合成生物学 ›› 2021, Vol. 2 ›› Issue (2): 222-233.DOI: 10.12211/2096-8280.2020-048
合成甲基营养细胞工厂同化甲醇的研究进展及未来展望
张卉1,2, 袁姚梦3,4, 张翀3,4, 杨松1,2,5, 邢新会3,4
- 1.青岛农业大学生命科学学院,山东 青岛 266109
2.青岛农业大学,山东省应用真菌重点实验室,山东 青岛 266109
3.清华大学化学工程系生物化工研究所,工业生物催化教育部重点实验室,北京 100084
4.清华大学合成与系统生物学研究中心,北京 100084
5.青岛农业大学,青岛生物沼气环境微生物国际合作基地,山东 青岛 266109
-
收稿日期:2020-06-15修回日期:2021-01-11出版日期:2021-04-30发布日期:2021-04-30 -
通讯作者:杨松,邢新会 -
作者简介:张卉 (1996─),女,在读硕士。主要研究方向为微生物代谢工程。E-mail:zhanghui_96@126.com
杨松(1977─),男,博士,教授。主要研究方向为代谢工程和系统组学。E-mail:yangsong1209@163.com
邢新会(1963─),男,博士,教授。主要研究方向为生物化工,合成生物学,生物育种。E-mail:xhxing@tsinghua.edu.cn -
基金资助:国家重点研发计划(2018YFA0901500)
Research progresses and future prospects of synthetic methylotrophic cell factory for methanol assimilation
ZHANG Hui1,2, YUAN Yaomeng3,4, ZHANG Chong3,4, YANG Song1,2,5, XING Xinhui3,4
- 1.School of Life Sciences,Qingdao Agricultural University,Qingdao 266109,Shandong,China
2.Shandong Province Key Laboratory of Applied Mycology,Qingdao Agricultural University,Qingdao 266109,Shandong,China
3.Key Lab of Industrial Biocatalysis,Ministry of Education,Institute of Biochemical Engineering,Department of Chemical Engineering,Tsinghua University,Beijing 100084,China
4.Center for Synthetic and Systems Biology,Tsinghua University,Beijing 100084,China
5.Qingdao International Center on Microbes Utilizing Biogas,Qingdao Agricultural University,Qingdao 266109,Shandong,China
-
Received:2020-06-15Revised:2021-01-11Online:2021-04-30Published:2021-04-30 -
Contact:YANG Song, XING Xinhui
摘要:
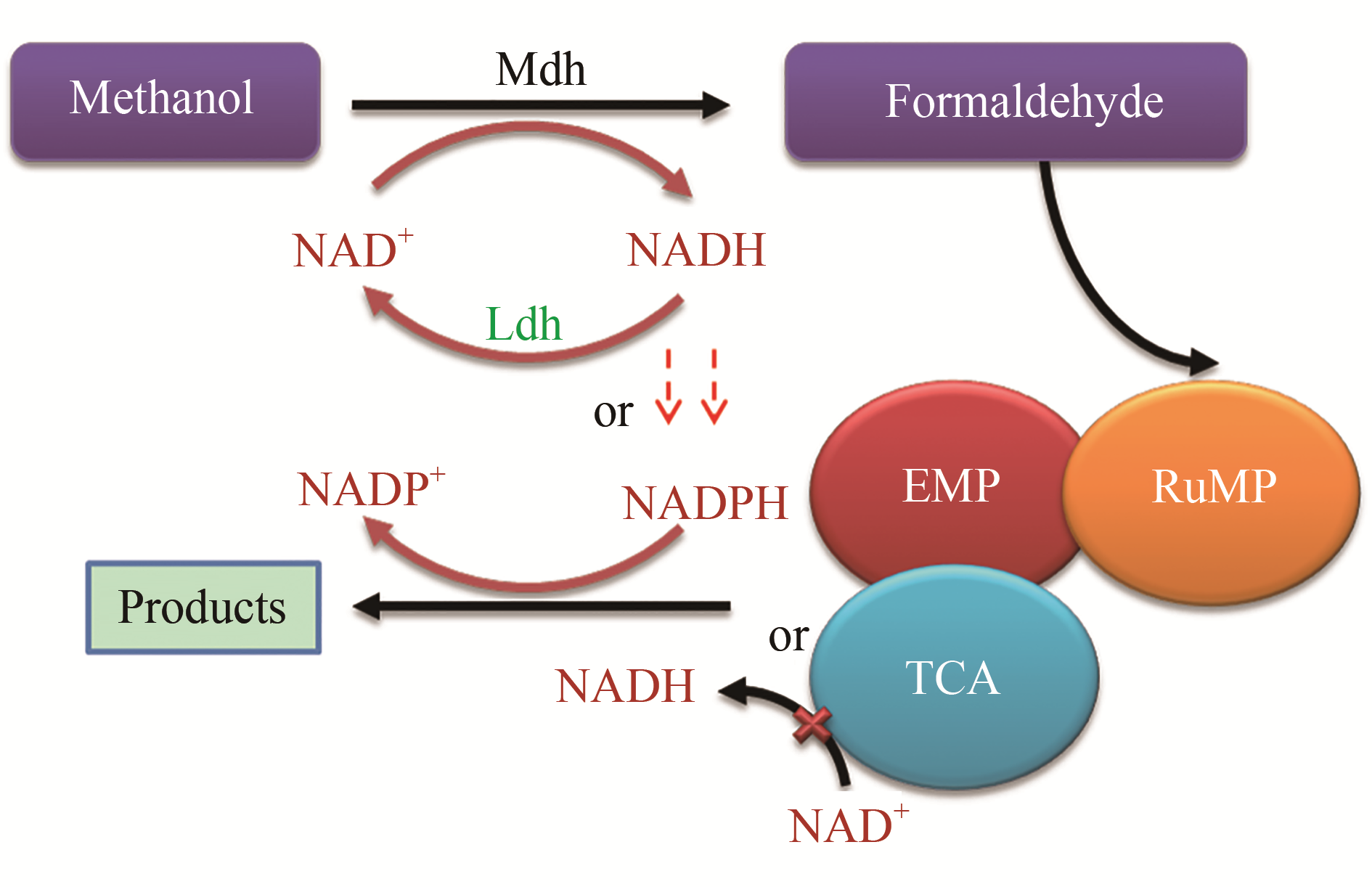
甲醇具有来源广、易储存运输、原料价格竞争力强等优势,被视为极具潜力的生物制造非糖基碳源资源。常用的模式底盘微生物研究历史长、认知清楚、操作工具多,在工程化改造中具有显著优势。近年来,通过借鉴天然甲基营养型微生物的甲醇利用途径对模式底盘进行改造,获得具备高效利用甲醇能力的合成甲基营养细胞工厂的研究日益受到关注。本文系统综述合成甲基营养细胞工厂的甲醇氧化、同化基因及其调控元件,和以共用糖基碳源结合适应性进化策略为主的核酮糖单磷酸途径重构及甲醇同化途径设计构建的研究现状,进而结合合成甲基营养细胞工厂面临的挑战进行展望,提出基于全基因组靶向基因编辑技术结合实验室适应性进化策略构建更高效合成甲基营养细胞工厂的研究方向。
中图分类号:
引用本文
张卉, 袁姚梦, 张翀, 杨松, 邢新会. 合成甲基营养细胞工厂同化甲醇的研究进展及未来展望[J]. 合成生物学, 2021, 2(2): 222-233.
ZHANG Hui, YUAN Yaomeng, ZHANG Chong, YANG Song, XING Xinhui. Research progresses and future prospects of synthetic methylotrophic cell factory for methanol assimilation[J]. Synthetic Biology Journal, 2021, 2(2): 222-233.
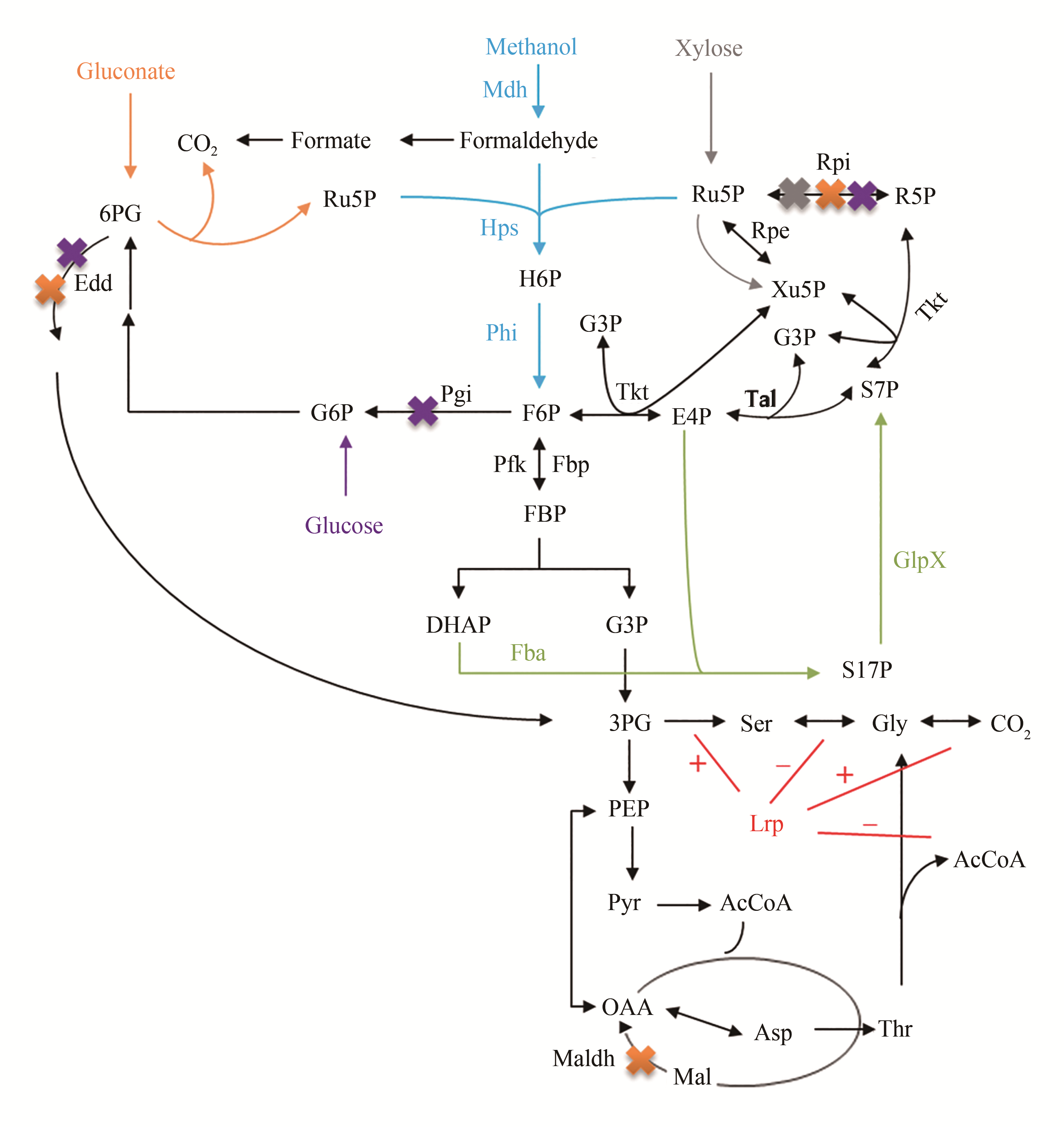
图1 基于共用糖基碳源的合成甲基营养细胞工厂构建的代谢途径[36-38,41-45](蓝色指示代表RuMP途径;橘色,紫色和灰色号分别指示在合成甲基营养细胞工厂构建中敲除edd、rpi和maldh,并以葡萄糖酸为共用糖基的代谢途径,敲除edd、rpi和pgi并以葡萄糖为共用糖基的代谢途径以及敲除rpi以木糖为共用糖基的代谢途径;绿色指示代表异源表达glpx和fba的基因;红色指示代表亮氨酸响应调控蛋白的调控,其中“+”代表激活途径,“-”代表抑制途径)
Fig. 1 The construction of the synthetic methylotrophic cell factory based on the sugar as the co-substrate[36-38, 41-45][The blue represents the Rump pathway; the orange, purple and gray represent the metabolic pathway of knocking out genes (edd, rpi and maldh) based on the gluconate as the co-substrate, knocking out genes (edd, rpi and pgi) using glucose as the the co-substrate, knocking out the gene rpi based on the xylose as the co-substrate, respectively; the green represents the heterologous expression of genes glpx and fba. The red represents the regulation of leucine-responsive protein, in which "+" represents the activation pathway and "-" represents the inhibition pathway]
| 碳源 | 宿主 | Mdh、Hps和Phi来源 | 甲醇同化进展 | 文献 |
|---|---|---|---|---|
| 甲醇和酵母抽提物 | 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 39%三羧酸循环中间产物和53%糖酵解中间产物被13C甲醇标记,实现柚皮素合成 | [ |
| 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 增强核酮糖-5-磷酸再生和甲醇同化,增加3-磷酸甘油酸和磷酸烯醇式丙酮酸的13C甲醇标记量 | [ | |
| 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | P frm 控制动态调控酶的活性,增加大肠杆菌在甲醇中的生长速率 | [ | |
甲醇和 葡萄糖 | 谷氨酸棒状杆菌 | 甲醇芽孢杆菌,枯草芽孢杆菌,枯草芽孢杆菌 | 甲醇消耗速率为1.7 mmol/(L·h) | [ |
| 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 增加甲醇进入中间代谢物的碳通量 | [ | |
| 甲醇和葡萄糖酸盐 | 大肠杆菌 | 甲醇芽孢杆菌,甲基鞭毛杆菌,甲基鞭毛杆菌 | 通过实验室适应性进化,24%甲醇进入中心碳代谢,甲醇可作为主要生长碳源 | [ |
甲醇和 木糖 | 大肠杆菌 | 钩虫贪铜菌,甲醇芽孢杆菌,甲基鞭毛杆菌 | 通过实验室适应性进化,甲醇和木糖大约以1∶1的摩尔比共同消耗,进化菌株的生长速率为(0.17±0.006) h-1,并利用甲醇生产乙醇和丁醇 | [ |
| 大肠杆菌 | 甲醇芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 进化的Mdh2突变体最大反应速度增加3.5倍,甲醇进入突变菌株中心代谢物的碳通量是未突变菌株的2倍 | [ | |
| 谷氨酸棒状杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 高达63%的13C甲醇进入细胞内代谢物 | [ | |
甲醇和 核糖 | 大肠杆菌 | 甲醇芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 40%的甲醇进入中心碳代谢途径 | [ |
| 谷氨酸棒状杆菌 | 甲醇芽孢杆菌,枯草芽孢杆菌,枯草芽孢杆菌 | 25%的13C标记出现在糖酵解和磷酸戊糖途径中间代谢物 | [ |
表1 共用糖基碳源增强甲醇同化
Tab. 1 Enhancement of methanol assimilation based on the sugar as the co-substrate
| 碳源 | 宿主 | Mdh、Hps和Phi来源 | 甲醇同化进展 | 文献 |
|---|---|---|---|---|
| 甲醇和酵母抽提物 | 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 39%三羧酸循环中间产物和53%糖酵解中间产物被13C甲醇标记,实现柚皮素合成 | [ |
| 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 增强核酮糖-5-磷酸再生和甲醇同化,增加3-磷酸甘油酸和磷酸烯醇式丙酮酸的13C甲醇标记量 | [ | |
| 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | P frm 控制动态调控酶的活性,增加大肠杆菌在甲醇中的生长速率 | [ | |
甲醇和 葡萄糖 | 谷氨酸棒状杆菌 | 甲醇芽孢杆菌,枯草芽孢杆菌,枯草芽孢杆菌 | 甲醇消耗速率为1.7 mmol/(L·h) | [ |
| 大肠杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 增加甲醇进入中间代谢物的碳通量 | [ | |
| 甲醇和葡萄糖酸盐 | 大肠杆菌 | 甲醇芽孢杆菌,甲基鞭毛杆菌,甲基鞭毛杆菌 | 通过实验室适应性进化,24%甲醇进入中心碳代谢,甲醇可作为主要生长碳源 | [ |
甲醇和 木糖 | 大肠杆菌 | 钩虫贪铜菌,甲醇芽孢杆菌,甲基鞭毛杆菌 | 通过实验室适应性进化,甲醇和木糖大约以1∶1的摩尔比共同消耗,进化菌株的生长速率为(0.17±0.006) h-1,并利用甲醇生产乙醇和丁醇 | [ |
| 大肠杆菌 | 甲醇芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 进化的Mdh2突变体最大反应速度增加3.5倍,甲醇进入突变菌株中心代谢物的碳通量是未突变菌株的2倍 | [ | |
| 谷氨酸棒状杆菌 | 嗜热脂肪芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 高达63%的13C甲醇进入细胞内代谢物 | [ | |
甲醇和 核糖 | 大肠杆菌 | 甲醇芽孢杆菌,甲醇芽孢杆菌,甲醇芽孢杆菌 | 40%的甲醇进入中心碳代谢途径 | [ |
| 谷氨酸棒状杆菌 | 甲醇芽孢杆菌,枯草芽孢杆菌,枯草芽孢杆菌 | 25%的13C标记出现在糖酵解和磷酸戊糖途径中间代谢物 | [ |
| 宿主 | 基因名称 | 基因功能 | 基因突变类型 | 依赖甲醇生长的预测功能 | 文献 |
|---|---|---|---|---|---|
| 大肠杆菌 | frmA | 谷胱甘肽依赖的甲醛脱氢酶 | 移码突变 | 促进甲醛氧化为CO2,提供额外的NADH | [ |
| fdoG | 甲酸脱氢酶 | 移码突变 | 促进甲酸氧化为CO2,提供额外的NADH | ||
| gltA | 柠檬酸合成酶 | 转座子插入 | 降低三羧酸循环的碳通量,插入转座子元件 | ||
| ptsH | 编码Hpr的蛋白酶 | 转座子插入 | 破坏磷酸糖转移酶系统,插入转座子元件 | ||
| pgi | 磷酸葡萄糖异构酶 | 基因内缺失 | 提高磷酸葡萄糖异构酶酶活,增加细胞生长所需的NADPH | ||
| 大肠杆菌 | gntR | DNA结合转录抑制子 | 碱基置换 | 改变葡萄糖酸摄取量 | [ |
| frmA | 谷胱甘肽依赖的甲醛脱氢酶 | 移码突变 | 失活甲醛氧化途径,甲醛用于同化途径 | ||
| nadR | DNA结合转录抑制子/烟酰胺单核苷酸腺苷转移酶 | 碱基置换 | 改变NAD+/NADH比值 | ||
| 大肠杆菌 | ptsI | 组成磷酸转移酶系统的酶I | 基因内缺失 | 降低葡萄糖消耗速率 | [ |
| icd | 异柠檬酸脱氢酶 | 基因内缺失 | 降低三羧酸循环的碳通量,以维持细胞氧化还原平衡 | ||
| 大肠杆菌 | zwf | 6-磷酸葡萄糖脱氢酶 | 碱基置换 | 调低Entner-Doudoroff途径通量 | [ |
| pykF | 丙酮酸激酶I | 基因内插入 | 调低糖酵解途径通量 | ||
| cyaA | 腺苷酸环化酶 | 基因内插入 | 降低三羧酸循环相关酶的转录水平,改变NAD+/NADH比值 | ||
| deoD | 嘌呤核苷磷酸化酶 | 基因内插入 | 提供甲醛固定受体 | ||
| frmAB, yaiO | 甲醛脱毒操纵子,外膜蛋白 | 基因间缺失 | 增加细胞内甲醛浓度 | ||
| 谷氨酸棒状杆菌 | atlR | 碳水化合物代谢的多功能调节子 | 碱基置换 | 提高醇脱氢酶和木糖激酶的催化活性以强化甲醇和木糖的利用效率 | [ |
| metY | 邻乙酰高丝氨酸巯基化酶 | 碱基置换 | 提高宿主对甲醇的耐受性 | ||
| mtrA | 参与细胞形态、抗生素易感性、渗透保护在内的基因功能的多重调控子 | 碱基置换 | 通过调节类谷氧还蛋白和NAD+合成酶,参与维持细胞内氧化还原状态 | ||
| cgl2030, ctaE | 预测的ATP激酶,细胞色素C氧化酶亚基 蛋白酶 | 碱基置换 | 改变能量代谢 |
表2 甲醇依赖菌株通过ALE获得的有利突变
Tab. 2 Summary of mutations obtained by adaptive laboratory evolution in synthetic methanol-dependent strains
| 宿主 | 基因名称 | 基因功能 | 基因突变类型 | 依赖甲醇生长的预测功能 | 文献 |
|---|---|---|---|---|---|
| 大肠杆菌 | frmA | 谷胱甘肽依赖的甲醛脱氢酶 | 移码突变 | 促进甲醛氧化为CO2,提供额外的NADH | [ |
| fdoG | 甲酸脱氢酶 | 移码突变 | 促进甲酸氧化为CO2,提供额外的NADH | ||
| gltA | 柠檬酸合成酶 | 转座子插入 | 降低三羧酸循环的碳通量,插入转座子元件 | ||
| ptsH | 编码Hpr的蛋白酶 | 转座子插入 | 破坏磷酸糖转移酶系统,插入转座子元件 | ||
| pgi | 磷酸葡萄糖异构酶 | 基因内缺失 | 提高磷酸葡萄糖异构酶酶活,增加细胞生长所需的NADPH | ||
| 大肠杆菌 | gntR | DNA结合转录抑制子 | 碱基置换 | 改变葡萄糖酸摄取量 | [ |
| frmA | 谷胱甘肽依赖的甲醛脱氢酶 | 移码突变 | 失活甲醛氧化途径,甲醛用于同化途径 | ||
| nadR | DNA结合转录抑制子/烟酰胺单核苷酸腺苷转移酶 | 碱基置换 | 改变NAD+/NADH比值 | ||
| 大肠杆菌 | ptsI | 组成磷酸转移酶系统的酶I | 基因内缺失 | 降低葡萄糖消耗速率 | [ |
| icd | 异柠檬酸脱氢酶 | 基因内缺失 | 降低三羧酸循环的碳通量,以维持细胞氧化还原平衡 | ||
| 大肠杆菌 | zwf | 6-磷酸葡萄糖脱氢酶 | 碱基置换 | 调低Entner-Doudoroff途径通量 | [ |
| pykF | 丙酮酸激酶I | 基因内插入 | 调低糖酵解途径通量 | ||
| cyaA | 腺苷酸环化酶 | 基因内插入 | 降低三羧酸循环相关酶的转录水平,改变NAD+/NADH比值 | ||
| deoD | 嘌呤核苷磷酸化酶 | 基因内插入 | 提供甲醛固定受体 | ||
| frmAB, yaiO | 甲醛脱毒操纵子,外膜蛋白 | 基因间缺失 | 增加细胞内甲醛浓度 | ||
| 谷氨酸棒状杆菌 | atlR | 碳水化合物代谢的多功能调节子 | 碱基置换 | 提高醇脱氢酶和木糖激酶的催化活性以强化甲醇和木糖的利用效率 | [ |
| metY | 邻乙酰高丝氨酸巯基化酶 | 碱基置换 | 提高宿主对甲醇的耐受性 | ||
| mtrA | 参与细胞形态、抗生素易感性、渗透保护在内的基因功能的多重调控子 | 碱基置换 | 通过调节类谷氧还蛋白和NAD+合成酶,参与维持细胞内氧化还原状态 | ||
| cgl2030, ctaE | 预测的ATP激酶,细胞色素C氧化酶亚基 蛋白酶 | 碱基置换 | 改变能量代谢 |
| 1 | CLOMBURG J M, CRUMBLEY A M, GONZALEZ R. Industrial biomanufacturing: the future of chemical production[J]. Science, 2017, 355(6320): 38. |
| 2 | ZHU T, ZHAO T, BANKEFA O E, et al. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: challenges and opportunities[J]. Biotechnology Advances, 2020, 39: 107467. |
| 3 | MARTENS J A, BOGAERTS A, DE KIMPE N, et al. The chemical route to a carbon dioxide neutral world[J]. Chemsuschem, 2017, 10(6): 1039-1055. |
| 4 | OLAH G A. Beyond oil and gas: the methanol economy[J]. Angewandte Chemie International Edition, 2005, 44(18): 2636-2639. |
| 5 | ZHANG M, YUAN X J, ZHANG C, et al. Bioconversion of methanol into value-added chemicals in native and synthetic methylotrophs[J]. Current Issues Molecular Biology, 2019, 33: 225-236. |
| 6 | SCHRADER J, SCHILLING M, HOLTMANN D, et al. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria[J]. Trends in Biotechnology, 2009, 27(2):107-115. |
| 7 | ZHANG W M, SONG M, YANG Q, et al. Current advance in bioconversion of methanol to chemicals[J]. Biotechnology for Biofuels, 2018, 11: 260. |
| 8 | CHISTOSERDOVA L, KALYUZHNAYA M G. Current trends in methylotrophy[J]. Trends in Microbiology, 2018, 26(8): 703-714. |
| 9 | CUI J, GOOD N M, HU B, et al. Metabolomics revealed an association of metabolite changes and defective growth in Methylobacterium extorquens AM1 overexpressing ecm during growth on methanol[J]. PLoS One, 2016, 11(4): e0154043. |
| 10 | ANTHONY C, GHOSH M. The structure and function of the PQQ-containing quinoprotein dehydrogenases[J]. Progress in Biophysics and Molecular Biology, 1998, 69(1): 1-21. |
| 11 | KROG A, HEGGESET T M, MÜLLER J E, et al. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties[J]. PLoS One, 2013, 8(3): e59188. |
| 12 | GUNKEL K, DIJK R VAN, VEENHUIS M, et al. Routing of Hansenula polymorpha alcohol oxidase: an alternative peroxisomal protein-sorting machinery[J]. Molecular Biology of the Cell, 2004, 15(3): 1347-1355. |
| 13 | ZHU W L, CUI J Y, CUI L Y, et al. Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway[J]. Applied Microbiology and Biotechnology, 2016, 100(5): 2171-2182. |
| 14 | LIANG W F, CUI L Y, CUI J Y, et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39: 159-168. |
| 15 | YANG Y M, CHEN W J, YANG J, et al. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route[J]. Microbial Cell Factories, 2017, 16(1): 179. |
| 16 | SONNTAG F, KRONER C, LUBUTA P, et al. Engineering Methylobacterium extorquens for de novo synthesis of the sesquiterpenoid α-humulene from methanol[J]. Metabolic Engineering, 2015, 32: 82-94. |
| 17 | LENNART S V B, REMUS-EMSERMANN M, WEISHAUPT R, et al. A set of versatile brick vectors and promoters for the assembly, expression, and integration of synthetic operons in Methylobacterium extorquens AM1 and other alphaproteobacteria[J]. ACS Synthetic Biology, 2015, 4(4): 430. |
| 18 | CARRILLO M, WAGNER M, PETIT F, et al. Design and control of extrachromosomal elements in Methylorubrum extorquens AM1[J]. ACS Synthetic Biology, 2019, 8(11): 2451-2456. |
| 19 | CHISTOSERDOVA L. Modularity of methylotrophy, revisited[J]. Environmental Microbiology, 2011, 13(10): 2603-2622. |
| 20 | MÜLLER J E, MEYER F, LITSANOV B, et al. Core pathways operating during methylotrophy of Bacillus methanolicus MGA3 and induction of a bacillithiol-dependent detoxification pathway upon formaldehyde stress[J]. Molecular Microbiology, 2015, 98(6): 1089-1100. |
| 21 | WHITAKER W B, SANDOVAL N R, BENNETT R K, et al. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization[J]. Current Opinion in Biotechnology, 2015, 33: 165-175. |
| 22 | WHITAKER W B, JONES J A, BENNETT R K, et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli [J]. Metabolic Engineering, 2017, 39: 49-59. |
| 23 | WOOLSTON B M, KING J R, REITER M, et al. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli [J]. Nature Communications, 2018, 9(1): 2387. |
| 24 | CHEN F Y H, JUNG H W, TSUEI C Y, et al. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol[J]. Cell, 2020, 182(4): 933-946. e14. |
| 25 | WITTHOFF S, SCHMITZ K, NIEDENFÜHR S, et al. Metabolic engineering of Corynebacterium glutamicum for methanol metabolism[J]. Applied and Environmental Microbiology, 2015, 81(6): 2215-2225. |
| 26 | MÜLLER J E N, MEYER F, LITSANOV B, et al. Engineering Escherichia coli for methanol conversion[J]. Metabolic Engineering, 2015, 28: 190-201. |
| 27 | WU T Y, CHEN C T, LIU J T J, et al. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1[J]. Applied Microbiology and Biotechnology, 2016, 100(11): 4969-4983. |
| 28 | ROTH T B, WOOLSTON B M, STEPHANOPOULOS G, et al. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2[J]. ACS Synthetic Biology, 2019, 8(4): 796-806. |
| 29 | SIEGEL J B, SMITH A L, POUST S, et al. Computational protein design enables a novel one-carbon assimilation pathway[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(12): 3704-3709. |
| 30 | LU X, LIU Y, YANG Y, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design[J]. Nature Communications, 2019, 10(1): 1378. |
| 31 | PRICE J V, CHEN L, WHITAKER W B, et al. Scaffoldless engineered enzyme assembly for enhanced methanol utilization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(45): 12691-12696. |
| 32 | FAN L, WANG Y, TUYISHIME P, et al. Engineering artificial fusion proteins for enhanced methanol bioconversion[J]. 2018, 19(23): Chembiochem, 2465-2471. |
| 33 | OSMAN D, PIERGENTILI C, CHEN J, et al. The effectors and sensory sites of formaldehyde-responsive regulator FrmR and metal-sensing variant[J]. The J ournal of Biological Chemistry, 2016, 291(37): 19502-19516. |
| 34 | DENBY K J, IWIG J, BISSON C, et al. The mechanism of a formaldehyde-sensing transcriptional regulator[J]. Scientific Reports, 2016, 6: 38879. |
| 35 | ROHLHILL J, SANDOVAL N R, PAPOUTSAKIS E T. Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol[J]. ACS Synthetic Biology, 2017, 6(8): 1584-1595. |
| 36 | BENNETT R K, GONZALEZ J E, WHITAKER W B, et al. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph[J]. Metabolic Engineering, 2018, 45: 75-85. |
| 37 | BRAUTASET T, JAKOBSEN M Ø M, FLICKINGER M C, et al. Plasmid-dependent methylotrophy in thermotolerant Bacillus methanolicus [J]. Journal of Bacteriology, 2004, 186(5): 1229-1238. |
| 38 | MÜLLER J E, HEGGESET T M, WENDISCH V F, et al. Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol[J]. Applied Microbiology and Biotechnology, 2015, 99(2): 535-551. |
| 39 | ROHLHILL J, GERALD HAR J R, ANTONIEWICZ M R, et al. Improving synthetic methylotrophy via dynamic formaldehyde regulation of pentose phosphate pathway genes and redox perturbation[J]. Metabolic Engineering, 2020, 57: 247-255. |
| 40 | KELLER P, NOOR E, MEYER F, et al. Methanol-dependent Escherichia coli strains with a complete ribulose monophosphate cycle[J]. Nature Communications, 2020, 11(1): 5403. |
| 41 | MEYER F, KELLER P, HARTL J, et al. Methanol-essential growth of Escherichia coli [J]. Nature Communications, 2018, 9(1): 1508. |
| 42 | BENNETT R K, DILLON M, GERALD HAR J R, et al. Engineering Escherichia coli for methanol-dependent growth on glucose for metabolite production[J]. Metabolic Engineering, 2020, 60: 45-55. |
| 43 | CHEN C T, CHEN F Y, BOGORAD I W, et al. Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production[J]. Metabolic Engineering, 2018, 49: 257-266. |
| 44 | TUYISHIME P, WANG Y, FAN L, et al. Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production[J]. Metabolic Engineering, 2018, 49: 220-231. |
| 45 | GONZALEZ J E, BENNETT R K, PAPOUTSAKIS E T, et al. Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein[J]. Metabolic Engineering, 2018, 45: 67-74. |
| 46 | LEßMEIER L, PFEIFENSCHNEIDER J, CARNICER M, et al. Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate[J]. Applied Microbiology and Biotechnology, 2015, 99(23): 10163-10176. |
| 47 | WANG X, WANG X L, LU X L, et al. Methanol fermentation increases the production of NAD(P)H-dependent chemicals in synthetic methylotrophic Escherichia coli [J]. Biotechnology for Biofuels, 2019, 12(1): 17. |
| 48 | WANG C, REN J, ZHOU L, et al. An aldolase-catalyzed new metabolic pathway for the assimilation of formaldehyde and methanol to synthesize 2-keto-4-hydroxybutyrate and 1, 3-propanediol in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8(11): 2483-2493. |
| 49 | YU H, LIAO J C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9(1): 3992. |
| 50 | HE H, HOPER R, DODENHOFT M, et al. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E. coli [J]. Metabolic Engineering, 2020, 60: 1-13. |
| 51 | DAI Z, GU H, ZHANG S, et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae [J]. Bioresource Technology, 2017, 245(pt b): 1407-1412. |
| 52 | BOGORAD I W, CHEN C T, THEISEN M K, et al. Building carbon-carbon bonds using a biocatalytic methanol condensation cycle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(45): 15928-15933. |
| 53 | CHOU A, CLOMBURG J M, QIAN S, et al. 2-Hydroxyacyl-CoA lyase catalyzes acyloin condensation for one-carbon bioconversion[J]. Nature Chemical Biology, 2019, 15(9): 900-906. |
| 54 | YANG X, YUAN Q, LUO H, et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways[J]. Metabolic Engineering, 2019, 56: 142-153. |
| 55 | ZHANG J, JENSEN M K, KEASLING J D. Development of biosensors and their application in metabolic engineering[J]. Current Opinion in Chemical Biology, 2015, 28: 1-8. |
| 56 | ROGERS J K, TAYLOR N D, CHURCH G M. Biosensor-based engineering of biosynthetic pathways[J]. Current Opinion in Biotechnology, 2016, 42: 84-91. |
| 57 | CHEN N H, DJOKO K Y, VEYRIER F J, et al. Formaldehyde stress responses in bacterial pathogens[J]. Frontiers in Microbiology, 2016, 7: 257. |
| 58 | CUI L Y, WANG S S, GUAN C G, et al. Breeding of methanol-tolerant Methylobacterium extorquens AM1 by atmospheric and room temperature plasma mutagenesis combined with adaptive laboratory evolution[J]. Biotechnology Journal, 2018, 13(6): e1700679. |
| 59 | HU B, YANG Y M, BECK D A, et al. Comprehensive molecular characterization of Methylobacterium extorquens AM1 adapted for 1-butanol tolerance[J]. Biotechnology for Biofuels, 2016, 9: 84. |
| 60 | HU B, LIDSTROM M E. Metabolic engineering of Methylobacterium extorquens AM1 for 1-butanol production[J]. Biotechnology for Biofuels, 2014, 7(1): 1-10. |
| 61 | WANG T M, GUAN C G, GUO J H, et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance[J]. Nature Communications, 2018, 9(1): 2475. |
| 62 | MO X H, ZHANG H, WANG T M, et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis[J]. Applied Microbiology and Biotechnology, 2020, 104(10): 4515-4532. |
| 63 | GLEIZER S, BEN-NISSAN R, BAR-ON Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 [J]. Cell, 2019, 179(6): 1255-1263. |
| 64 | FABARIUS J T, WEGAT V, ROTH A, SIEBER V. Synthetic methylotrophy in yeasts: towards a circular bioeconomy[J]. Trends in Biotechnology, 2020, 39(4):348-358. |
| 65 | DONG C, FONTANA J, PATEL A, et al. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria[J]. Nature Communications, 2018, 9(1): 2489. |
| 66 | GARST A D, BASSALO M C, PINES G, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering[J]. Nature Biotechnology, 2017, 35(1): 48-55. |
| 67 | WONG B G, MANCUSO C P, KIRIAKOV S, et al. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER[J]. Nature Biotechnology, 2018, 36(7): 614-623. |
| 68 | JIAN X J, GUO X J, WANG J, et al. Microbial microdroplet culture system (MMC): An integrated platform for automated, high-throughput microbial cultivation and adaptive evolution[J]. Biotechnology and Bioengineering, 2020, 117(6): 1724-1737. |
| [1] | 王俊婷, 郭潇佳, 李青, 万里, 赵宗保. 创制非天然辅酶偏好型甲醇脱氢酶[J]. 合成生物学, 2021, 2(4): 651-661. |
| [2] | 王也, 王昊晨, 晏明皓, 胡冠华, 汪小我. 生物分子序列的人工智能设计[J]. 合成生物学, 2021, 2(1): 1-14. |
| [3] | 高教琪, 周雍进. 甲醇生物转化的机遇与挑战[J]. 合成生物学, 2020, 1(2): 158-173. |
| [4] | 张博, 马永硕, 尚轶, 黄三文. 植物合成生物学研究进展[J]. 合成生物学, 2020, 1(2): 121-140. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||