合成生物学 ›› 2023, Vol. 4 ›› Issue (3): 422-443.DOI: 10.12211/2096-8280.2023-004
基于人工智能和计算生物学的合成生物学元件设计
王晟1, 王泽琛1,2, 陈威华1, 陈珂1, 彭向达1, 欧发芬1, 郑良振1,3, 孙瑨原1,4, 沈涛1, 赵国屏3
- 1.上海智峪生物科技有限公司,上海 200030
2.山东大学,山东 济南 250100
3.中国科学院深圳先进技术研究院,广东 深圳 518055
4.中国科学院微生物研究所,北京 100101
-
收稿日期:2023-01-11修回日期:2023-04-03出版日期:2023-06-30发布日期:2023-07-05 -
通讯作者:王晟 -
作者简介:王晟 (1983—),男,博士,上海智峪生物科技有限公司CEO,中国科学院深圳先进技术研究院客座研究员。研究方向为基于深度学习的蛋白质结构预测、基于人工智能的合成生物学。 E-mail:wangsheng@zelixir.com王泽琛 (1997—),男,博士研究生。研究方向为基于深度学习的蛋白质-配体相互作用预测和虚拟筛选。 E-mail:wangzechen@mail.sdu.edu.cn陈威华 (1982—),男,博士研究生。研究方向为合成生物学方向,基因合成与组装。 E-mail:chenweihua@zelixir.com -
基金资助:中国科学院国际大科学计划培育专项(153D31KYSB20170121)
Design of synthetic biology components based on artificial intelligence and computational biology
WANG Sheng1, WANG Zechen1,2, CHEN Weihua1, CHEN Ke1, PENG Xiangda1, OU Fafen1, ZHENG Liangzhen1,3, SUN Jinyuan1,4, SHEN Tao1, ZHAO Guoping3
- 1.Shanghai Zelixir Biotech Company Ltd. ,Shanghai 200030,China
2.Shandong University,Jinan 250100,Shandong,China
3.Shenzhen Institute of Advanced Technology,Chinese Academey of Sciences,Shenzhen 518055,Guangdong,China
4.Institute of Microbiology,Chinese Academey of Sciences,Beijing 100101,China
-
Received:2023-01-11Revised:2023-04-03Online:2023-06-30Published:2023-07-05 -
Contact:WANG Sheng
摘要:
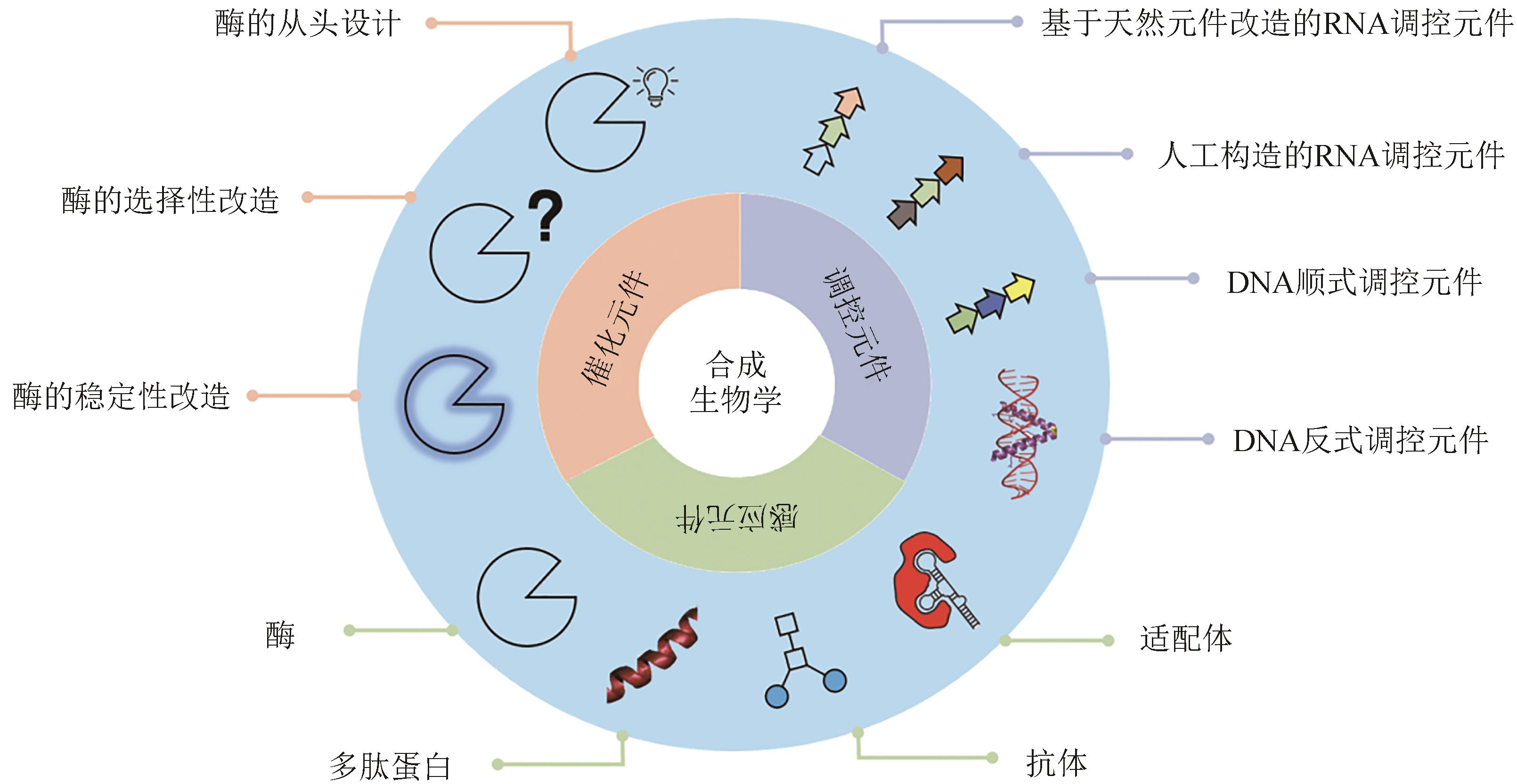
合成生物学是按照一定的规律综合已有的信息设计和构建全新的生物元件、装置和系统,或者重新设计已有的天然生物系统。合成生物学的核心在于设计、改造、重建或制造生物元件、生物反应系统、代谢途径与过程,乃至创造具有生命活动能力的细胞和生物个体,为解决人类发展在环境、资源、能源等方面面临的若干重大挑战提供新技术方案。毫无疑问,从DNA重组到基因电路设计,合成生物学的发展为众多领域带来全新的解决方案,优良的催化与调控元件是设计高效、鲁棒的系统的基础。然而,合成生物学的元件通常是天然的生物大分子,其固有的复杂性限制了对其工程化改造,导致合成生物技术的潜力未能得到充分发掘。随着人工智能(artificial intelligence,AI)与计算生物学的兴起和发展,有望助力该技术更好地发挥其价值。本文主要介绍了基于AI与计算生物学的不同类型的元件设计,聚焦催化元件、调控元件、传感元件三类元件的设计和前沿进展以及生物元件改造在合成生物学研究领域中的应用方面的研究进展。
中图分类号:
引用本文
王晟, 王泽琛, 陈威华, 陈珂, 彭向达, 欧发芬, 郑良振, 孙瑨原, 沈涛, 赵国屏. 基于人工智能和计算生物学的合成生物学元件设计[J]. 合成生物学, 2023, 4(3): 422-443.
WANG Sheng, WANG Zechen, CHEN Weihua, CHEN Ke, PENG Xiangda, OU Fafen, ZHENG Liangzhen, SUN Jinyuan, SHEN Tao, ZHAO Guoping. Design of synthetic biology components based on artificial intelligence and computational biology[J]. Synthetic Biology Journal, 2023, 4(3): 422-443.
| 名称 | 地址 | 用途与原理 | 参考文献 |
|---|---|---|---|
| FireProt | https://loschmidt.chemi.muni.cz/fireprotweb/ | 通过结合进化的保守性和基于结构的能量计算,进行多点突变稳定性设计 | [ |
| GRAPE-WEB | https://nmdc.cn/grape-web/ | 结合基于物理能量和统计能量的蛋白质设计力场,进行单点突变稳定性设计,利用结构特征和实验结果使用机器学习算法组合突变分类 | — |
| PROSS | https://pross.weizmann.ac.il/step/pross-terms/ | 基于Rosetta能量函数,设计可以提高稳定性的组合突变 | [ |
| Funclib | https://funclib.weizmann.ac.il/bin/steps | 通过单点突变在进化中的概率和结构计算的能量筛选用于组合突变的候选,使用Rosetta能量函数评价组合突变的稳定性,设计突变组合突变改变底物谱、改善可溶性表达 | [ |
| ABACUS2 | https://biocomp.ustc.edu.cn/servers/abacus-design.php | 基于从蛋白质晶体结构中统计得到的能量函数,进行单点突变能量计算和序列骨架适配度打分 | [ |
| Swiss-Model | https://swissmodel.expasy.org/ | 基于序列搜索,以同源的晶体结构为模版,进行同源建模 | [ |
| ConSurf | https://consurf.tau.ac.il/consurf_index.php | 基于序列搜索算法,分析保守性 | [ |
| ROSIE | https://rosie.graylab.jhu.edu/ | 基于Rosetta的结构relax,分子对接,突变预测等一系列任务 | [ |
表1 基于计算生物工具的在线服务器
Table 1 Online servers based on computational biology tools
| 名称 | 地址 | 用途与原理 | 参考文献 |
|---|---|---|---|
| FireProt | https://loschmidt.chemi.muni.cz/fireprotweb/ | 通过结合进化的保守性和基于结构的能量计算,进行多点突变稳定性设计 | [ |
| GRAPE-WEB | https://nmdc.cn/grape-web/ | 结合基于物理能量和统计能量的蛋白质设计力场,进行单点突变稳定性设计,利用结构特征和实验结果使用机器学习算法组合突变分类 | — |
| PROSS | https://pross.weizmann.ac.il/step/pross-terms/ | 基于Rosetta能量函数,设计可以提高稳定性的组合突变 | [ |
| Funclib | https://funclib.weizmann.ac.il/bin/steps | 通过单点突变在进化中的概率和结构计算的能量筛选用于组合突变的候选,使用Rosetta能量函数评价组合突变的稳定性,设计突变组合突变改变底物谱、改善可溶性表达 | [ |
| ABACUS2 | https://biocomp.ustc.edu.cn/servers/abacus-design.php | 基于从蛋白质晶体结构中统计得到的能量函数,进行单点突变能量计算和序列骨架适配度打分 | [ |
| Swiss-Model | https://swissmodel.expasy.org/ | 基于序列搜索,以同源的晶体结构为模版,进行同源建模 | [ |
| ConSurf | https://consurf.tau.ac.il/consurf_index.php | 基于序列搜索算法,分析保守性 | [ |
| ROSIE | https://rosie.graylab.jhu.edu/ | 基于Rosetta的结构relax,分子对接,突变预测等一系列任务 | [ |
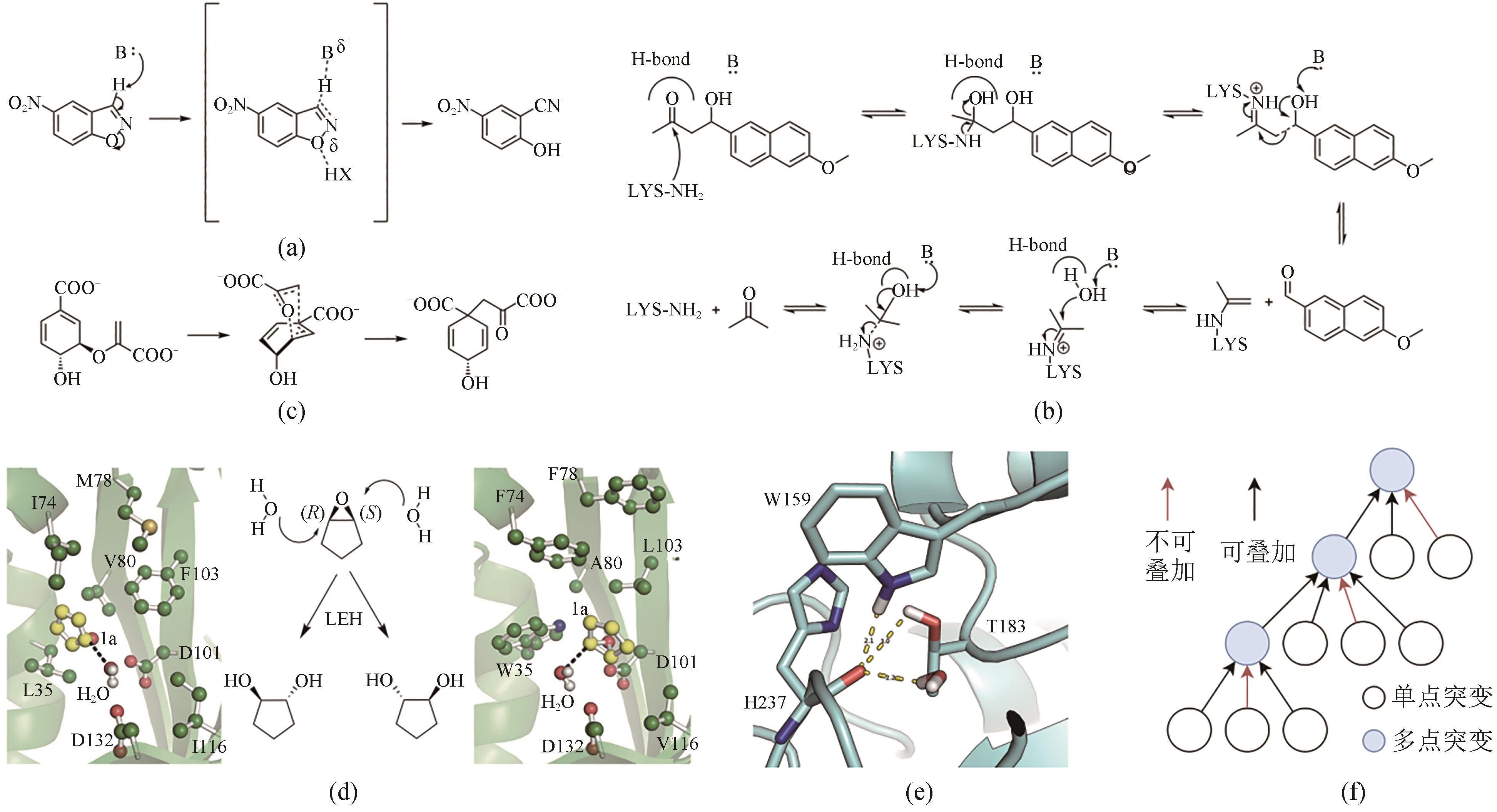
图2 基于计算生物学的催化元件设计(a)Kemp消除反应机制;(b)Retro-Aldo反应机制;(c)分支酸变位酶催化机制;(d)柠檬烯环氧水解的两种不同的近攻击态构象示意图,左侧为pro-RR,右侧为pro-SS,结构示意图修改自文献[17];(e)能量不利的未饱和氢键供体示意图,结构为IsPETase(PDB ID:5XJH),一个水分子和W159已经占据了H237的羰基可以形成的氢键(图中黄色虚线),T183的侧链羟基距离更远,难以形成氢键;(f)分组贪婪叠加策略示意图
Fig. 2 Design of the catalytic components based on computational biology(a) Kemp elimination reaction mechanism. (b) Retro-Aldo reaction mechanism. (c) Mechanism of branching acid translocase catalysis. (d) Schematic diagram of two different near-attack state conformations of limonene epoxide hydrolysis with pro-RR on the left and pro-SS on the right, which were modified from reference [17] with permission. (e) Schematic diagram of the energetically unfavorable unsaturated hydrogen bond donor with the structure of IsPETase (PDB ID: 5XJH), where a water molecule and W159 already occupy the hydrogen bond that can be formed by the carbonyl group of H237 (yellow dashed line in the figure), and the side chain hydroxyl group of T183 is much further away and difficult to form a hydrogen bond. (f) Schematic diagram of the grouped greedy stacking strategy
| 名称 | 地址 | 用途 | 参考文献 |
|---|---|---|---|
| Protein Data Bank | https://www.rcsb.org/ | 蛋白质实验解析的结构数据库 | [ |
| UniProt | https://www.uniprot.org/ | 蛋白质序列数据库 | [ |
| AlphaFold DB | https://alphafold.ebi.ac.uk/ | AlphaFold预测的蛋白质结构数据库 | [ |
| CATH | https://www.cathdb.info/ | 蛋白质结构域分类数据库 | [ |
| InterPro | https://www.ebi.ac.uk/interpro | 蛋白质家族分类数据库 | [ |
| BRENDA | https://www.brenda-enzymes.org/ | 综合性酶学数据库 | [ |
| VariBench | http://structure.bmc.lu.se/VariBench/index.php | 突变体实验测定数据库 | [ |
| Meltome | http://meltomeatlas.proteomics.wzw.tum.de:5003/ | 蛋白质熔融温度数据库 | [ |
| FireProt-DB | https://loschmidt.chemi.muni.cz/fireprotdb/ | 蛋白质点突变稳定性数据库 | [ |
| SABIO-RK | http://sabio.h-its.org/ | 酶动力学性质数据库 | [ |
表2 可用于机器学习模型训练的数据库
Table 2 Databases available for machine learning model training
| 名称 | 地址 | 用途 | 参考文献 |
|---|---|---|---|
| Protein Data Bank | https://www.rcsb.org/ | 蛋白质实验解析的结构数据库 | [ |
| UniProt | https://www.uniprot.org/ | 蛋白质序列数据库 | [ |
| AlphaFold DB | https://alphafold.ebi.ac.uk/ | AlphaFold预测的蛋白质结构数据库 | [ |
| CATH | https://www.cathdb.info/ | 蛋白质结构域分类数据库 | [ |
| InterPro | https://www.ebi.ac.uk/interpro | 蛋白质家族分类数据库 | [ |
| BRENDA | https://www.brenda-enzymes.org/ | 综合性酶学数据库 | [ |
| VariBench | http://structure.bmc.lu.se/VariBench/index.php | 突变体实验测定数据库 | [ |
| Meltome | http://meltomeatlas.proteomics.wzw.tum.de:5003/ | 蛋白质熔融温度数据库 | [ |
| FireProt-DB | https://loschmidt.chemi.muni.cz/fireprotdb/ | 蛋白质点突变稳定性数据库 | [ |
| SABIO-RK | http://sabio.h-its.org/ | 酶动力学性质数据库 | [ |
| 名称 | 描述 | 链接 | 年份 | 参考文献 |
|---|---|---|---|---|
| EPDNew | 真核启动子数据库 | http://epd.vital-it.ch/ | 2015 | [ |
| dbSUPER | 包含小鼠和人类超级增强子信息 | http://asntech.org/dbsuper/ | 2016 | [ |
| SEA | 包含人类、小鼠等多种生物的超级增强子信息 | http://sea.edbc.org/ | 2016 | [ |
| DiseaseEnhancer | 包含143种人类疾病中的847种疾病相关的增强子信息 | http://biocc.hrbmu.edu.cn/DiseaseEnhancer/ | 2018 | [ |
| HEDD | 人类增强子疾病数据库,包含约280万人类增强子的基因组信息 | https://zdzlab.einsteinmed.edu/1/hedd.php | 2018 | [ |
| SEdb | 人类超级增强子数据库,注释了超级增强子在基因调控中的功能 | http://www.licpathway.net/sedb/ | 2019 | [ |
| PlantPAN3.0 | 从78种植物中收集了17 230个转录因子,部分包含结合位点信息 | http://plantpan.itps.ncku.edu.tw/ | 2019 | [ |
| REDfly | 包含实验验证的果蝇的CRM信息 | http://redfly.ccr.buffalo.edu/ | 2019 | [ |
| EnhancerAtlas2.0 | 包含586种组织/细胞中的13 494 603个增强子 | http://www.enhanceratlas.org/indexv2.php | 2016 | [ |
| UCSC Genome Browser database | 提供了人类、小鼠和SARS-Cov-2的基因组数据 | http://genome.ucsc.edu | 2021 | [ |
| SilencerDB | 包含33 060个试验确定的沉默子和5 045 547个机器学习算法预测的沉默子 | http://health.tsinghua.edu.cn/silencerdb/ | 2021 | [ |
表3 顺式调控元件相关的数据库
Table 3 Databases for cis-regulatory elements
| 名称 | 描述 | 链接 | 年份 | 参考文献 |
|---|---|---|---|---|
| EPDNew | 真核启动子数据库 | http://epd.vital-it.ch/ | 2015 | [ |
| dbSUPER | 包含小鼠和人类超级增强子信息 | http://asntech.org/dbsuper/ | 2016 | [ |
| SEA | 包含人类、小鼠等多种生物的超级增强子信息 | http://sea.edbc.org/ | 2016 | [ |
| DiseaseEnhancer | 包含143种人类疾病中的847种疾病相关的增强子信息 | http://biocc.hrbmu.edu.cn/DiseaseEnhancer/ | 2018 | [ |
| HEDD | 人类增强子疾病数据库,包含约280万人类增强子的基因组信息 | https://zdzlab.einsteinmed.edu/1/hedd.php | 2018 | [ |
| SEdb | 人类超级增强子数据库,注释了超级增强子在基因调控中的功能 | http://www.licpathway.net/sedb/ | 2019 | [ |
| PlantPAN3.0 | 从78种植物中收集了17 230个转录因子,部分包含结合位点信息 | http://plantpan.itps.ncku.edu.tw/ | 2019 | [ |
| REDfly | 包含实验验证的果蝇的CRM信息 | http://redfly.ccr.buffalo.edu/ | 2019 | [ |
| EnhancerAtlas2.0 | 包含586种组织/细胞中的13 494 603个增强子 | http://www.enhanceratlas.org/indexv2.php | 2016 | [ |
| UCSC Genome Browser database | 提供了人类、小鼠和SARS-Cov-2的基因组数据 | http://genome.ucsc.edu | 2021 | [ |
| SilencerDB | 包含33 060个试验确定的沉默子和5 045 547个机器学习算法预测的沉默子 | http://health.tsinghua.edu.cn/silencerdb/ | 2021 | [ |
| 名称 | 描述 | 链接 | 年份 | 参考文献 |
|---|---|---|---|---|
| GTRD | 包含试验确定的人类和小鼠转录因子结合位点 | http://gtrd.biouml.org/ | 2017 | [ |
| HOCOMOCO | 包含人类和小鼠转录因子结合位点 | https://hocomoco11.autosome.org/ | 2016 | [ |
| MeDReaders | 包含人类和小鼠中731个转录因子与甲基化DNA序列结合的信息 | http://medreader. org/ | 2018 | [ |
| TRRUST | 人类TF-靶标相互作用数据库 | https://www.grnpedia.org/trrust/ | 2015 | [ |
| AnimalTFDB 3.0 | 包含97个动物基因组中125 135个TF基因和80 060个转录辅助因子基因 | http://bioinfo.life.hust.edu.cn/AnimalTFDB/#!/ | 2019 | [ |
| SalMotifDB | 包含5个鲑鱼基因组中的转录因子及其顺式调控结合位点 | https://salmobase.org/SalMotifDB/ | 2019 | [ |
| hTFtarget | 包含人类转录因子及其靶体 | http://bioinfo.life.hust.edu.cn/hTFtarget#!/ | 2020 | [ |
| KnockTF | 包含人类组织/细胞中转录因子及其靶基因 | http://www.licpathway.net/KnockTF/ | 2020 | [ |
| JASPAR 2022 | 包含真核生物转录因子结合位点 | https://jaspar.genereg.net/ | 2022 | [ |
| PCRMS | 提供了人类和小鼠基因组中CRM和转录因子结合位点的预测数据 | https://cci-bioinfo.uncc.edu/ | 2022 | [ |
表4 转录因子相关的数据库
Table 4 Databases for transcription factor
| 名称 | 描述 | 链接 | 年份 | 参考文献 |
|---|---|---|---|---|
| GTRD | 包含试验确定的人类和小鼠转录因子结合位点 | http://gtrd.biouml.org/ | 2017 | [ |
| HOCOMOCO | 包含人类和小鼠转录因子结合位点 | https://hocomoco11.autosome.org/ | 2016 | [ |
| MeDReaders | 包含人类和小鼠中731个转录因子与甲基化DNA序列结合的信息 | http://medreader. org/ | 2018 | [ |
| TRRUST | 人类TF-靶标相互作用数据库 | https://www.grnpedia.org/trrust/ | 2015 | [ |
| AnimalTFDB 3.0 | 包含97个动物基因组中125 135个TF基因和80 060个转录辅助因子基因 | http://bioinfo.life.hust.edu.cn/AnimalTFDB/#!/ | 2019 | [ |
| SalMotifDB | 包含5个鲑鱼基因组中的转录因子及其顺式调控结合位点 | https://salmobase.org/SalMotifDB/ | 2019 | [ |
| hTFtarget | 包含人类转录因子及其靶体 | http://bioinfo.life.hust.edu.cn/hTFtarget#!/ | 2020 | [ |
| KnockTF | 包含人类组织/细胞中转录因子及其靶基因 | http://www.licpathway.net/KnockTF/ | 2020 | [ |
| JASPAR 2022 | 包含真核生物转录因子结合位点 | https://jaspar.genereg.net/ | 2022 | [ |
| PCRMS | 提供了人类和小鼠基因组中CRM和转录因子结合位点的预测数据 | https://cci-bioinfo.uncc.edu/ | 2022 | [ |
| 计算方法 | 优点 | 缺点 | 在生物传感器领域的应用 |
|---|---|---|---|
| QM方法 | 精确、可以计算化学反应或电荷转移 | 计算成本高、难以直接应用于生物大分子的计算 | 高精度的评估结合能;计算化学反应(结合QM/MM);计算量子电导 |
| MD方法 | 在一定范围内有着可靠的精度,计算效率比QM高数个数量级 | 当电子运动不可忽略时,精度不够可靠;面临优化序列空间的问题是计算效率依然不够高 | 对生物传感器的作用机制或分子的构象变化做机理分析;评估配体和受体之间的亲和力;在一定范围内做序列优化 |
| 分子对接和虚拟筛选 | 高的计算效率 | 精度低于MD方法 | 高效评估配体和受体的结合状态和结合模式;从打数据库或序列空间寻找潜在的配体或受体待优化对象 |
表5 计算方法比较
Table 5 Comparison of the computation methods
| 计算方法 | 优点 | 缺点 | 在生物传感器领域的应用 |
|---|---|---|---|
| QM方法 | 精确、可以计算化学反应或电荷转移 | 计算成本高、难以直接应用于生物大分子的计算 | 高精度的评估结合能;计算化学反应(结合QM/MM);计算量子电导 |
| MD方法 | 在一定范围内有着可靠的精度,计算效率比QM高数个数量级 | 当电子运动不可忽略时,精度不够可靠;面临优化序列空间的问题是计算效率依然不够高 | 对生物传感器的作用机制或分子的构象变化做机理分析;评估配体和受体之间的亲和力;在一定范围内做序列优化 |
| 分子对接和虚拟筛选 | 高的计算效率 | 精度低于MD方法 | 高效评估配体和受体的结合状态和结合模式;从打数据库或序列空间寻找潜在的配体或受体待优化对象 |
| 1 | LV X Q, HUESO-GIL A, BI X Y, et al. New synthetic biology tools for metabolic control[J]. Current Opinion in Biotechnology, 2022, 76: 102724. |
| 2 | FAULON J L, FAURE L. In silico, in vitro, and in vivo machine learning in synthetic biology and metabolic engineering[J]. Current Opinion in Chemical Biology, 2021, 65: 85-92. |
| 3 | LAWSON C E, MARTÍ J M, RADIVOJEVIC T, et al. Machine learning for metabolic engineering: a review[J]. Metabolic Engineering, 2021, 63: 34-60. |
| 4 | MADHAVAN A, ARUN K B, BINOD P, et al. Design of novel enzyme biocatalysts for industrial bioprocess: harnessing the power of protein engineering, high throughput screening and synthetic biology[J]. Bioresource Technology, 2021, 325: 124617. |
| 5 | DE JONGH R P H, VAN DIJK A D J, JULSING M K, et al. Designing eukaryotic gene expression regulation using machine learning[J]. Trends in Biotechnology, 2020, 38(2): 191-201. |
| 6 | ALFORD R F, LEAVER-FAY A, JELIAZKOV J R, et al. The Rosetta all-atom energy function for macromolecular modeling and design[J]. Journal of Chemical Theory and Computation, 2017, 13(6): 3031-3048. |
| 7 | MUSIL M, STOURAC J, BENDL J, et al. FireProt: web server for automated design of thermostable proteins[J]. Nucleic Acids Research, 2017, 45(W1): W393-W399. |
| 8 | GOLDENZWEIG A, GOLDSMITH M, HILL S E, et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability[J]. Molecular Cell, 2016, 63(2): 337-346. |
| 9 | KHERSONSKY O, LIPSH R, AVIZEMER Z, et al. Automated design of efficient and functionally diverse enzyme repertoires[J]. Molecular Cell, 2018, 72(1): 178-186.e5. |
| 10 | XIONG P, HU X H, HUANG B, et al. Increasing the efficiency and accuracy of the ABACUS protein sequence design method[J]. Bioinformatics, 2020, 36(1): 136-144. |
| 11 | SCHWEDE T, KOPP J, GUEX N, et al. SWISS-MODEL: an automated protein homology-modeling server[J]. Nucleic Acids Research, 2003, 31(13): 3381-3385. |
| 12 | ASHKENAZY H, EREZ E, MARTZ E, et al. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids[J]. Nucleic Acids Research, 2010, 38(): W529-W533. |
| 13 | MORETTI R, LYSKOV S, DAS R, et al. Web-accessible molecular modeling with Rosetta: the Rosetta Online Server that Includes Everyone (ROSIE)[J]. Protein Science, 2018, 27(1): 259-268. |
| 14 | RÖTHLISBERGER D, KHERSONSKY O, WOLLACOTT A M, et al. Kemp elimination catalysts by computational enzyme design[J]. Nature, 2008, 453(7192): 190-195. |
| 15 | JIANG L, ALTHOFF E A, CLEMENTE F R, et al. De novo computational design of Retro-Aldol enzymes[J]. Science, 2008, 319(5868): 1387-1391. |
| 16 | RUSS W P, FIGLIUZZI M, STOCKER C, et al. An evolution-based model for designing chorismate mutase enzymes[J]. Science, 2020, 369(6502): 440-445. |
| 17 | WIJMA H J, FLOOR R J, BJELIC S, et al. Enantioselective enzymes by computational design and in silico screening[J]. Angewandte Chemie International Edtion, 2015, 54(12): 3726-3730. |
| 18 | LI R F, WIJMA H J, SONG L, et al. Computational redesign of enzymes for regio- and enantioselective hydroamination[J]. Nature Chemical Biology, 2018, 14(7): 664-670. |
| 19 | CUI Y L, WANG Y H, TIAN W Y, et al. Development of a versatile and efficient C-N lyase platform for asymmetric hydroamination via computational enzyme redesign[J]. Nature Catalysis, 2021, 4(5): 364-373. |
| 20 | MENG Q L, CAPRA N, PALACIO C M, et al. Robust ω-transaminases by computational stabilization of the subunit interface[J]. ACS Catalysis, 2020, 10(5): 2915-2928. |
| 21 | BEDNAR D, BEERENS K, SEBESTOVA E, et al. FireProt: energy- and evolution-based computational design of thermostable multiple-point mutants[J]. PLoS Computational Biology, 2015, 11(11): e1004556. |
| 22 | WIJMA H J, FLOOR R J, JEKEL P A, et al. Computationally designed libraries for rapid enzyme stabilization[J]. Protein Engineering, Design and Selection, 2014, 27(2): 49-58. |
| 23 | DELGADO J, RADUSKY L G, CIANFERONI D, et al. FoldX 5.0: working with RNA, small molecules and a new graphical interface[J]. Bioinformatics, 2019, 35(20): 4168-4169. |
| 24 | WU B, WIJMA H J, SONG L, et al. Versatile peptide C-terminal functionalization via a computationally engineered peptide amidase[J]. ACS Catalysis, 2016, 6(8): 5405-5414. |
| 25 | CUI Y L, CHEN Y C, LIU X Y, et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy[J]. ACS Catalysis, 2021, 11(3): 1340-1350. |
| 26 | UNIPROT CONSORTIUM THE. UniProt: a worldwide hub of protein knowledge[J]. Nucleic Acids Research 2019, 47(D1), D506-D515. |
| 27 | VARADI M, ANYANGO S, DESHPANDE M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models[J]. Nucleic Acids Research, 2022, 50(D1): D439-D444. |
| 28 | ORENGO C A, MICHIE A D, JONES S, et al. CATH—a hierarchic classification of protein domain structures[J]. Structure, 1997, 5(8): 1093-1108. |
| 29 | BLUM M, CHANG H Y, CHUGURANSKY S, et al. The InterPro protein families and domains database: 20 years on[J]. Nucleic Acids Research, 2021, 49(D1): D344-D354. |
| 30 | CHANG A, JESKE L, ULBRICH S, et al. BRENDA, the ELIXIR core data resource in 2021: new developments and updates[J]. Nucleic Acids Research, 2021, 49(D1): D498-D508. |
| 31 | SARKAR A, YANG Y, VIHINEN M. Variation benchmark datasets: update, criteria, quality and applications[J]. Database, 2020, 2020: baz117. |
| 32 | JARZAB A, KURZAWA N, HOPF T, et al. Meltome atlas—thermal proteome stability across the tree of life[J]. Nature Methods, 2020, 17(5): 495-503. |
| 33 | STOURAC J, DUBRAVA J, MUSIL M, et al. FireProtDB: dataBase of manually curated protein stability data[J]. Nucleic Acids Research, 2021, 49(D1): D319-D324. |
| 34 | WITTIG U, REY M, WEIDEMANN A, et al. SABIO-RK: an updated resource for manually curated biochemical reaction kinetics[J]. Nucleic Acids Research, 2018, 46(D1): D656-D660. |
| 35 | REPECKA D, JAUNISKIS V, KARPUS L, et al. Expanding functional protein sequence spaces using generative adversarial networks[J]. Nature Machine Intelligence, 2021, 3(4): 324-333. |
| 36 | WANG J, LISANZA S, JUERGENS D, et al. Scaffolding protein functional sites using deep learning[J]. Science, 2022, 377(6604): 387-394. |
| 37 | ANISHCHENKO I, PELLOCK S J, CHIDYAUSIKU T M, et al. De novo protein design by deep network hallucination[J]. Nature, 2021, 600(7889): 547-552. |
| 38 | DAUPARAS J, ANISHCHENKO I, BENNETT N, et al. Robust deep learning-based protein sequence design using ProteinMPNN[J]. Science, 2022, 378(6615): 49-56. |
| 39 | SUGIKI S, NIIDE T, TOYA Y, et al. Logistic regression-guided identification of cofactor specificity-contributing residues in enzyme with sequence datasets partitioned by catalytic properties[J]. ACS Synthetic Biology, 2022, 11(12): 3973-3985. |
| 40 | HU R Y, FU L H, CHEN Y C, et al. Protein engineering via Bayesian optimization-guided evolutionary algorithm and robotic experiments[J]. Briefings in Bioinformatics, 2023, 24(1): bbac570. |
| 41 | SHROFF R, COLE A W, DIAZ D J, et al. Discovery of novel gain-of-function mutations guided by structure-based deep learning[J]. ACS Synthetic Biology, 2020, 9(11): 2927-2935. |
| 42 | LU H Y, DIAZ D J, CZARNECKI N J, et al. Machine learning-aided engineering of hydrolases for PET depolymerization[J]. Nature, 2022, 604(7907): 662-667. |
| 43 | BARBER-ZUCKER S, MINDEL V, GARCIA-RUIZ E, et al. Stable and functionally diverse versatile peroxidases designed directly from sequences[J]. Journal of the American Chemical Society, 2022, 144(8): 3564-3571. |
| 44 | DOERR S, MAJEWSKI M, PÉREZ A, et al. TorchMD: a deep learning framework for molecular simulations[J]. Journal of Chemical Theory and Computation, 2021, 17(4): 2355-2363. |
| 45 | GREEN P J, PINES O, INOUYE M. The role of antisense RNA in gene regulation[J]. Annual Review of Biochemistry, 1986, 55: 569-597. |
| 46 | PAPENFORT K, VANDERPOOL C K. Target activation by regulatory RNAs in bacteria[J]. FEMS Microbiology Reviews, 2015, 39(3): 362-378. |
| 47 | PANG B X, VAN WEERD J H, HAMOEN F L, et al. Identification of non-coding silencer elements and their regulation of gene expression[J]. Nature Reviews Molecular Cell Biology, 2022: 1-13. |
| 48 | DREOS R, AMBROSINI G, PÉRIER R C, et al. The eukaryotic promoter database: expansion of EPDnew and new promoter analysis tools[J]. Nucleic Acids Research, 2015, 43(Database issue): D92-D96. |
| 49 | KHAN A, ZHANG X G. dbSUPER: a database of super-enhancers in mouse and human genome[J]. Nucleic Acids Research, 2016, 44(D1): D164-D171. |
| 50 | WEI Y J, ZHANG S M, SHANG S P, et al. SEA: a super-enhancer archive[J]. Nucleic Acids Research, 2016, 44(D1): D172-D179. |
| 51 | ZHANG G X, SHI J, ZHU S W, et al. DiseaseEnhancer: a resource of human disease-associated enhancer catalog[J]. Nucleic Acids Research, 2018, 46(D1): D78-D84. |
| 52 | WANG Z, ZHANG Q W, ZHANG W, et al. HEDD: human enhancer disease database[J]. Nucleic Acids Research, 2018, 46(D1): D113-D120. |
| 53 | JIANG Y, QIAN F C, BAI X F, et al. SEdb: a comprehensive human super-enhancer database[J]. Nucleic Acids Research, 2019, 47(D1): D235-D243. |
| 54 | CHOW C N, LEE T Y, HUNG Y C, et al. PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants[J]. Nucleic Acids Research, 2019, 47(D1): D1155-D1163. |
| 55 | RIVERA J, KERÄNEN S V E, GALLO S M, et al. REDfly: the transcriptional regulatory element database for Drosophila[J]. Nucleic Acids Research, 2019, 47(D1): D828-D834. |
| 56 | GAO T S, HE B, LIU S, et al. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types[J]. Bioinformatics, 2016, 32(23): 3543-3551. |
| 57 | GONZALEZ J N, ZWEIG A S, SPEIR M L, et al. The UCSC genome browser database: 2021 update[J]. Nucleic Acids Research, 2021, 49(D1): D1046-D1057. |
| 58 | ZENG W W, CHEN S Q, CUI X J, et al. SilencerDB: a comprehensive database of silencers[J]. Nucleic Acids Research, 2021, 49(D1): D221-D228. |
| 59 | UMAROV R, LI Y, ARAKAWA T, et al. ReFeaFi: genome-wide prediction of regulatory elements driving transcription initiation[J]. PLoS Computational Biology, 2021, 17(9): e1009376. |
| 60 | ZRIMEC J, BÖRLIN C S, BURIC F, et al. Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure[J]. Nature Communications, 2020, 11: 6141. |
| 61 | ZRIMEC J, FU X Z, MUHAMMAD A S, et al. Controlling gene expression with deep generative design of regulatory DNA[J]. Nature Communications, 2022, 13: 5099. |
| 62 | MENG H L, MA Y F, MAI G Q, et al. Construction of precise support vector machine based models for predicting promoter strength[J]. Quantitative Biology, 2017, 5(1): 90-98. |
| 63 | CAZIER A P, BLAZECK J. Advances in promoter engineering: novel applications and predefined transcriptional control[J]. Biotechnology Journal, 2021, 16(10): e2100239. |
| 64 | OUBOUNYT M, LOUADI Z, TAYARA H, et al. DeePromoter: robust promoter predictor using deep learning[J]. Frontiers in Genetics, 2019, 10: 286. |
| 65 | WANG Y, WANG H C, WEI L, et al. Synthetic promoter design in Escherichia coli based on a deep generative network[J]. Nucleic Acids Research, 2020, 48(12): 6403-6412. |
| 66 | MENON S, PIRAMANAYAKAM S, AGARWAL G. Computational identification of promoter regions in prokaryotes and eukaryotes[J]. EPRA International Journal of Agriculture and Rural Economic Research, 2021, 9(7): 21-28. |
| 67 | CATARINO R R, STARK A. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation[J]. Genes & Development, 2018, 32(3/4): 202-223. |
| 68 | ANDERSSON R, SANDELIN A. Determinants of enhancer and promoter activities of regulatory elements[J]. Nature Reviews Genetics, 2020, 21(2): 71-87. |
| 69 | ANDERSSON R, REFSING ANDERSEN P, VALEN E, et al. Nuclear stability and transcriptional directionality separate functionally distinct RNA species[J]. Nature Communications, 2014, 5: 5336. |
| 70 | DIAO Y R, FANG R X, LI B, et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells[J]. Nature Methods, 2017, 14(6): 629-635. |
| 71 | KIM T K, HEMBERG M, GRAY J M, et al. Widespread transcription at neuronal activity-regulated enhancers[J]. Nature, 2010, 465(7295): 182-187. |
| 72 | FULCO C P, NASSER J, JONES T R, et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations[J]. Nature Genetics, 2019, 51(12): 1664-1669. |
| 73 | MAURANO M T, HUMBERT R, RYNES E, et al. Systematic localization of common disease-associated variation in regulatory DNA[J]. Science, 2012, 337(6099): 1190-1195. |
| 74 | KHANAL J, TAYARA H, CHONG K T. Identifying enhancers and their strength by the integration of word embedding and convolution neural network[J]. IEEE Access, 2020, 8: 58369-58376. |
| 75 | MIN X P, YE C M, LIU X R, et al. Predicting enhancer-promoter interactions by deep learning and matching heuristic[J]. Briefings in Bioinformatics, 2021, 22(4): bbaa254. |
| 76 | DE ALMEIDA B P, REITER F, PAGANI M, et al. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers[J]. Nature Genetics, 2022, 54(5): 613-624. |
| 77 | FENG C Q, ZHANG Z Y, ZHU X J, et al. iTerm-PseKNC: a sequence-based tool for predicting bacterial transcriptional terminators[J]. Bioinformatics, 2019, 35(9): 1469-1477. |
| 78 | YEVSHIN I, SHARIPOV R, VALEEV T, et al. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments[J]. Nucleic Acids Research, 2017, 45(D1): D61-D67. |
| 79 | KULAKOVSKIY I V, VORONTSOV I E, YEVSHIN I S, et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis[J]. Nucleic Acids Research, 2018, 46(D1): D252-D259. |
| 80 | KULAKOVSKIY I V, VORONTSOV I E, YEVSHIN I S, et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models[J]. Nucleic Acids Research, 2016, 44(D1): D116-D125. |
| 81 | WANG G H, LUO X M, WANG J N, et al. MeDReaders: a database for transcription factors that bind to methylated DNA[J]. Nucleic Acids Research, 2018, 46(D1): D146-D151. |
| 82 | HAN H, CHO J W, LEE S, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions[J]. Nucleic Acids Research, 2018, 46(D1): D380-D386. |
| 83 | HAN H, SHIM H, SHIN D, et al. TRRUST: a reference database of human transcriptional regulatory interactions[J]. Scientific Reports, 2015, 5: 11432. |
| 84 | HU H, MIAO Y R, JIA L H, et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors[J]. Nucleic Acids Research, 2019, 47(D1): D33-D38. |
| 85 | MULUGETA T D, NOME T, TO T H, et al. SalMotifDB: a tool for analyzing putative transcription factor binding sites in salmonid genomes[J]. BMC Genomics, 2019, 20(1): 694. |
| 86 | ZHANG Q, LIU W, ZHANG H M, et al. hTFtarget: a comprehensive database for regulations of human transcription factors and their targets[J]. Genomics, Proteomics & Bioinformatics, 2020, 18(2): 120-128. |
| 87 | FENG C C, SONG C, LIU Y J, et al. KnockTF: a comprehensive human gene expression profile database with knockdown/knockout of transcription factors[J]. Nucleic Acids Research, 2020, 48(D1): D93-D100. |
| 88 | CASTRO-MONDRAGON J A, RIUDAVETS-PUIG R, RAULUSEVICIUTE I, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles[J]. Nucleic Acids Research, 2022, 50(D1): D165-D173. |
| 89 | NI P Y, SU Z C. PCRMS: a database of predicted cis-regulatory modules and constituent transcription factor binding sites in genomes[J]. Database, 2022, 2022: baac024. |
| 90 | FU L Y, ZHANG L H, DOLLINGER E, et al. Predicting transcription factor binding in single cells through deep learning[J]. Science Advances, 2020, 6(51): eaba9031. |
| 91 | KIM G B, GAO Y, PALSSON B O, et al. DeepTFactor: a deep learning-based tool for the prediction of transcription factors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(2): e2021171118. |
| 92 | ZHANG Q H, WANG S G, CHEN Z H, et al. Locating transcription factor binding sites by fully convolutional neural network[J]. Briefings in Bioinformatics, 2021, 22(5): bbaa435. |
| 93 | ZHENG A, LAMKIN M, ZHAO H Q, et al. Deep neural networks identify sequence context features predictive of transcription factor binding[J]. Nature Machine Intelligence, 2021, 3(2): 172-180. |
| 94 | NAKASHIMA N, TAMURA T, GOOD L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli [J]. Nucleic Acids Research, 2006, 34(20): e138. |
| 95 | NA D, YOO S M, CHUNG H, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs[J]. Nature Biotechnology, 2013, 31(2): 170-174. |
| 96 | ZHANG R H, ZHANG Y, WANG J, et al. Development of antisense RNA-mediated quantifiable inhibition for metabolic regulation[J]. Metabolic Engineering Communications, 2021, 12: e00168. |
| 97 | GREEN A A, SILVER P A, COLLINS J J, et al. Toehold switches: de-novo-designed regulators of gene expression[J]. Cell, 2014, 159(4): 925-939. |
| 98 | CHAPPELL J, WESTBROOK A, VEROSLOFF M, et al. Computational design of small transcription activating RNAs for versatile and dynamic gene regulation[J]. Nature Communications, 2017, 8: 1051. |
| 99 | CHAPPELL J, TAKAHASHI M K, LUCKS J B. Creating small transcription activating RNAs[J]. Nature Chemical Biology, 2015, 11(3): 214-220. |
| 100 | JANG S H, JANG S Y, YANG J N, et al. RNA-based dynamic genetic controllers: development strategies and applications[J]. Current Opinion in Biotechnology, 2018, 53: 1-11. |
| 101 | ZADEH J N, STEENBERG C D, BOIS J S, et al. NUPACK: analysis and design of nucleic acid systems[J]. Journal of Computational Chemistry, 2011, 32(1): 170-173. |
| 102 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 103 | CHEN T, CHENG G Y, AHMED S, et al. New methodologies in screening of antibiotic residues in animal-derived foods: Biosensors[J]. Talanta, 2017, 175: 435-442. |
| 104 | SONGA E A, OKONKWO J O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: a review[J]. Talanta, 2016, 155: 289-304. |
| 105 | PUIU M, BALA C. Peptide-based biosensors: from self-assembled interfaces to molecular probes in electrochemical assays[J]. Bioelectrochemistry, 2018, 120: 66-75. |
| 106 | SHARMA S, BYRNE H, O'KENNEDY R J. Antibodies and antibody-derived analytical biosensors[J]. Essays in Biochemistry, 2016, 60(1): 9-18. |
| 107 | WANG R E, ZHANG Y, CAI J, et al. Aptamer-based fluorescent biosensors[J]. Current Medicinal Chemistry, 2011, 18(27): 4175-4184. |
| 108 | BLIND M, BLANK M. Aptamer selection technology and recent advances[J]. Molecular Therapy Nucleic Acids, 2015, 4(1): e223. |
| 109 | HONG P T K, JANG C H. Sensitive and label-free liquid crystal-based optical sensor for the detection of malathion[J]. Analytical Biochemistry, 2020, 593: 113589. |
| 110 | KIM H S, AN Z F, JANG C H. Label-free optical detection of thrombin using a liquid crystal-based aptasensor[J]. Microchemical Journal, 2018, 141: 71-79. |
| 111 | O'NEILL M, KELLY S M. Liquid crystals for charge transport, luminescence, and photonics[J]. Advanced Materials, 2003, 15(14): 1135-1146. |
| 112 | BYKHOVSKI A, ZHANG W D, JENSEN J, et al. Analysis of electronic structure, binding, and vibrations in biotin-streptavidin complexes based on density functional theory and molecular mechanics[J]. The Journal of Physical Chemistry B, 2013, 117(1): 25-37. |
| 113 | PAULLA K K, FARAJIAN A A. Concentration effects of carbon oxides on sensing by graphene nanoribbons: ab initio modeling[J]. The Journal of Physical Chemistry C, 2013, 117(24): 12815-12825. |
| 114 | KUMAR N, SEMINARIO J M. Design of nanosensors for fissile materials in nuclear waste water[J]. The Journal of Physical Chemistry C, 2013, 117(45): 24033-24041. |
| 115 | NEZHADALI A, MOJARRA M. Computational study and multivariate optimization of hydrochlorothiazide analysis using molecularly imprinted polymer electrochemical sensor based on carbon nanotube/polypyrrole film[J]. Sensors and Actuators B: Chemical, 2014, 190: 829-837. |
| 116 | LIU Q Y, ZUO F, ZHAO Z G, et al. Molecular dynamics investigations of an indicator displacement assay mechanism in a liquid crystal sensor[J]. Physical Chemistry Chemical Physics: PCCP, 2017, 19(35): 23924-23933. |
| 117 | CAUSIN P, SACCO R, VERRI M. A multiscale approach in the computational modeling of the biophysical environment in artificial cartilage tissue regeneration[J]. Biomechanics and Modeling in Mechanobiology, 2013, 12(4): 763-780. |
| 118 | ZHANG W J, DU Y Q, CRANFORD S W, et al. Biosensor design through molecular dynamics simulation[J]. World Academy of Science, Engineering and Technology, International Journal of Biomedical and Biological Engineering 2016, 10(1): 10-14. |
| 119 | KHOSHBIN Z, HOUSAINDOKHT M R, IZADYAR M, et al. Theoretical design and experimental study of new aptamers with the improved target-affinity: new insights into the Pb2+-specific aptamers as a case study[J]. Journal of Molecular Liquids, 2019, 289: 111159. |
| 120 | ZHUANG S L, WANG H F, DING K K, et al.. Interactions of benzotriazole UV stabilizers with human serum albumin: atomic insights revealed by biosensors, spectroscopies and molecular dynamics simulations[J]. Chemosphere, 2016, 144: 1050-1059. |
| 121 | KOLLMAN P A, MASSOVA I, REYES C, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models[J]. Accounts of Chemical Research, 2000, 33(12): 889-897. |
| 122 | KHOSHBIN Z, HOUSAINDOKHT M R. Computer-aided aptamer design for sulfadimethoxine antibiotic: step by step mutation based on MD simulation approach[J]. Journal of Biomolecular Structure & Dynamics, 2021, 39(9): 3071-3079. |
| 123 | DO P C, LEE E H, LE L. Steered molecular dynamics simulation in rational drug design[J]. Journal of Chemical Information and Modeling, 2018, 58(8): 1473-1482. |
| 124 | THYPARAMBIL A A, ABRAMYAN T M, BAZIN I, et al. Site of tagging influences the ochratoxin recognition by peptide NFO4: a molecular dynamics study[J]. Journal of Chemical Information and Modeling, 2017, 57(8): 2035-2044. |
| 125 | THYPARAMBIL A A, BAZIN I, GUISEPPI-ELIE A. Evaluation of ochratoxin recognition by peptides using explicit solvent molecular dynamics[J]. Toxins, 2017, 9(5): 164. |
| 126 | SALMASO V, STURLESE M, CUZZOLIN A, et al. Combining self- and cross-docking as benchmark tools: the performance of DockBench in the D3R grand challenge 2[J]. Journal of Computer-Aided Molecular Design, 2018, 32(1): 251-264. |
| 127 | LIU Q J, WANG H, LI H L, et al. Impedance sensing and molecular modeling of an olfactory biosensor based on chemosensory proteins of honeybee[J]. Biosensors & Bioelectronics, 2013, 40(1): 174-179. |
| 128 | ABOLHASAN R, MEHDIZADEH A, RASHIDI M R, et al. Application of hairpin DNA-based biosensors with various signal amplification strategies in clinical diagnosis[J]. Biosensors & Bioelectronics, 2019, 129: 164-174. |
| 129 | CROSS S, BARONI M, CAROSATI E, et al. FLAP: GRID molecular interaction fields in virtual screening. validation using the DUD data set[J]. Journal of Chemical Information and Modeling, 2010, 50(8): 1442-1450. |
| 130 | MA D L, CHAN D S H, LEE P, et al. Molecular modeling of drug-DNA interactions: virtual screening to structure-based design[J]. Biochimie, 2011, 93(8): 1252-1266. |
| 131 | PIZZONI D, MASCINI M, COMPAGNONE D, et al. Virtual screening peptide selection for a peptide based gas sensors array[C/OL]//Proceedings of the Second National Conference on Sensors, Rome, Italy, 19-21 February, 2014, 2015: 89-93 [2023-01-01]. . |
| 132 | MASCINI M, PIZZONI D, PEREZ G, et al. Tailoring gas sensor arrays via the design of short peptides sequences as binding elements[J]. Biosensors & Bioelectronics, 2017, 93: 161-169. |
| 133 | FRANCA E F, LEITE F L, CUNHA R A, et al. Designing an enzyme-based nanobiosensor using molecular modeling techniques[J]. Physical Chemistry Chemical Physics: PCCP, 2011, 13(19): 8894-8899. |
| 134 | HONG ENRIQUEZ R P, PAVAN S, BENEDETTI F, et al. Designing short peptides with high affinity for organic molecules: A combined docking, molecular dynamics, and Monte Carlo approach[J]. Journal of Chemical Theory and Computation, 2012, 8(3): 1121-1128. |
| 135 | SHCHERBININ D S, GNEDENKO O V, KHMELEVA S A, et al. Computer-aided design of aptamers for cytochrome P450[J]. Journal of Structural Biology, 2015, 191(2): 112-119. |
| 136 | PRANDI I G, RAMALHO T C, FRANÇA T C C. Esterase 2 as a fluorescent biosensor for the detection of organophosphorus compounds: docking and electronic insights from molecular dynamics[J]. Molecular Simulation, 2019, 45(17): 1432-1436. |
| 137 | SHAHBAAZ M, KANCHI S, SABELA M, et al. Structural basis of pesticide detection by enzymatic biosensing: a molecular docking and MD simulation study[J]. Journal of Biomolecular Structure & Dynamics, 2018, 36(6): 1402-1416. |
| 138 | CHAKRAVORTY D K, PARKER T M, GUERRA A J, et al. Energetics of zinc-mediated interactions in the allosteric pathways of metal sensor proteins[J]. Journal of the American Chemical Society, 2013, 135(1): 30-33. |
| 139 | GROENHOF G. Introduction to QM/MM simulations[J]. Methods in Molecular Biology, 2013, 924: 43-66. |
| 140 | PAPAMICHAEL E M, STAMATIS H, STERGIOU P Y, et al. Enzyme kinetics and modeling of enzymatic systems[M/OL]. Advances in Enzyme Technology, Amsterdam: Elsevier, 2019: 71-104[2023-01-01]. . |
| 141 | RYDE U. QM/MM calculations on proteins[J]. Methods in Enzymology, 2016, 577: 119-158. |
| 142 | WONG M W, XIE H F, KWA S T. Anion recognition by azophenol thiourea-based chromogenic sensors: a combined DFT and molecular dynamics investigation[J]. Journal of Molecular Modeling, 2013, 19(1): 205-213. |
| 143 | CHARCHAR P, CHRISTOFFERSON A J, TODOROVA N, et al. Understanding and designing the gold-bio interface: insights from simulations[J]. Small, 2016, 12(18): 2395-2418. |
| 144 | ZHU C, LI L S, YANG G, et al. High-efficiency selection of aptamers for bovine lactoferrin by capillary electrophoresis and its aptasensor application in milk powder[J]. Talanta, 2019, 205: 120088. |
| 145 | YARIZADEH K, BEHBAHANI M, MOHABATKAR H, et al. Computational analysis and optimization of carcinoembryonic antigen aptamers and experimental evaluation[J]. Journal of Biotechnology, 2019, 306: 1-8. |
| 146 | KHAVANI M, IZADYAR M, HOUSAINDOKHT M R. Theoretical design and experimental study on the gold nanoparticles based colorimetric aptasensors for detection of neomycin B[J]. Sensors and Actuators B: Chemical, 2019, 300: 126947. |
| 147 | MITCHLER M M, GARCIA J M, MONTERO N E, et al. Transcription factor-based biosensors: a molecular-guided approach for natural product engineering[J]. Current Opinion in Biotechnology, 2021, 69: 172-181. |
| 148 | HOSSAIN G S, SAINI M, MIYAKE R, et al. Genetic biosensor design for natural product biosynthesis in microorganisms[J]. Trends in Biotechnology, 2020, 38(7): 797-810. |
| 149 | LIANG W F, CUI L Y, CUI J Y, et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39: 159-168. |
| 150 | KASEY C M, ZERRAD M, LI Y W, et al. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology[J]. ACS Synthetic Biology, 2018, 7(1): 227-239. |
| 151 | DE PAEPE B, MAERTENS J, VANHOLME B, et al. Chimeric LysR-type transcriptional biosensors for customizing ligand specificity profiles toward flavonoids[J]. ACS Synthetic Biology, 2019, 8(2): 318-331. |
| 152 | LIANG M D, LI Z L, WANG W S, et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules[J]. Nature Communications, 2019, 10: 3672. |
| 153 | MCCANN J J, PIKE D H, BROWN M C, et al. Computational design of a sensitive, selective phase-changing sensor protein for the VX nerve agent[J]. Science Advances, 2022, 8(27): eabh3421. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [11] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [12] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||