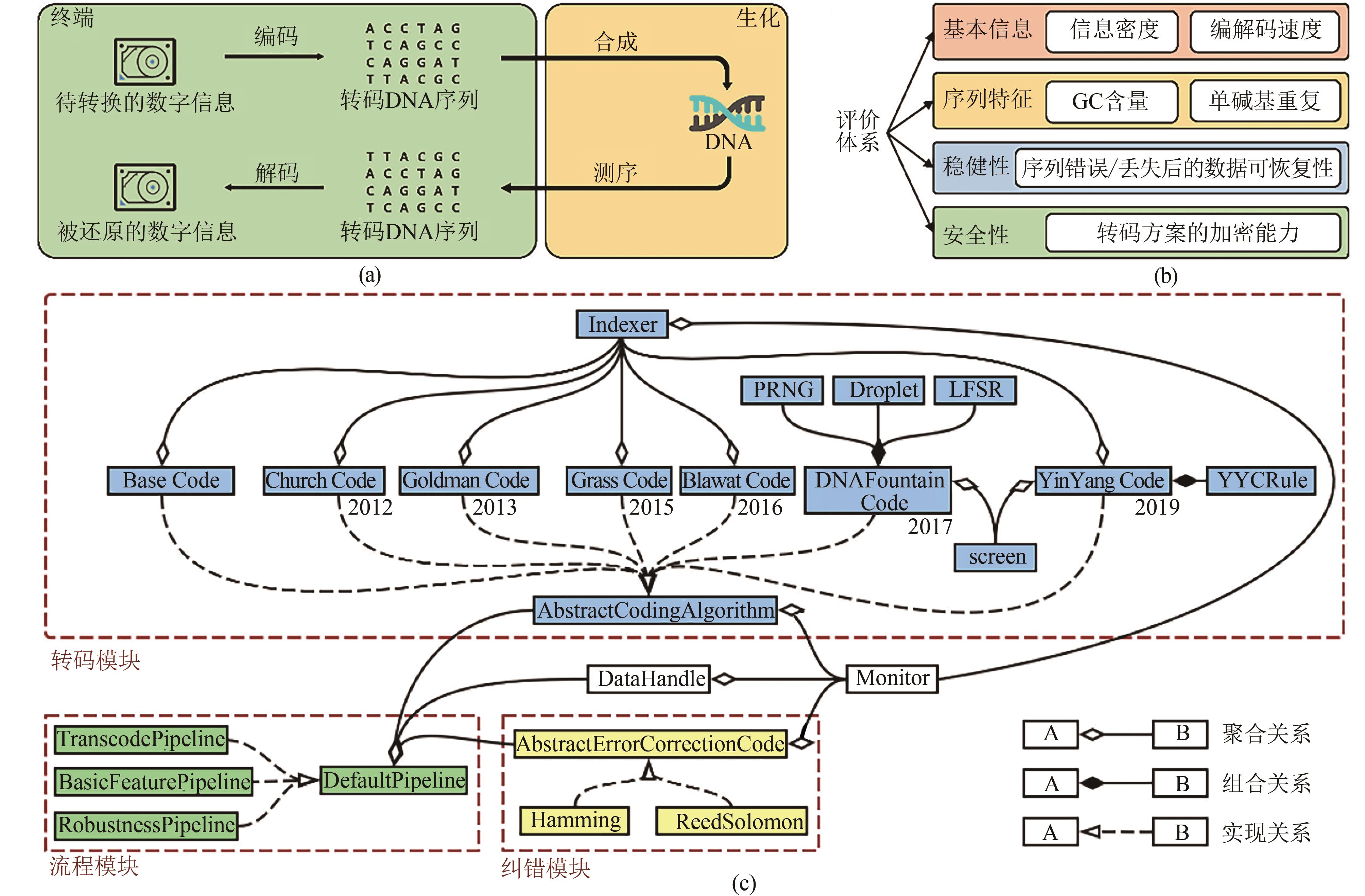
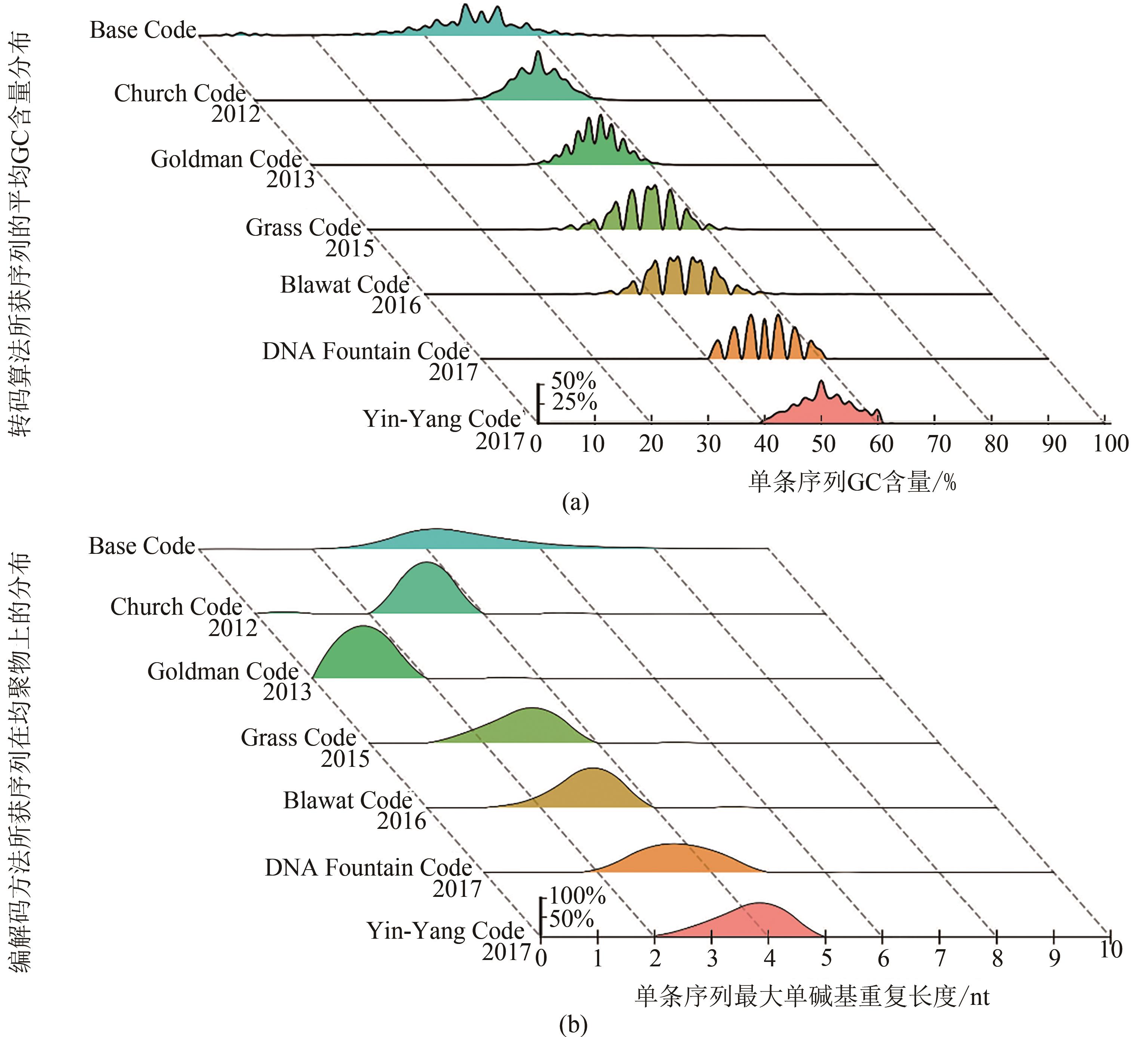
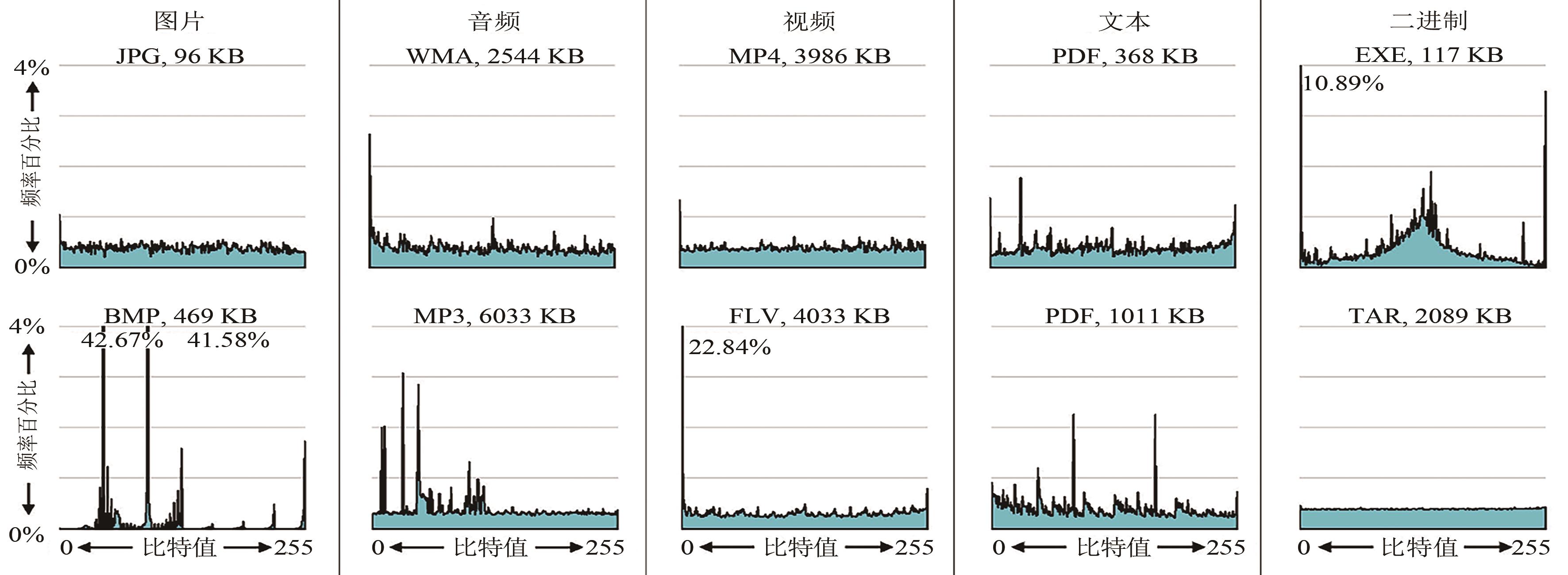
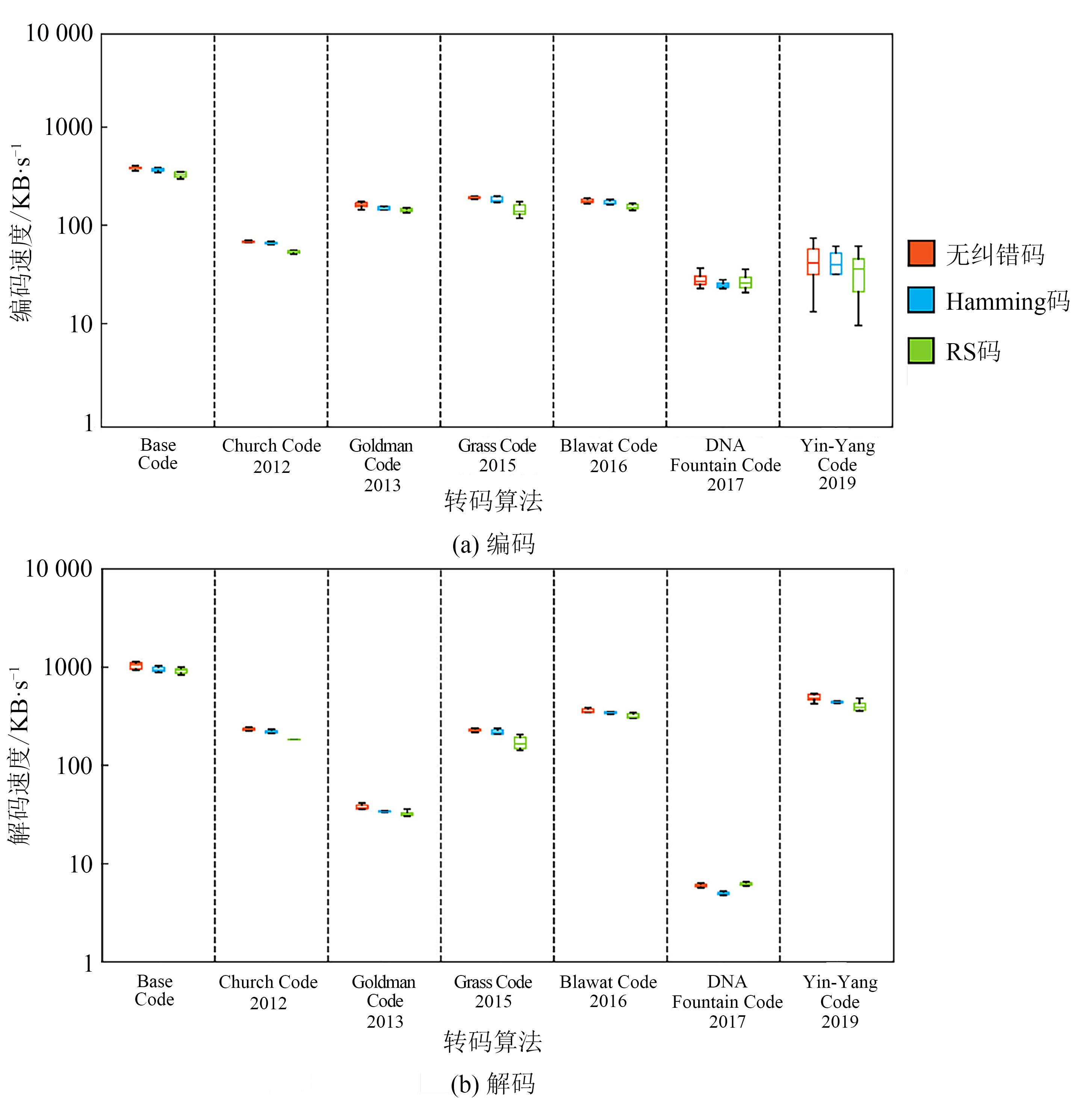
| 1 |
PING Z, MA D Z, HUANG X L, et al. Carbon-based archiving: current progress and future prospects of DNA-based data storage [J]. Gigascience, 2019, 8(6): gizo75.
|
| 2 |
DONG Y M, SUN F J, PING Z, et al. DNA storage: research landscape and future prospects [J]. National Science Review, 2020, 7(6): 1092-1107.
|
| 3 |
CHURCH G M, GAO Y, KOSURI S, Next-generation digital information storage in DNA [J]. Science, 2012, 337(6102): 1628.
|
| 4 |
GOLDMAN N, BERTONE P, CHEN S, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA [J]. Nature, 2013, 494(7435): 77-80.
|
| 5 |
GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes [J]. Angewandte Chemie International Edition, 2015, 54(8): 2552-2555.
|
| 6 |
ERLICH Y, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture [J]. Science, 2017, 355(6328): 950-954.
|
| 7 |
PING Z, CHEN S, HUANG X, et al. Towards practical and robust DNA-based data archiving by codec system named 'Yin-Yang' [EB/OL]. [2021-05-26]. .
|
| 8 |
HAO M, QIAO H, GAO Y, et al. A mixed culture of bacterial cells enables an economic DNA storage on a large scale [J]. Communications Biology, 2020, 3(1): 416.
|
| 9 |
PRESS W H, HAWKINS J A, JONES S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints [J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(31): 18489.
|
| 10 |
WANG Y X, NOOR-A-RAHIM M, GUNAWAN E, et al. Construction of bio-constrained code for DNA data storage [J]. IEEE Communications Letters, 2019, 23(6): 963-966.
|
| 11 |
NGUYEN T T, CAI K, IMMINK K A S, et al. Constrained coding with error control for DNA-based data storage [C]//2020 IEEE International Symposium on Information Theory (ISIT). 2020.
|
| 12 |
BLAWAT M, GAEDKE K, HUETTER I, et al. Forward error correction for DNA data storage [J]. Procedia Computer Science, 2016, 80: 1011-1022.
|
| 13 |
FOWLER M. Refactoring: improving the design of existing code [M]. Boston: Addison-Wesley Professional, 2018.
|
| 14 |
COX B J. Object-oriented programming: an evolutionary approach [M]. Boston: Addison-Wesley, 1986.
|
| 15 |
KOCH J, GANTENBEIN S, MASANIA K, et al. A DNA-of-things storage architecture to create materials with embedded memory [J]. Nature Biotechnology, 2020, 38(1): 39-43.
|
| 16 |
TANENBAUM A S BOS H. Modern operating systems [M]. London: Pearson, 2015.
|
| 17 |
SAYOOD K. Introduction to data compression [M]. Burlington: Morgan Kaufmann, 2017.
|
| 18 |
FENG L, FOH C H, JIANFEI C, et al. LT codes decoding: design and analysis [C]//2009 IEEE International Symposium on Information Theory. 2009.
|
| 19 |
YAZDI S H T, GABRYS R, MILENKOVIC O. Portable and error-free DNA-based data storage [J]. Scientific Reports, 2017, 7(1): 1-6.
|
| 20 |
ORGANICK L, CHEN Y J, ANG S D, et al. Probing the physical limits of reliable DNA data retrieval [J]. Nature Communications, 2020, 11(1): 1-7.
|
| 21 |
HECKEL R, MIKUTIS G, GRASS R N. A characterization of the DNA data storage channel [J]. Scientific Reports, 2019, 9(1): 9663.
|
| 22 |
MACKAY D J MAC KAY D J. Information theory, inference and learning algorithms [M]. Cambridge: Cambridge University Press, 2003.
|
| 23 |
KOSURI S, CHURCH G M. Large-scale de novo DNA synthesis: technologies and applications [J]. Nature Methods, 2014, 11(5): 499-507.
|
| 24 |
KULSKI J K. Next generation sequencing-advances, applications and challenges[M]. London: Intech Open, 2016: 3-60.
|
| 25 |
CHEN Y J, TAKAHASHI C N, ORGANICK L, et al. Quantifying molecular bias in DNA data storage [J]. Nature Communications, 2020, 11(1). DOI:http://doi.org/10.1038/s41467-020-16958-3 .
|
| 26 |
MOON T K. Error correction coding: mathematical methods and algorithms [M]. Hoboken: John Wiley & Sons, 2005.
|
| 27 |
STINSON D R, PATERSON M. Cryptography: theory and practice[M]. Boca Raton: CRC Press, 2018.
|
| 28 |
PASCHKE J, BURKERT J, FEHRIBACH R. Computing and estimating the number of n-ary Huffman sequences of a specified length [J]. Discrete Mathematics, 2011, 311(1): 1-7.
|
| 29 |
KOSHY T. Catalan numbers with applications [M]. Oxford: Oxford University Press, 2008.
|
| 30 |
AVAL J C. Multivariate fuss-catalan numbers [J]. Discrete Mathematics, 2008, 308(20): 4660-4669.
|
| 31 |
WEST D B. Introduction to graph theory [M]. Hoboken : Prentice Hall, 1996.
|
| 32 |
COMPEAU P E, PEVZNER P A, TESLER G. How to apply de Bruijn graphs to genome assembly [J]. Nature Biotechnology, 2011, 29(11): 987-991.
|
| 33 |
BOUCHET A. Greedy algorithm and symmetric matroids [J]. Mathematical Programming, 1987, 38(2): 147-159.
|
| 34 |
MILO R, SHEN-ORR S, ITZKOVITZ S, et al. Network motifs: simple building blocks of complex networks [J]. Science, 2002, 298(5594): 824-827.
|
| 35 |
BORNHOLT J, LOPEZ R, CARMEAN D M, et al. A DNA-based archival storage system [C]//Proceedings of the Twenty-First International Conference on Architectural Support for Programming Languages and Operating Systems. 2016.
|
| 36 |
CAFFERTY B J, TEN A S, FINK M J, et al. Storage of information using small organic molecules [J]. ACS Central Science, 2019, 5(5): 911-916.
|
| 37 |
KENNEDY E, ARCADIA C E, GEISER J, et al. Encoding information in synthetic metabolomes [J]. PLoS One, 2019, 14(7): e0217364.
|