合成生物学 ›› 2023, Vol. 4 ›› Issue (2): 301-317.DOI: 10.12211/2096-8280.2022-058
合成生物学在疾病信息记录与实时监测中的应用潜力
马孟丹1,2,3, 刘宇辰1,2
- 1.深圳大学第一附属医院,深圳市第二人民医院,深圳转化医学研究院,广东 深圳 518035
2.广东省泌尿生殖肿瘤系统生物学与合成生物学重点实验室,广东 深圳 518035
3.汕头大学医学院,广东 汕头 515041
-
收稿日期:2022-10-21修回日期:2022-12-29出版日期:2023-04-30发布日期:2023-04-27 -
通讯作者:刘宇辰 -
作者简介:马孟丹 (1997—),女,硕士研究生。主要研究方向为医学合成生物学。E-mail:mamengdan7@163.com刘宇辰 (1988—),男,副研究员,研究生导师、博士后合作导师。主要研究方向:(1)肿瘤生物治疗和医学合成生物学;(2)创新肿瘤治疗新方法;(3)构建人工基因线路,肿瘤精准治疗。E-mail:liuyuchenmdcg@163.com -
基金资助:国家重点研发计划(2021YFA0911600)
Potential application of synthetic biology in disease information recording and real-time monitoring
MA Mengdan1,2,3, LIU Yuchen1,2
- 1.The first Affiliated Hospital of Shenzhen University,Shenzhen Second People’s Hospital,Shenzhen Institute of Translational Medicine,Shenzhen 518035,Guangdong,China
2.Guangdong Provincial Key Laboratory of Systems Biology and Synthetic Biology for Urogenital Tumors,Shenzhen 518035,Guangdong,China
3.Shantou University Medical College,Shantou 515041,Guangdong,China
-
Received:2022-10-21Revised:2022-12-29Online:2023-04-30Published:2023-04-27 -
Contact:LIU Yuchen
摘要:
中图分类号:
引用本文
马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317.
MA Mengdan, LIU Yuchen. Potential application of synthetic biology in disease information recording and real-time monitoring[J]. Synthetic Biology Journal, 2023, 4(2): 301-317.
| 记录装置系统 | 种群分布vs单细胞记录 | 写入周期 | 记录能力 | 发生顺序 | 持续时间 | 灵敏度 | 保真度 | |
|---|---|---|---|---|---|---|---|---|
| 双稳态开关 | 种群 | 短 | 一般 | 否 | 是 | 低 | 低 | |
| DNA重组酶技术 | 种群 | 长 | 一般 | 否 | 否 | 低 | 低 | |
| ssDNA编辑技术 | HiSCRIBE[ | 种群 | 长 | 强 | 否 | 是 | 一般 | 高 |
| CRISPR系统 | Record-seq[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 |
| CAMERA[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 较高 | |
| DNA typewriter[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 | |
| LINNAEUS[ | 种群 | 短 | 弱 | 否 | 否 | 一般 | 低 | |
| mSCRIBE[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| iTracer[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| DOMINO[ | 种群 | 长 | 强 | 否 | 否 | 一般 | 一般 | |
表1 基于细胞的记录装置系统技术汇总
Table 1 Summary for real-time monitoring and recording systems in cell
| 记录装置系统 | 种群分布vs单细胞记录 | 写入周期 | 记录能力 | 发生顺序 | 持续时间 | 灵敏度 | 保真度 | |
|---|---|---|---|---|---|---|---|---|
| 双稳态开关 | 种群 | 短 | 一般 | 否 | 是 | 低 | 低 | |
| DNA重组酶技术 | 种群 | 长 | 一般 | 否 | 否 | 低 | 低 | |
| ssDNA编辑技术 | HiSCRIBE[ | 种群 | 长 | 强 | 否 | 是 | 一般 | 高 |
| CRISPR系统 | Record-seq[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 |
| CAMERA[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 较高 | |
| DNA typewriter[ | 种群 | 长 | 强 | 是 | 是 | 高 | 高 | |
| LINNAEUS[ | 种群 | 短 | 弱 | 否 | 否 | 一般 | 低 | |
| mSCRIBE[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| iTracer[ | 单细胞 | 长 | 强 | 是 | 是 | 高 | 高 | |
| DOMINO[ | 种群 | 长 | 强 | 否 | 否 | 一般 | 一般 | |
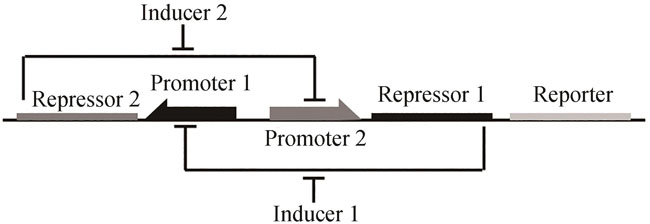
图1 双稳态开关设计[48]抑制物1抑制启动子1的转录,由诱导物1诱导;抑制物2抑制启动子2的转录,由诱导物2诱导
Fig. 1 Design for toggle switch[48]Repressors 1 and 2 inhibit transcription driven by Promoters 1 and 2, respectively, which is induced by Inducers 1 and 2 correspondingly
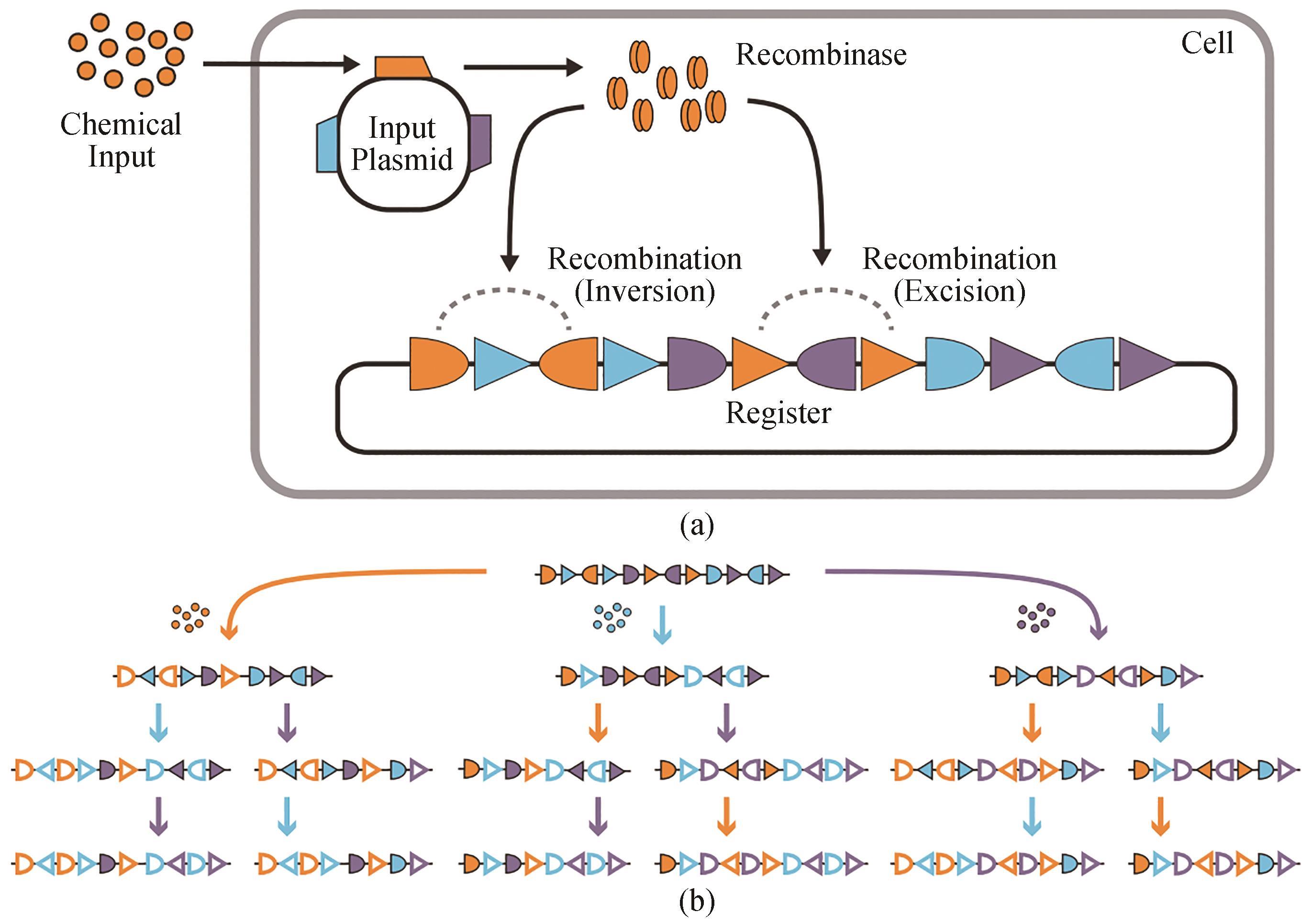
图2 三输入、16状态的RSM图示[55](a)RSM机制。化学输入诱导重组酶的表达(来自输入质粒上的基因),由重叠和正交的重组酶识别位点组成的DNA登记表。不同的重组酶可以由不同的输入来控制。这些重组靶向多个正交对的同源识别位点(显示为三角形和半椭圆形),以催化反转(当这些位点是反对齐的)或切除(当这些位点是对齐的)。(b)登记表的设计是为每种身份和输入顺序采用不同的DNA状态。三种不同的输入(橙色、蓝色和紫色)由彩色箭头表示,每个箭头表示一个不同的重组酶。未重组的识别位点有阴影;重组的识别位点有轮廓
Fig. 2 Summary for three-input and 16-state RSM[55](a) RSM mechanism. A chemical input induces the expression of a recombinase encoded by a gene on the input plasmid, which modifies a DNA register with overlapping and orthogonal recombinase recognition sites. Specific recombinases can be controlled by corresponding inputs. Each of these recombinases can target multiple orthogonal pairs of their cognate recognition sites (shown as triangles and half-ovals) to catalyze inversion (when the sites are anti-aligned) or excision (when the sites are aligned). (b) The register is designed to adopt a specific DNA state for every identity and order of inputs. Three different inputs are represented by colored arrows (orange, blue, and purple), each of which expresses a specific recombinase. Unrecombined recognition sites are shown by solid symbols, and symbols without filling highlight recombined recognition sites
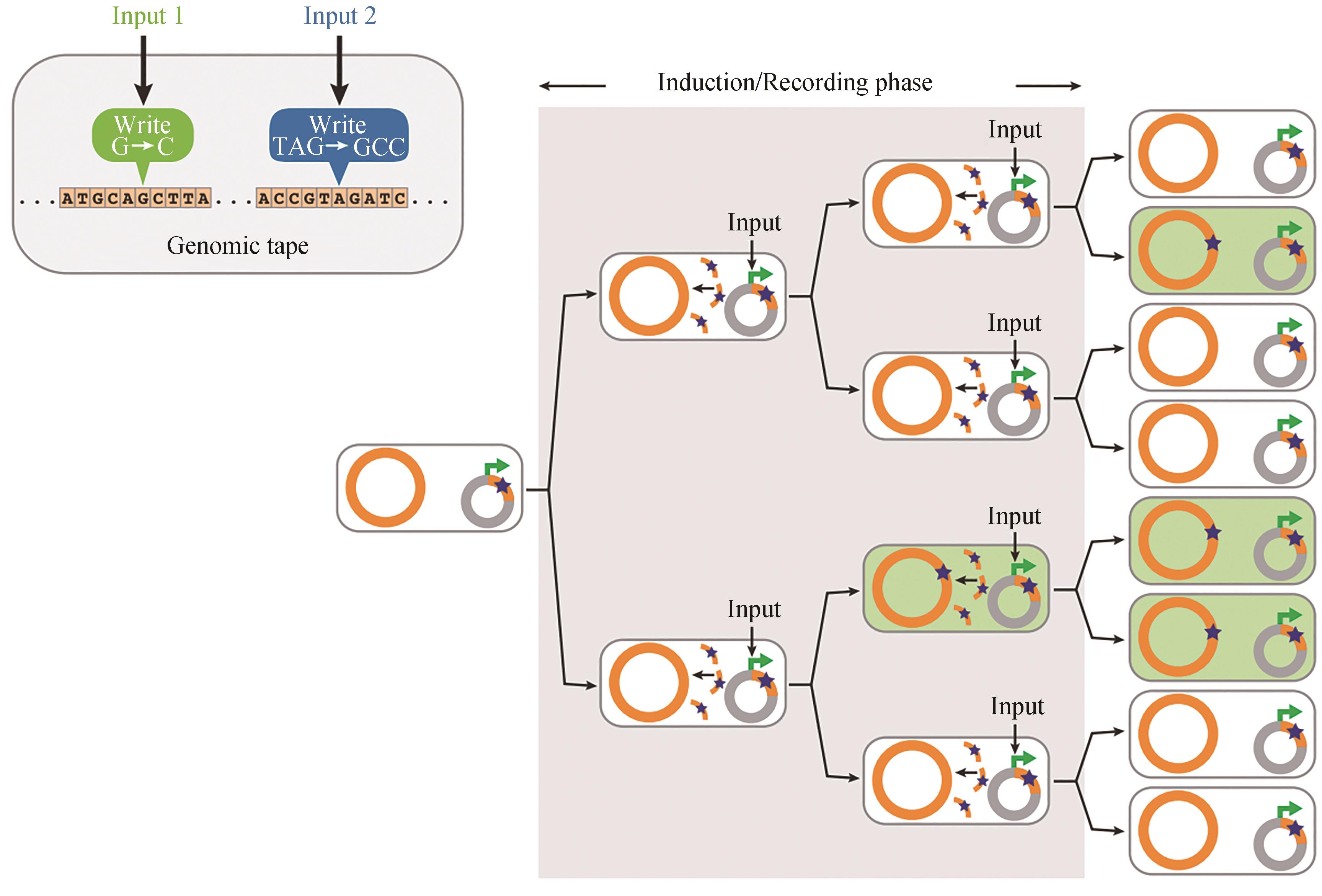
图3 SCRIBE实现了分布式基因组编码记忆[25]在输入存在的情况下,ssDNA(橙色曲线)从质粒携带的盒(灰色圆圈)中产生,并重组成基于序列同源性的特定基因组位点(橙色圆圈)。这就导致了精确突变的积累(绿色细胞中的星星),产生输入信号的大小和持续时间的相关函数
Fig. 3 SCRIBE-based distributed encoded memory at genome levels [25]In the presence of an input, ssDNAs (orange curved lines) are produced from a plasmid-borne cassette (gray circles) and recombined into specific genomic loci (orange circles) that are targeted on the basis of sequence homology. This results in the accumulation of precise mutations (stars in green cells) as a function of the magnitude and duration of exposure to the input
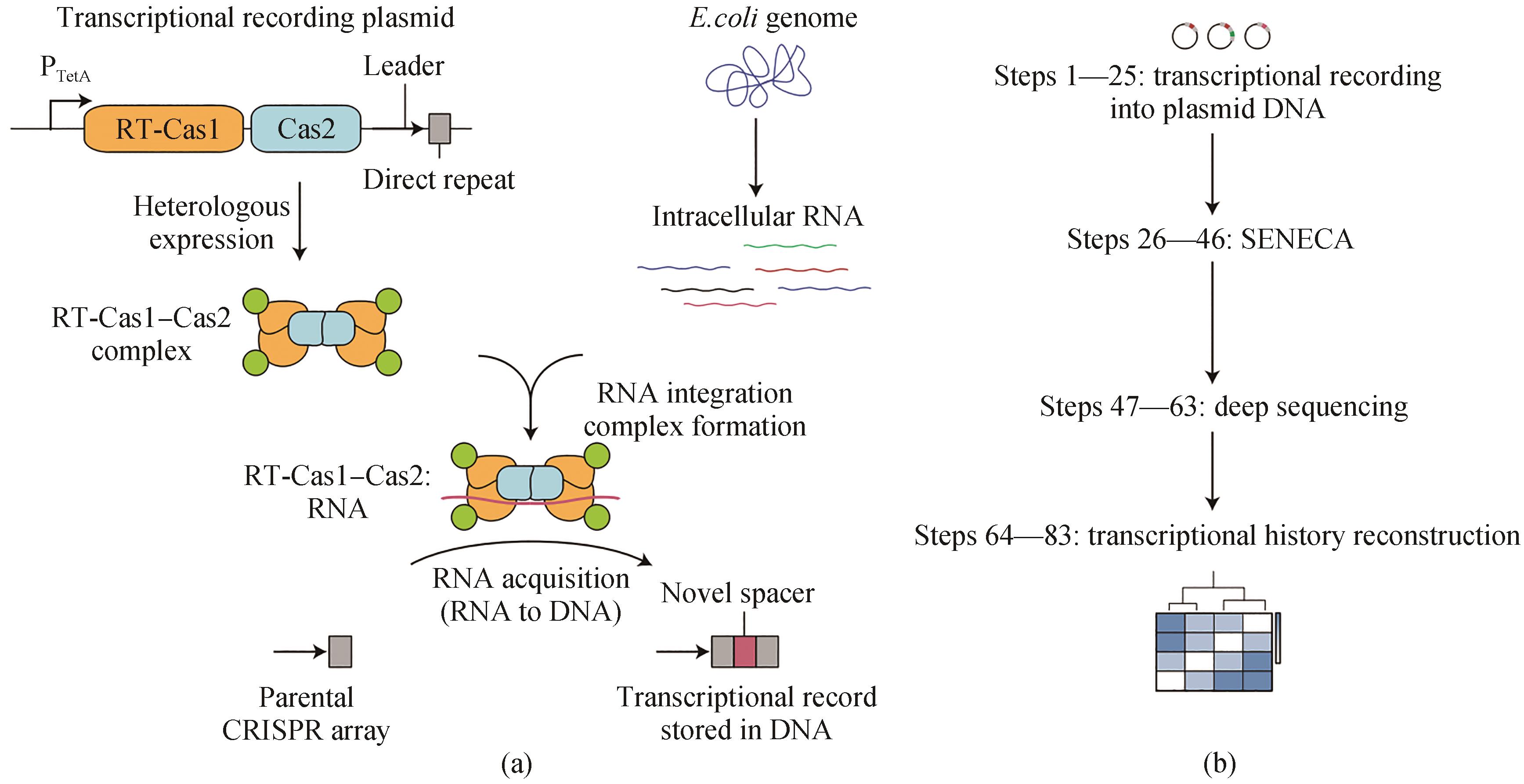
图4 从RNA中获取CRISPR间隔区的转录记录[59](a)Record-Seq使用从食糖梭菌获得RNA的RT-Cas1-Cas2复合体将转录信息编码到质粒携带的CRISPR序列中。转录记录是通过直接从细胞内RNA获得CRISPR间隔区,然后通过FsRT-Cas1-Cas2的RT结构域逆转录RNA原间隔区来产生的。(b)提取质粒DNA,然后选择性扩增扩展CRISPR阵列(SENECA)和深度测序,进行转录历史的重建
Fig. 4 Transcriptional record of RNA extracted from CRISPR spacer acquisition[59](a) Record-seq uses the RNA-acquiring RT-Cas1-Cas2 complex from Fusicatenibacter saccharivorans to encode transcriptional information into plasmid-borne CRISPR arrays. The transcriptional record is generated by CRISPR spacer acquisition directly from intracellular RNAs followed by reverse transcription of RNA protospacers through the RT domain of FsRT-Cas1-Cas2. (b) Extraction of plasmid DNA followed by the selective amplification of expanded CRISPR arrays (SENECA) and deep sequencing enable the reconstruction of transcriptional histories
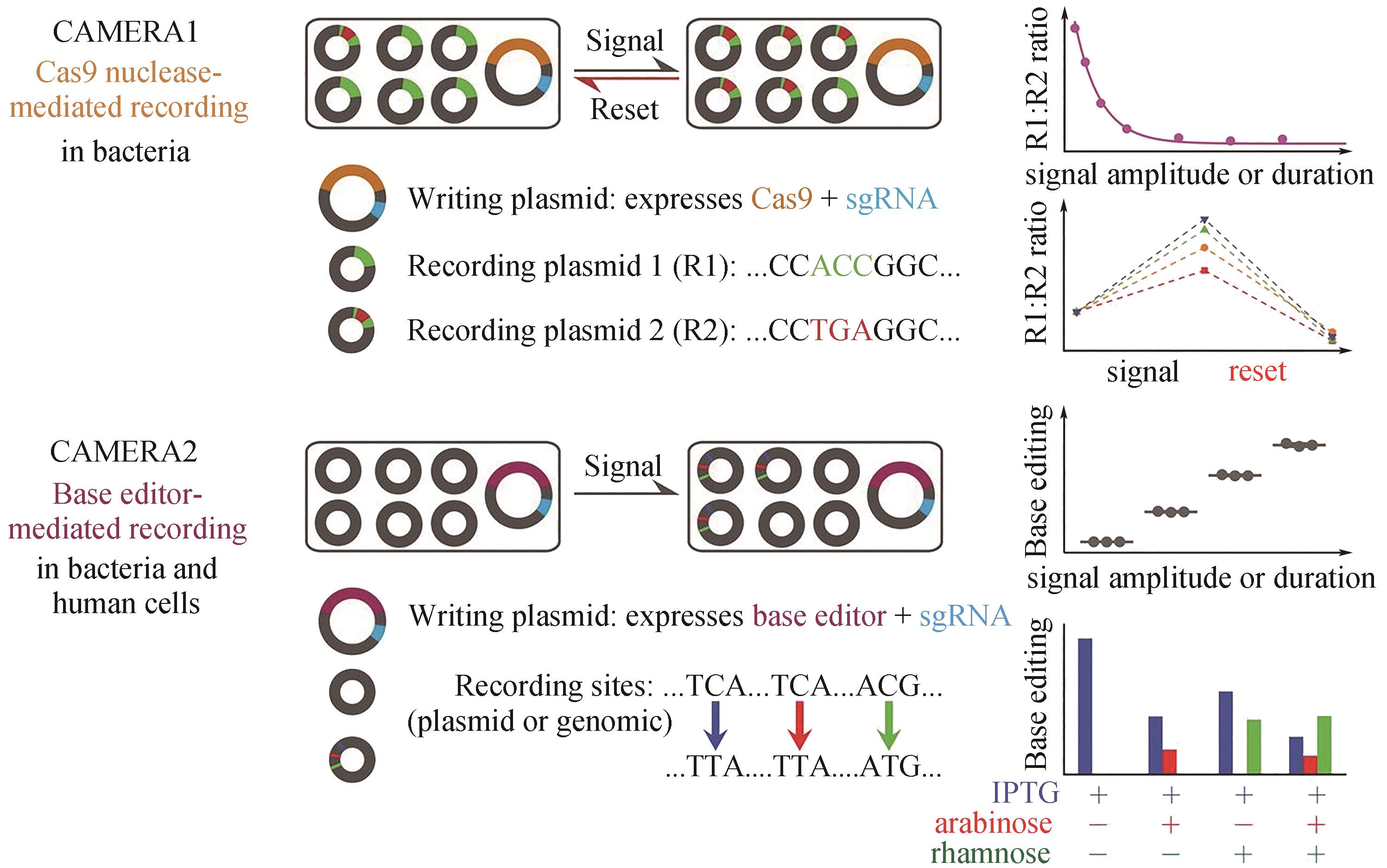
图5 CAMERA在细菌和哺乳动物细胞中的多路模拟细胞记录[39]CAMERA 1将刺激记录为相互排斥的DNA序列比例的变化。CAMERA 2使用碱基编辑器记录单核苷酸变化时信号的持续时间或振幅。这两个系统都可以复用,独立记录多个事件,包括暴露于抗生素、营养物质、病毒、光以及Wnt信号
Fig. 5 Multiple analog of cellular recording by CAMERA systems in bacteria and mammalian cells[39]CAMERA 1 records stimuli as changes in the ratio of mutually exclusive DNA sequences. CAMERA 2 uses base editors to record the duration or amplitude of signals as single-nucleotide changes. Both systems can be repeatedly used to independently record multiple events, including exposure to antibiotics, nutrients, viruses, and light, as well as Wnt signaling
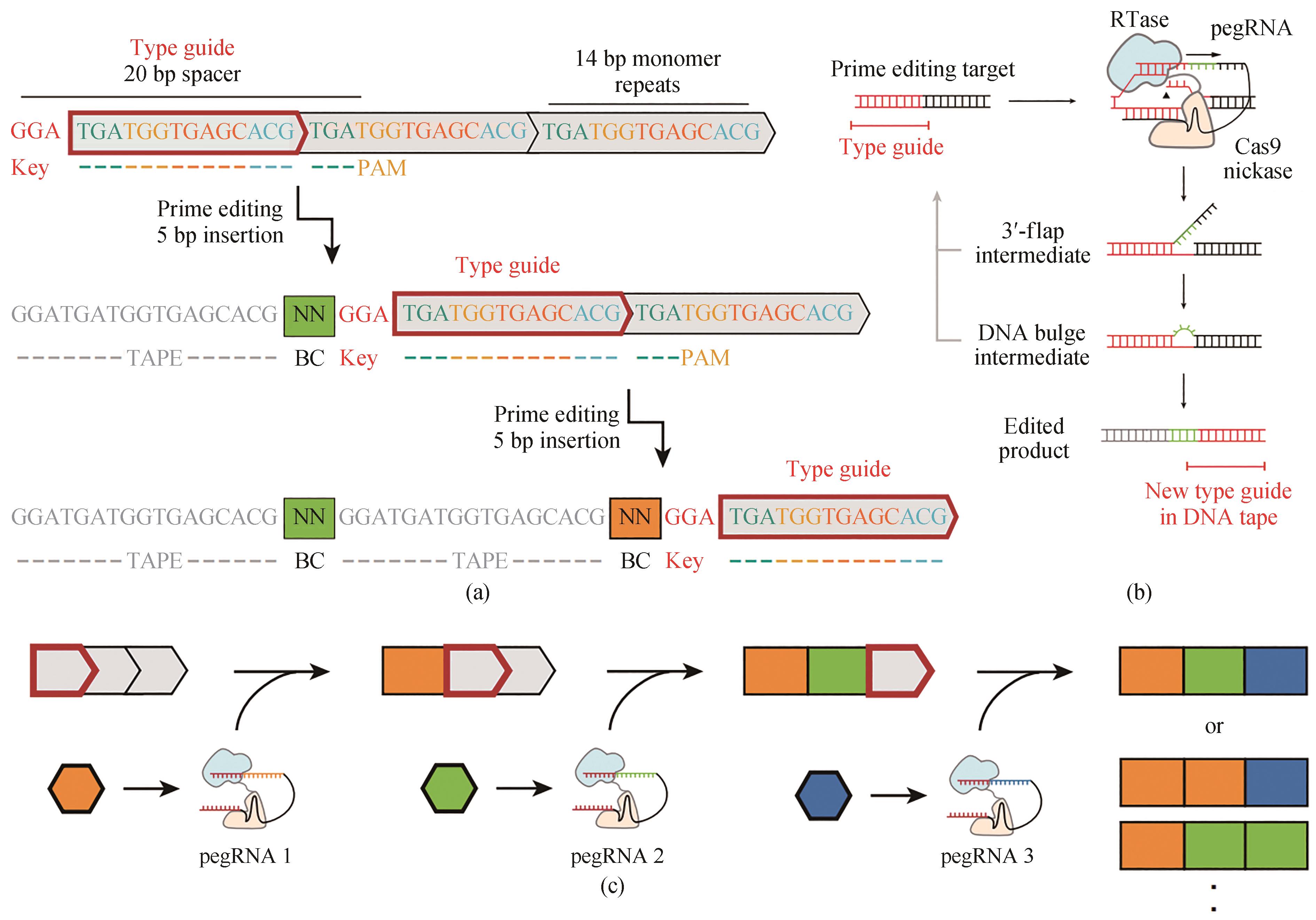
图6 用DNA typewriter进行序列基因组编辑[40](a)Type guide上连续两个编辑序列件的示意图,它随着每个编辑序列的位置移动。DNA带由CRISPR-Cas9靶点(灰色方框)的串联序列组成,除第一个外,所有的靶点在其5′端被截断,因此不起作用。5 bp的插入包括一个2 bp的pegRNA特异条形码以及一个3 bp的key来激活下一个单体。因为基因组编辑在这个系统中是按顺序的,所以记录事件的时间顺序可以简单地通过它们沿着序列的物理顺序来读出。(b)用DNA typewriter引导编辑的示意图,引导编辑识别CRISPR-Cas9目标,并使用pegRNA指定的编辑对其进行修改。对于DNA typewriter,插入编辑序列会在随后的单体上生成新的编辑目标。(c)DNA typewriter顺序记录原理图,单个pegRNA与PE2酶一起,可以是事件驱动或者结构性表达的
Fig. 6 Sequential genome editing with DNA typewriter[40](a) Schematic of two successive editing events at the type guide, which shifts in position with each editing event. The DNA tape consists of a tandem array of CRISPR–Cas9 target sites (grey boxes), all but the first of which are truncated at their 5′ ends and therefore inactive. The 5-bp insertion includes a 2-bp pegRNA-specific barcode as well as a 3-bp key that activates the next monomer. Because genome editing is sequential in this scheme, the temporal order of recorded events can simply be read out by their physical order along the array. (b) Schematic of prime editing with DNA typewriter. Prime editing recognizes a CRISPR–Cas9 target and modifies it with the edit specified by the pegRNA. With DNA typewriter, an insertional editing event generates a new prime editing target at the subsequent monomer. (c) Schematic of ordered recording via DNA typewriter. Individual pegRNAs are potentially event-driven or constitutively expressed, together with the PE2 enzyme
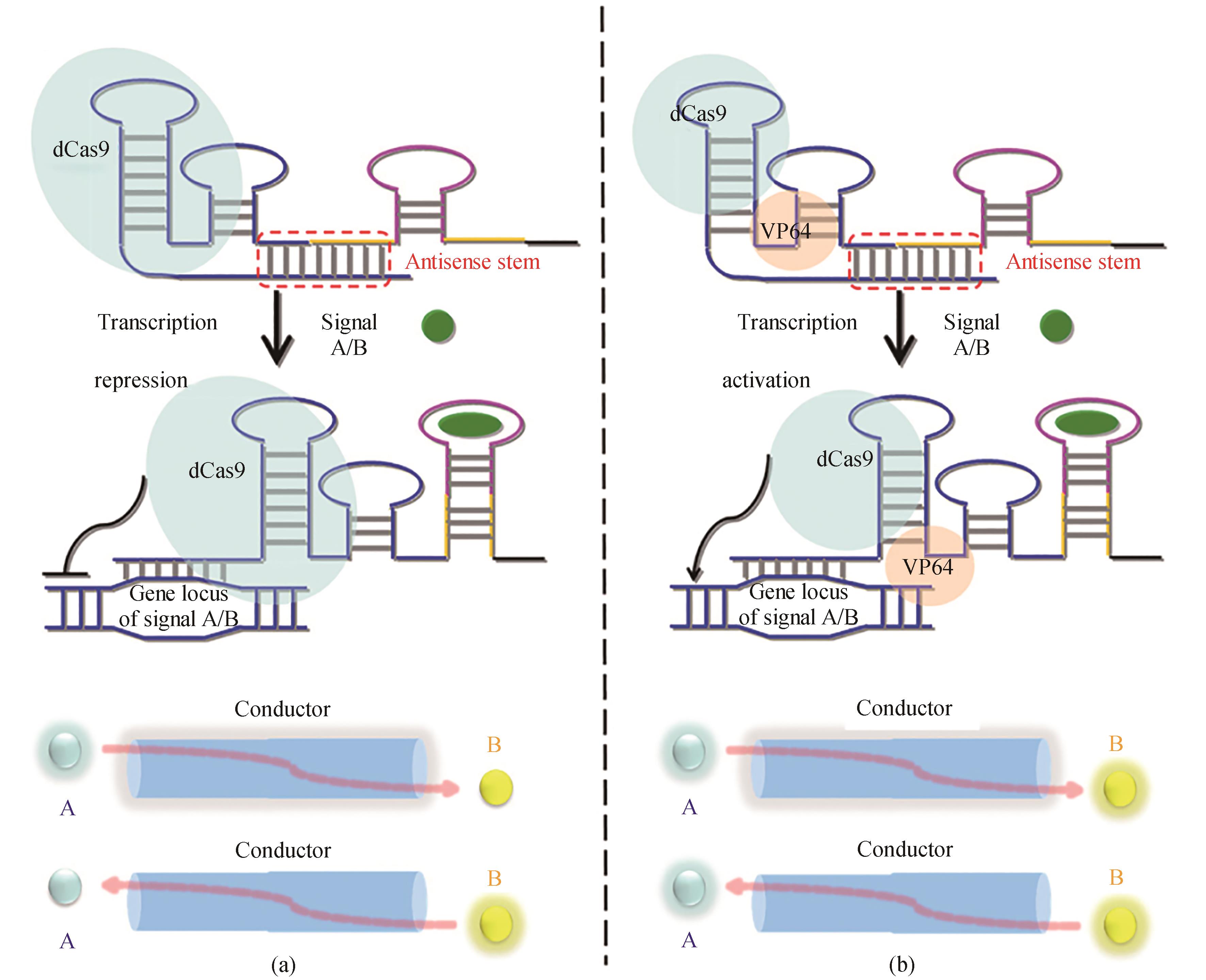
图7 基于CRISPR-Cas9的信号导体的设计[70]重新设计的sgRNAs用来灭活(a)或激活(b)基因表达的链置换机制的说明。sgRNA的反义序列显示为蓝色,适配子茎显示为黄色。在没有信号A/B的情况下,sgRNA的引导区在反义茎内配对,sgRNA处于关闭状态。在信号A/B的存在下,重新设计的sgRNA的构象被切换到“ON”状态。在这种状态下,sgRNA的导向区与其目标DNA结合,从而分别通过dCas9-VP64融合蛋白和dCas9蛋白启动和关闭信号B/A的产生
Fig. 7 Design of the CRISPR-Cas9-based signal conductor to link one signal with another[70]General illustration of the strand-displacement mechanism by which the redesigned sgRNA acts to deactivate (a) or activate (b) gene expression. The antisense sequence of the sgRNA is shown in blue, and the aptamer stem is shown in yellow. In the absence of signal A/B, the guide region of sgRNA is paired within the antisense stem and the sgRNA is in the 'off' state. In the presence of signal A/B, the conformation of the redesigned sgRNA is switched to the 'on' state. In this state, the guide region of the sgRNA binds to its target DNA, and thus turns the production of signal B/A on and off through the dCas9-VP64 fusion protein and the dCas9 protein, respectively.
| 70 | LIU Y C, ZHAN Y H, CHEN Z C, et al. Directing cellular information flow via CRISPR signal conductors[J]. Nature Methods, 2016, 13(11): 938-944. |
| 71 | SLOMOVIC S, PARDEE K, COLLINS J J. Synthetic biology devices for in vitro and in vivo diagnostics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(47): 14429-14435. |
| 72 | HAELLMAN V, FUSSENEGGER M. Synthetic biology—toward therapeutic solutions[J]. Journal of Molecular Biology, 2016, 428(5 Pt B): 945-962. |
| 73 | WALTER J G, STAHL F. Aptazymes: expanding the specificity of natural catalytic nucleic acids by application of in vitro selected oligonucleotides[J]. Advances in Biochemical Engineering/Biotechnology, 2020, 170: 107-119. |
| 74 | YOKOBAYASHI Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells[J]. Current Opinion in Chemical Biology, 2019, 52: 72-78. |
| 75 | WANG X Y, DONG K L, KONG D Q, et al. A far-red light-inducible CRISPR-Cas12a platform for remote-controlled genome editing and gene activation[J]. Science Advances, 2021, 7(50): eabh2358. |
| 76 | KAVITA K, BREAKER R R. Discovering riboswitches: the past and the future[J]. Trends in Biochemical Sciences, 2023, 48(2): 119-141. |
| 77 | LIU Y C, HAN J H, CHEN Z C, et al. Engineering cell signaling using tunable CRISPR–Cpf1-based transcription factors[J]. Nature Communications, 2017, 8: 2095. |
| 78 | KIM T, LU T K. CRISPR/Cas-based devices for mammalian synthetic biology[J]. Current Opinion in Chemical Biology, 2019, 52: 23-30. |
| 79 | WIELAND M, FUSSENEGGER M. Engineering molecular circuits using synthetic biology in mammalian cells[J]. Annual Review of Chemical and Biomolecular Engineering, 2012, 3: 209-234. |
| 80 | INNISS M C, SILVER P A. Building synthetic memory[J]. Current Biology, 2013, 23(17): R812-R816. |
| 81 | ZHAN H J, ZHOU Q, GAO Q J, et al. Multiplexed promoterless gene expression with CRISPReader[J]. Genome Biology, 2019, 20(1): 113. |
| 1 | CAMERON D E, BASHOR C J, COLLINS J J. A brief history of synthetic biology[J]. Nature Reviews Microbiology, 2014, 12(5): 381-390. |
| 2 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age[J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 3 | MOON T S, LOU C B, TAMSIR A, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491(7423): 249-253. |
| 4 | STRICKER J, COOKSON S, BENNETT M R, et al. A fast, robust and tunable synthetic gene oscillator[J]. Nature, 2008, 456(7221): 516-519. |
| 5 | FRIEDLAND A E, LU T K, WANG X, et al. Synthetic gene networks that count[J]. Science, 2009, 324(5931): 1199-1202. |
| 6 | WEI P, WONG W W, PARK J S, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells[J]. Nature, 2012, 488(7411): 384-388. |
| 7 | ROYBAL K T, RUPP L J, MORSUT L, et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits[J]. Cell, 2016, 164(4): 770-779. |
| 8 | MAYS Z J, NAIR N U. Synthetic biology in probiotic lactic acid bacteria: at the frontier of living therapeutics[J]. Current Opinion in Biotechnology, 2018, 53: 224-231. |
| 9 | SLOMOVIC S, COLLINS J J. DNA sense-and-respond protein modules for mammalian cells[J]. Nature Methods, 2015, 12(11): 1085-1090. |
| 10 | COURBET A, ENDY D, RENARD E, et al. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates[J]. Science Translational Medicine, 2015, 7(289): 289ra83. |
| 11 | FENNO L E, MATTIS J, RAMAKRISHNAN C, et al. Targeting cells with single vectors using multiple-feature Boolean logic[J]. Nature Methods, 2014, 11(7): 763-772. |
| 12 | BOGORAD I W, LIN T S, LIAO J C. Synthetic non-oxidative glycolysis enables complete carbon conservation[J]. Nature, 2013, 502(7473): 693-697. |
| 13 | BERNABÉ-ORTS J M, QUIJANO-RUBIO A, VAZQUEZ-VILAR M, et al. A memory switch for plant synthetic biology based on the phage ϕC31 integration system[J]. Nucleic Acids Research, 2020, 48(6): 3379-3394. |
| 14 | JIN S, BAE J Y, SONG Y, et al. Synthetic biology on acetogenic bacteria for highly efficient conversion of C1 gases to biochemicals[J]. International Journal of Molecular Sciences, 2020, 21(20): 7639. |
| 15 | BIRD L J, KUNDU B B, TSCHIRHART T, et al. Engineering wired life: synthetic biology for electroactive bacteria[J]. ACS Synthetic Biology, 2021, 10(11): 2808-2823. |
| 16 | CUBILLOS-RUIZ A, GUO T X, SOKOLOVSKA A, et al. Engineering living therapeutics with synthetic biology[J]. Nature Reviews Drug Discovery, 2021, 20(12): 941-960. |
| 17 | LIU Z H, WANG J Y, NIELSEN J. Yeast synthetic biology advances biofuel production[J]. Current Opinion in Microbiology, 2022, 65: 33-39. |
| 18 | SHENDURE J, JI H. Next-generation DNA sequencing[J]. Nature Biotechnology, 2008, 26(10): 1135-1145. |
| 19 | HEATHER J M, CHAIN B. The sequence of sequencers: the history of sequencing DNA[J]. Genomics, 2016, 107(1): 1-8. |
| 20 | FARZADFARD F, LU T K. Emerging applications for DNA writers and molecular recorders[J]. Science, 2018, 361(6405): 870-875. |
| 21 | SONG X, REIF J. Nucleic acid databases and molecular-scale computing[J]. ACS Nano, 2019, 13(6): 6256-6268. |
| 22 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20(8): 456-466. |
| 23 | HAM T S, LEE S K, KEASLING J D, et al. Design and construction of a double inversion recombination switch for heritable sequential genetic memory[J]. PLoS One, 2008, 3(7): e2815. |
| 24 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337(6102): 1628. |
| 82 | ANZALONE A V, KOBLAN L W, LIU D R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature Biotechnology, 2020, 38(7): 824-844. |
| 83 | LIENERT F, LOHMUELLER J J, GARG A, et al. Synthetic biology in mammalian cells: next generation research tools and therapeutics[J]. Nature Reviews Molecular Cell Biology, 2014, 15(2): 95-107. |
| 84 | LU T K, COLLINS J J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(12): 4629-4634. |
| 85 | ORTIZ M E, ENDY D. Engineered cell-cell communication via DNA messaging[J]. Journal of Biological Engineering, 2012, 6(1): 16. |
| 86 | ZHANG Y, PTACIN J L, FISCHER E C, et al. A semi-synthetic organism that stores and retrieves increased genetic information[J]. Nature, 2017, 551(7682): 644-647. |
| 87 | HO J M L, BENNETT M R. Improved memory devices for synthetic cells[J]. Science, 2018, 360(6385): 150-151. |
| 25 | FARZADFARD F, LU T K. Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations[J]. Science, 2014, 346(6211): 1256272. |
| 26 | OISHI K, KLAVINS E. Framework for engineering finite state machines in gene regulatory networks[J]. ACS Synthetic Biology, 2014, 3(9): 652-665. |
| 27 | AALIPOUR A, CHUANG H Y, MURTY S, et al. Engineered immune cells as highly sensitive cancer diagnostics[J]. Nature Biotechnology, 2019, 37(5): 531-539. |
| 28 | JIANG K Y, KOOB J, DAWN CHEN X, et al. Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR[J]. Nature Biotechnology, 2022: 1-10. |
| 29 | 杨璐, 吴楠, 白茸茸, 等. 基因回路型全细胞微生物传感器的设计、优化与应用[J]. 合成生物学, 2022, 3(6): 1061-1080. |
| YANG L, WU N, BAI R R, et al. Design, optimization and application of whole-cell microbial biosensors with engineered genetic circuits[J]. Synthetic Biology Journal, 2022, 3(6): 1061-1080. | |
| 30 | KRETZSCHMAR K, WATT F M. Lineage tracing[J]. Cell, 2012, 148(1/2): 33-45. |
| 31 | WAGNER D E, KLEIN A M. Lineage tracing meets single-cell omics: opportunities and challenges[J]. Nature Reviews Genetics, 2020, 21(7): 410-427. |
| 32 | MEACHAM C E, MORRISON S J. Tumour heterogeneity and cancer cell plasticity[J]. Nature, 2013, 501(7467): 328-337. |
| 33 | WEINREB C, RODRIGUEZ-FRATICELLI A, CAMARGO F D, et al. Lineage tracing on transcriptional landscapes links state to fate during differentiation[J]. Science, 2020, 367(6479): eaaw3381. |
| 34 | SNIPPERT H J, VAN DER FLIER L G, SATO T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells[J]. Cell, 2010, 143(1): 134-144. |
| 35 | MCKENNA A, FINDLAY G M, GAGNON J A, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing[J]. Science, 2016, 353(6298): aaf7907. |
| 36 | MO C Y, GUO J X, QIN J C, et al. Single-cell transcriptomics of LepR-positive skeletal cells reveals heterogeneous stress-dependent stem and progenitor pools[J]. The EMBO Journal, 2022, 41(4): e108415 |
| 37 | HE L J, LI Y, LI Y, et al. Enhancing the precision of genetic lineage tracing using dual recombinases[J]. Nature Medicine, 2017, 23(12): 1488-1498. |
| 38 | TANNA T, SCHMIDT F, CHEREPKOVA M Y, et al. Recording transcriptional histories using Record-seq[J]. Nature Protocols, 2020, 15(2): 513-539. |
| 39 | TANG W X, LIU D R. Rewritable multi-event analog recording in bacterial and mammalian cells[J]. Science, 2018, 360(6385): eaap8992. |
| 40 | CHOI J, CHEN W, MINKINA A, et al. A time-resolved, multi-symbol molecular recorder via sequential genome editing[J]. Nature, 2022, 608(7921): 98-107. |
| 41 | SPANJAARD B, HU B, MITIC N, et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars[J]. Nature Biotechnology, 2018, 36(5): 469-473. |
| 42 | PERLI S D, CUI C H, LU T K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells[J]. Science, 2016, 353(6304): aag0511. |
| 43 | HE Z S, MAYNARD A, JAIN A, et al. Lineage recording in human cerebral organoids[J]. Nature Methods, 2022, 19(1): 90-99. |
| 44 | FARZADFARD F, GHARAEI N, HIGASHIKUNI Y, et al. Single-nucleotide-resolution computing and memory in living cells[J]. Molecular Cell, 2019, 75(4): 769-780.e4. |
| 45 | SHETH R U, WANG H H. DNA-based memory devices for recording cellular events[J]. Nature Reviews Genetics, 2018, 19(11): 718-732. |
| 46 | AUSLÄNDER S, FUSSENEGGER M. Synthetic biology. Dynamic genome engineering in living cells[J]. Science, 2014, 346(6211): 813-814. |
| 47 | ATKINSON M R, SAVAGEAU M A, MYERS J T, et al. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli [J]. Cell, 2003, 113(5): 597-607. |
| 48 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 49 | BIANCALANI T, ASSAF M. Genetic toggle switch in the absence of cooperative binding: exact results[J]. Physical Review Letters, 2015, 115(20): 208101. |
| 50 | BHOMKAR P, MATERI W, WISHART D S. The bacterial nanorecorder: engineering E. coli to function as a chemical recording device[J]. PLoS One, 2011, 6(11): e27559. |
| 51 | KOTULA J W, KERNS S J, SHAKET L A, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(13): 4838-4843. |
| 52 | MIMEE M, TUCKER A C, VOIGT CA, et al. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota[J]. Cell Systems, 2015, 1(1): 62-71. |
| 53 | BONNET J, YIN P, ORTIZ M E, et al. Amplifying genetic logic gates[J]. Science, 2013, 340(6132): 599-603. |
| 54 | SIUTI P, YAZBEK J, LU T K. Synthetic circuits integrating logic and memory in living cells[J]. Nature Biotechnology, 2013, 31(5): 448-452. |
| 55 | ROQUET N, SOLEIMANY A P, FERRIS A C, et al. Synthetic recombinase-based state machines in living cells[J]. Science, 2016, 353(6297): aad8559. |
| 56 | LAMPSON B C, INOUYE M, INOUYE S. Retrons, msDNA, and the bacterial genome[J]. Cytogenetic and Genome Research, 2005, 110(1/2/3/4): 491-499. |
| 57 | LIM D B, MAAS W K. Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B[J]. Cell, 1989, 56(5): 891-904. |
| 58 | SCHMIDT F, CHEREPKOVA M Y, PLATT R J. Transcriptional recording by CRISPR spacer acquisition from RNA[J]. Nature, 2018, 562(7727): 380-385. |
| 59 | SCHMIDT F, ZIMMERMANN J, TANNA T, et al. Noninvasive assessment of gut function using transcriptional recording sentinel cells[J]. Science, 2022, 376(6594): eabm6038. |
| 60 | AMITAI G, SOREK R. CRISPR-Cas adaptation: insights into the mechanism of action[J]. Nature Reviews Microbiology, 2016, 14(2): 67-76. |
| 61 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 62 | DOUDNA J A, CHARPENTIER E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| 63 | MANGHWAR H, LINDSEY K, ZHANG X L, et al. CRISPR/cas system: recent advances and future prospects for genome editing[J]. Trends in Plant Science, 2019, 24(12): 1102-1125. |
| 64 | ZHAN H J, XIAO L L, LI A L, et al. Engineering cellular signal sensors based on CRISPR-sgRNA reconstruction approaches[J]. International Journal of Biological Sciences, 2020, 16(8): 1441-1449. |
| 65 | NISSIM L, PERLI S D, FRIDKIN A, et al. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells[J]. Molecular Cell, 2014, 54(4): 698-710. |
| 66 | ZHAN H J, LI A L, CAI Z M, et al. Improving transgene expression and CRISPR-Cas9 efficiency with molecular engineering-based molecules[J]. Clinical and Translational Medicine, 2020, 10(6): e194. |
| 67 | NIU Q W, LIN S S, REYES J L, et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance[J]. Nature Biotechnology, 2006, 24(11): 1420-1428. |
| 68 | HUANG X B, CHEN Z C, LIU Y C. RNAi-mediated control of CRISPR functions[J]. Theranostics, 2020, 10(15): 6661-6673. |
| 69 | LIU Y C, ZENG Y Y, LIU L, et al. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells[J]. Nature Communications, 2014, 5: 5393. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117. |
| [3] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [4] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [5] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [6] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [11] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [12] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [13] | 张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141. |
| [14] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [15] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
