
| [5] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [6] | 高琪, 肖文海. 酵母合成单萜类化合物的研究进展[J]. 合成生物学, 2025, 6(2): 357-372. |
| [7] | 张璐鸥, 徐丽, 胡晓旭, 杨滢. 合成生物学助力化妆品走进生物制造新时代[J]. 合成生物学, 2025, 6(2): 479-491. |
| [8] | 鲁锦畅, 武耀康, 吕雪芹, 刘龙, 陈坚, 刘延峰. 神经酰胺类鞘脂的绿色生物制造[J]. 合成生物学, 2025, 6(2): 422-444. |
| [9] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| [10] | 韦灵珍, 王佳, 孙新晓, 袁其朋, 申晓林. 黄酮类化合物生物合成及其在化妆品中应用的研究[J]. 合成生物学, 2025, 6(2): 373-390. |
| [11] | 肖森, 胡立涛, 石智诚, 王发银, 余思婷, 堵国成, 陈坚, 康振. 可控分子量透明质酸的生物合成研究进展[J]. 合成生物学, 2025, 6(2): 445-460. |
| [12] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [13] | 左一萌, 张姣姣, 连佳长. 酿酒酵母使能技术在化妆品原料合成中的应用[J]. 合成生物学, 2025, 6(2): 233-253. |
| [14] | 汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478. |
| [15] | 郭婷婷, 韩湘凝, 黄熙婷, 张婷婷, 孔健. 乳酸菌的合成生物学工具及在合成益肤因子中的应用[J]. 合成生物学, 2025, 6(2): 320-333. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||