|
||
|
Creation of non-natural cofactor-dependent methanol dehydrogenase
Synthetic Biology Journal
2021, 2 (4):
651-661.
DOI: 10.12211/2096-8280.2021-016
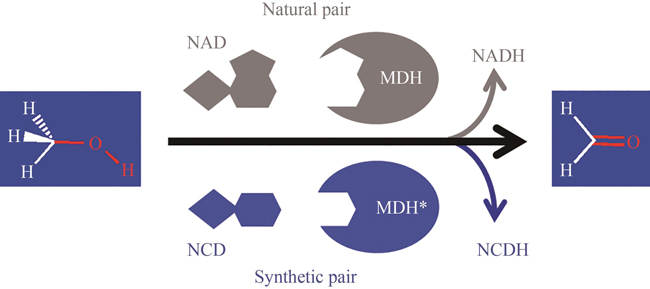
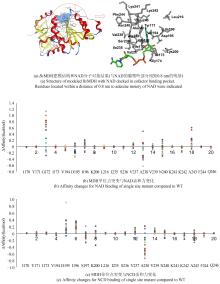
The C1 organic carbon compound methanol is a potential raw material for biorefinery. Recently, intensive efforts have been devoted to engineer cell factories for direct conversion of methanol into valuable metabolites. The oxidation of methanol into formaldehyde is the first committed step to provide useful substance for metabolism. While some methylotrophic microorganisms oxidize methanol with hydrogen peroxide as a co-product, many engineered systems are designed to co-produce NADH, the reduced nicotinamide adenine dinucleotide (NAD). The later route, normally catalyzed by NAD-dependent methanol dehydrogenase (MDH), is more attractive as NADH can be used as reducing power for cellular metabolism. However, NAD(H) are used by many redox enzymes and methanol oxidation-derived NADH can cause unpredictable biological effects. We recently engineered the cofactor preference of several NAD-dependent redox enzymes to favor a non-natural cofactor nicotinamide cytosine dinucleotide (NCD). By coupling these enzymes we demonstrated the construction of NCD-linked redox systems orthogonal to the natural cofactor NAD(H), which could be used for pathway-selective chemical energy transfer in Escherichia coli. By redesigning the cofactor binding pocket of MDH, it is possible to obtain mutants favoring NCD, and thus utilize methanol as a carbon source with co-producing reduced NCD for dedicated redox chemistry. In this paper, we first analyzed the cofactor-binding pocket of Bacillus stearothermophilus DSM2334 derived NAD-dependent MDH (UniProt: P42327.1). Through virtual screening and single-site mutation library screening, hot spots for cofactor binding were identified. More mutants were rationally generated based on insights into the volume of cofactor binding cavity and those were obtained with improved activities in the presence of NCD. The mutants were overexpressed in E. coli, purified, and their catalytic performance were analyzed. The results showed that the catalytic efficiency of the mutant 9D1 (MDH Y171R/I196V/V237T/N240E/K241A) with NCD reached 858 L/(mol·s), and its NCD preference was 13 000-fold higher than that of the wild-type protein. These MDH variants can be considered as new functional parts for the bioconversion of methanol and the construction of new redox metabolism.

Fig. 4
Results of activity assay of crude extracts of BsMDH and its variants
Other Images/Table from this Article
|