合成生物学 ›› 2025, Vol. 6 ›› Issue (1): 105-117.DOI: 10.12211/2096-8280.2023-068
CRISPR-Cas系统的小型化研究进展
董颖1, 马孟丹1, 黄卫人1,2
- 1.深圳大学第一附属医院泌尿外科,国家地方联合肿瘤基因组临床应用关键技术工程实验室,广东 深圳 518036
2.中国科学院深圳先进技术研究院合成生物学研究所,广东 深圳 518000
-
收稿日期:2023-09-21修回日期:2024-03-21出版日期:2025-02-28发布日期:2025-03-12 -
通讯作者:黄卫人 -
作者简介:董颖 (1998—),女,硕士研究生。研究方向为医学合成生物学。E-mail:dongyingdcc@163.com马孟丹 (1997—),女,博士研究生。研究方向为医学合成生物学。E-mail:mamengdan7@163.com黄卫人 (1980—),男,研究员,博士生导师。研究方向为肿瘤合成生物学,肿瘤类器官培养及精准医学。E-mail:pony8980@163.com
Progress in the miniaturization of CRISPR-Cas systems
DONG Ying1, MA Mengdan1, HUANG Weiren1,2
- 1.National Engineering Laboratory of Key Technologies for Clinical Application of Local Joint Tumor Genome,Department of Urology,The First Affiliated Hospital of Shenzhen University,Shenzhen 518036,Guangdong,China
2.Institute of Synthetic Biology,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518000,Guangdong,China
-
Received:2023-09-21Revised:2024-03-21Online:2025-02-28Published:2025-03-12 -
Contact:HUANG Weiren
摘要:
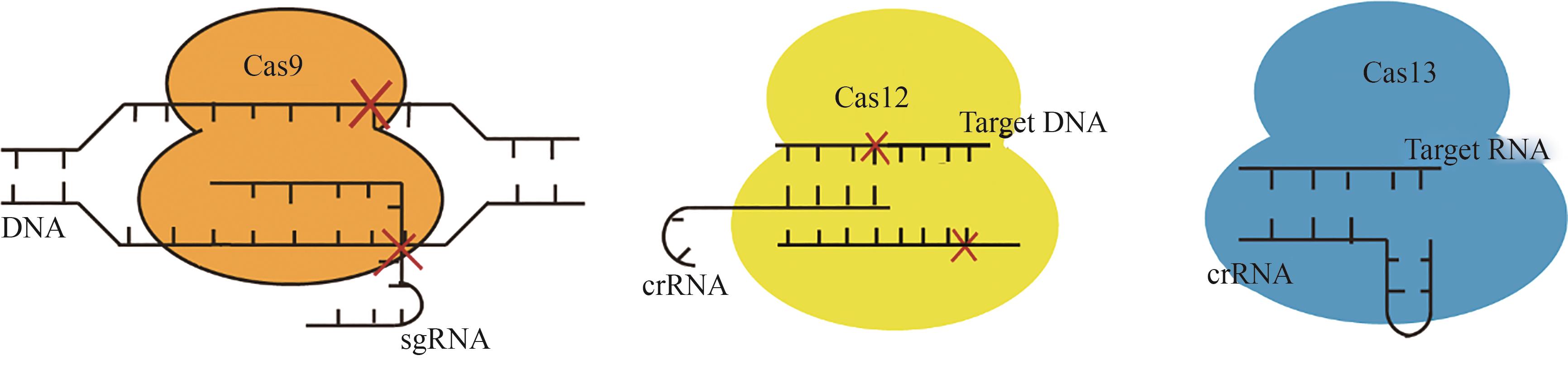
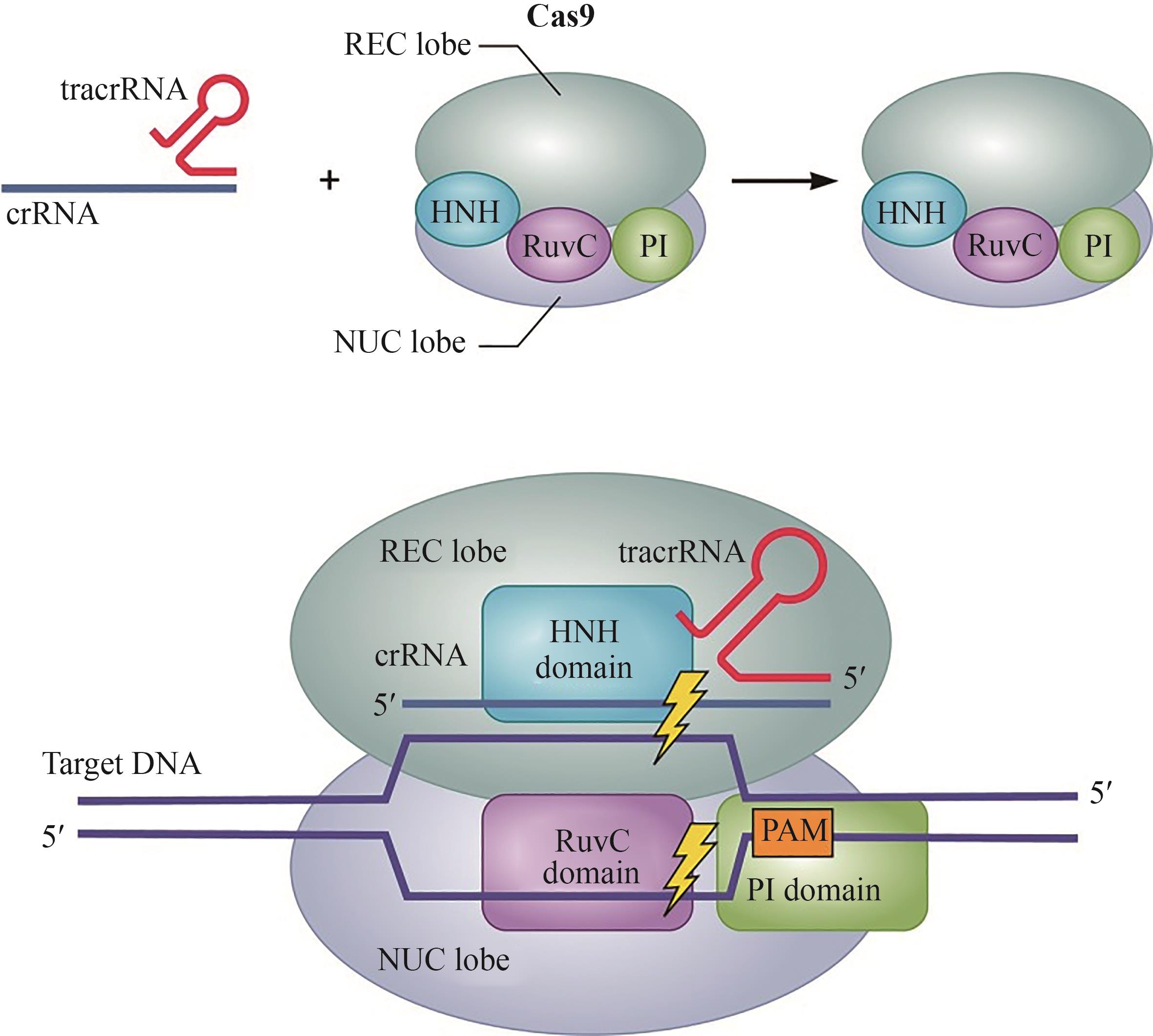
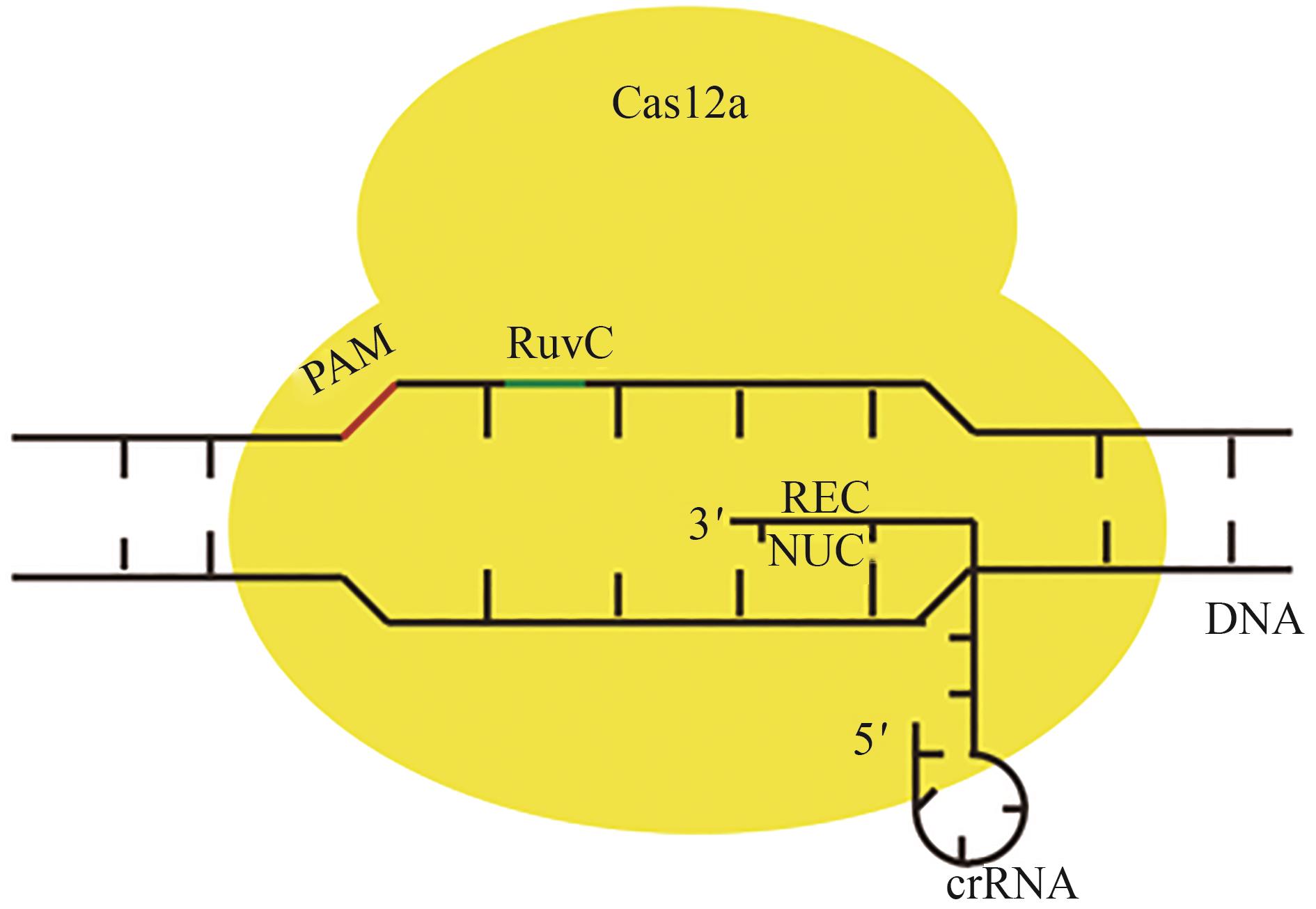
CRISPR-Cas基因编辑技术由于其简便性和高效性,已被广泛应用于生物学、医学、农学等领域的基础与应用研究。目前广泛使用的Cas核酸酶均具有较大的分子量(通常大于1000个氨基酸),而广泛应用于基因治疗中的腺相关病毒(AAV)载体的承载容量却十分有限,在容纳CRISPR核酸酶与gRNA的编码序列之余往往难以承载更多其他功能元件,如碱基编辑、转录调控、多基因编辑等相应元件,这严重限制了其在基因治疗等领域的应用。使用紧凑型Cas蛋白变体的CRISPR-Cas系统可能有助于用AAV产生和传递基因组编辑和调节工具到人类细胞。因此,小型化的CRISPR-Cas系统开发是解决这一技术难题的重要途径,本文主要概括了基于Cas9、Cas12和Cas13蛋白系统在小型化方面的研究进展,包括筛选新型Cas蛋白、缩减蛋白结构域以及引导RNA的改造等,旨在为开发微型精准基因编辑和调控工具提供新思路。目前小型化的CRISPR-Cas系统的局限性主要体现在蛋白分子量的大小和基因编辑的效率、特异性不可兼得上,在未来的研究中若能解决这一问题,获得更小型化的结构域,相信不仅能够优化该系统在体内的传递,更有望为临床带来高效率且低损害的治疗方法。
中图分类号:
引用本文
董颖, 马孟丹, 黄卫人. CRISPR-Cas系统的小型化研究进展[J]. 合成生物学, 2025, 6(1): 105-117.
DONG Ying, MA Mengdan, HUANG Weiren. Progress in the miniaturization of CRISPR-Cas systems[J]. Synthetic Biology Journal, 2025, 6(1): 105-117.
| 特点 | Cas9 | Cas12 | Cas13 |
|---|---|---|---|
| 靶标类型 | DNA | DNA | RNA |
| 切割模式 | 产生平末端的DNA双链断裂 | 产生黏性末端的DNA双链断裂(部分产生单链DNA切割断裂) | 产生单链RNA断裂 |
| PAM序列(用于识别并结合目标) | 通常需要一个特定的PAM序列 | 需要PAM序列(不同于Cas9) | 不需要PAM序列,直接靶向于RNA |
| 优势 | 广泛应用,工具资源丰富,操作简单 | 因其需要更小的PAM及其黏性末端而更容易进行编辑 | 针对RNA的编辑能力更强 |
| 适用场景 | 基因编辑及调控、染色质成像;DNA剪切、修复和替换;转录调控[ | 基因编辑及调控、染色质成像;特异性即时检测[ | RNA编辑、调控翻译水平、RNA修饰;基因沉默;抑制病毒RNA复制[ |
| 实际应用案例 | 输血依赖性β地中海贫血(TDT)和镰状细胞病(SCD)[ | 检测COVID-19[ | 开发SHERLOCK技术,检测寨卡和登革热病毒的特定菌株[ |
表1 Cas9、Cas12、Cas13的特点对比
Table 1 Comparison of the characteristics of Cas9, Cas12 and Cas13
| 特点 | Cas9 | Cas12 | Cas13 |
|---|---|---|---|
| 靶标类型 | DNA | DNA | RNA |
| 切割模式 | 产生平末端的DNA双链断裂 | 产生黏性末端的DNA双链断裂(部分产生单链DNA切割断裂) | 产生单链RNA断裂 |
| PAM序列(用于识别并结合目标) | 通常需要一个特定的PAM序列 | 需要PAM序列(不同于Cas9) | 不需要PAM序列,直接靶向于RNA |
| 优势 | 广泛应用,工具资源丰富,操作简单 | 因其需要更小的PAM及其黏性末端而更容易进行编辑 | 针对RNA的编辑能力更强 |
| 适用场景 | 基因编辑及调控、染色质成像;DNA剪切、修复和替换;转录调控[ | 基因编辑及调控、染色质成像;特异性即时检测[ | RNA编辑、调控翻译水平、RNA修饰;基因沉默;抑制病毒RNA复制[ |
| 实际应用案例 | 输血依赖性β地中海贫血(TDT)和镰状细胞病(SCD)[ | 检测COVID-19[ | 开发SHERLOCK技术,检测寨卡和登革热病毒的特定菌株[ |
| 蛋白 | 分子量/kDa | PAM | spacer/nt | tracrRNA | 直接重复 序列(DR) | 切割dsDNA | 切割ssDNA | 实验细胞/细菌 |
|---|---|---|---|---|---|---|---|---|
| SpCas9[ | 160 | 5′-NGG-3′ | 42 | Y | 36 | Y | N | HEK293T |
| Nme1Cas9[ | 162.4 | 5'-N4GAYW/N4GYTT-3'或5'-N4GTCT-3' | 24 | Y | 24 | Y | N | Neisseria meningitidis |
| SaCas9[ | 126 | 5′-NNGRRT-3′ | 21~23 | Y | 19-24 | Y | N | Staphylococcus aureus |
| CjCas9[ | 108 | 5′-N4RYAC-3′ | 22 | Y | 35 | Y | N | Campylobacter jejuni |
| mini-SaCas9[ | 100 | 5′-NNGRRT-3′ | 20 | N | 24 | N | N | Staphylococcus aureus |
| Nme2Cas9[ | 160 | 5′-N4CC-3′ | 22~24 | Y | 24 | Y | N | Neisseria meningitidis |
| SauriCas9[ | 118 | 5′-NNGG-3′ | 20 | Y | 36 | Y | N | Staphylococcus auricularis |
| BlatCas9[ | 120 | 5′-N4CNAA-3′ | 17~24 | Y | 24 | Y | N | HEK9T |
| MISER Cas9[ | 100 | 5′-NNRGAA-3′ | 42 | Y | 36 | N | N | Streptococcus pyogenes |
| SchCas9[ | 115 | 5′-NNGR-3′ | 21 | Y | 32 | Y | N | — |
| Nsp2Cas9[ | 117 | 5′-N4C-3′ | 22~26 | N | 23 | Y | N | — |
| IscB[ | 54 | 5′-NWRRNA-3′ | 20 | Y | 206 | Y | N | — |
表2 Cas9的小型化研究
Table 2 Miniaturization studies of Cas9
| 蛋白 | 分子量/kDa | PAM | spacer/nt | tracrRNA | 直接重复 序列(DR) | 切割dsDNA | 切割ssDNA | 实验细胞/细菌 |
|---|---|---|---|---|---|---|---|---|
| SpCas9[ | 160 | 5′-NGG-3′ | 42 | Y | 36 | Y | N | HEK293T |
| Nme1Cas9[ | 162.4 | 5'-N4GAYW/N4GYTT-3'或5'-N4GTCT-3' | 24 | Y | 24 | Y | N | Neisseria meningitidis |
| SaCas9[ | 126 | 5′-NNGRRT-3′ | 21~23 | Y | 19-24 | Y | N | Staphylococcus aureus |
| CjCas9[ | 108 | 5′-N4RYAC-3′ | 22 | Y | 35 | Y | N | Campylobacter jejuni |
| mini-SaCas9[ | 100 | 5′-NNGRRT-3′ | 20 | N | 24 | N | N | Staphylococcus aureus |
| Nme2Cas9[ | 160 | 5′-N4CC-3′ | 22~24 | Y | 24 | Y | N | Neisseria meningitidis |
| SauriCas9[ | 118 | 5′-NNGG-3′ | 20 | Y | 36 | Y | N | Staphylococcus auricularis |
| BlatCas9[ | 120 | 5′-N4CNAA-3′ | 17~24 | Y | 24 | Y | N | HEK9T |
| MISER Cas9[ | 100 | 5′-NNRGAA-3′ | 42 | Y | 36 | N | N | Streptococcus pyogenes |
| SchCas9[ | 115 | 5′-NNGR-3′ | 21 | Y | 32 | Y | N | — |
| Nsp2Cas9[ | 117 | 5′-N4C-3′ | 22~26 | N | 23 | Y | N | — |
| IscB[ | 54 | 5′-NWRRNA-3′ | 20 | Y | 206 | Y | N | — |
| 蛋白 | 分子量/kDa | PAM | spacer/nt | tracrRNA | 直接重复 序列(DR) | 切割dsDNA | 切割ssDNA | 实验细胞/细菌 |
|---|---|---|---|---|---|---|---|---|
| Cas12d(CasY)[ | 132 | 5′-TA-3′ | 17 | Y | 26 | Y | — | uncultivated microbes |
| Cas12e5(CasX)[ | 108 | 5′-TTCN-3′ | 20 | Y | 23 | Y | — | uncultivated microbes |
| FnCpf1(Cas12a)[ | 147 | 5′-KYTV-3′ | 20 | N | 14 | Y | Y | — |
| AaCas12b(C2c1)[ | 125 | 5′-TTN-3′ | 20 | Y | 37 | Y | Y | Alicyclobacillus acidiphilus |
| Cas12c(C2c3)[ | 133.09~146.3 | 5′-TG-3′或5′- TN-3′ | 17-18 | Y | 17 | Y | Y | HEK293T |
| Cas12g[ | 79.2~91.3 | 5′-NA-3′ | 30 | Y | 36 | N | Y | HEK293T |
| Cas12h[ | 95.7~101.64 | 5′- RTR-3′ | — | N | — | Y | Y | HEK293T |
| Cas12i[ | 113~120 | 5′ TTN-3′ | 28 | N | 24 | Y | Y | HEK293T |
| Cas12j[ | 70~80 | 5′-TBN-3′或5′-TTN-3′ | 14-20 | N | 26 | Y | Y | HEK293T |
| CasMINI[ | 58 | 5′-TTTR-3′ | 23 | Y | 26 | Y | Y | TRE3G-GFP HEK293T |
| UnCas12f[ | 44~77 | 5′-TTTA-3′ | 34-39 | Y | 37 | Y | Y | — |
表3 Cas12的小型化研究
Table 3 Miniaturization studies of Cas12
| 蛋白 | 分子量/kDa | PAM | spacer/nt | tracrRNA | 直接重复 序列(DR) | 切割dsDNA | 切割ssDNA | 实验细胞/细菌 |
|---|---|---|---|---|---|---|---|---|
| Cas12d(CasY)[ | 132 | 5′-TA-3′ | 17 | Y | 26 | Y | — | uncultivated microbes |
| Cas12e5(CasX)[ | 108 | 5′-TTCN-3′ | 20 | Y | 23 | Y | — | uncultivated microbes |
| FnCpf1(Cas12a)[ | 147 | 5′-KYTV-3′ | 20 | N | 14 | Y | Y | — |
| AaCas12b(C2c1)[ | 125 | 5′-TTN-3′ | 20 | Y | 37 | Y | Y | Alicyclobacillus acidiphilus |
| Cas12c(C2c3)[ | 133.09~146.3 | 5′-TG-3′或5′- TN-3′ | 17-18 | Y | 17 | Y | Y | HEK293T |
| Cas12g[ | 79.2~91.3 | 5′-NA-3′ | 30 | Y | 36 | N | Y | HEK293T |
| Cas12h[ | 95.7~101.64 | 5′- RTR-3′ | — | N | — | Y | Y | HEK293T |
| Cas12i[ | 113~120 | 5′ TTN-3′ | 28 | N | 24 | Y | Y | HEK293T |
| Cas12j[ | 70~80 | 5′-TBN-3′或5′-TTN-3′ | 14-20 | N | 26 | Y | Y | HEK293T |
| CasMINI[ | 58 | 5′-TTTR-3′ | 23 | Y | 26 | Y | Y | TRE3G-GFP HEK293T |
| UnCas12f[ | 44~77 | 5′-TTTA-3′ | 34-39 | Y | 37 | Y | Y | — |
| 蛋白 | 分子量 /kDa | PFS | spacer /nt | tracrRNA | 直接重复 序列(DR) | ssRNA | 实验细胞/细菌 |
|---|---|---|---|---|---|---|---|
| Cas13a(C2c2)[ | 138 | 3′-AT /C | 14~18 | N | 28 | Y | — |
| Cas13b(C2c6)[ | 128 | 5′-D和3′-UAA | 30 | Y | 36 | Y | — |
| Cas13c(C2c7)[ | 123 | 3′-U | 30 | N | 36 | Y | — |
| Cas13d[ | 102 | N | 30 | N | 36 | Y | Ruminococcus flavefaciens FD-1 |
| Cas13Rx(RfxCas13d)[ | 106 | 3′-A | 30 | N | 22 | Y | Ruminococcus flavefaciens FD-1 |
| Cas13X.1[ | 85 | 5′-A/G/C-3′ | 30 | N | 36 | Y | HEK293T |
| Cas13Y.1[ | 85 | N | 30 | N | 36 | Y | HEK293T |
| Cas13bt[ | 85~88 | 5ʹ-D | 30 | N | 30 | Y | HEK293T |
| Cas13e3[ | 84 | N | 27 | N | 30 | Y | — |
表4 Cas13的小型化研究
Table 4 Miniaturization studies of Cas13
| 蛋白 | 分子量 /kDa | PFS | spacer /nt | tracrRNA | 直接重复 序列(DR) | ssRNA | 实验细胞/细菌 |
|---|---|---|---|---|---|---|---|
| Cas13a(C2c2)[ | 138 | 3′-AT /C | 14~18 | N | 28 | Y | — |
| Cas13b(C2c6)[ | 128 | 5′-D和3′-UAA | 30 | Y | 36 | Y | — |
| Cas13c(C2c7)[ | 123 | 3′-U | 30 | N | 36 | Y | — |
| Cas13d[ | 102 | N | 30 | N | 36 | Y | Ruminococcus flavefaciens FD-1 |
| Cas13Rx(RfxCas13d)[ | 106 | 3′-A | 30 | N | 22 | Y | Ruminococcus flavefaciens FD-1 |
| Cas13X.1[ | 85 | 5′-A/G/C-3′ | 30 | N | 36 | Y | HEK293T |
| Cas13Y.1[ | 85 | N | 30 | N | 36 | Y | HEK293T |
| Cas13bt[ | 85~88 | 5ʹ-D | 30 | N | 30 | Y | HEK293T |
| Cas13e3[ | 84 | N | 27 | N | 30 | Y | — |
| 25 | HOU Z G, ZHANG Y, PROPSON N E, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(39): 15644-15649. |
| 26 | RAN F A, CONG L, YAN W X, et al. In vivo genome editing using Staphylococcus aureus Cas9[J]. Nature, 2015, 520(7546): 186-191. |
| 27 | ZHANG S Q, ZHANG Q, HOU X M, et al. Dynamics of Staphylococcus aureus Cas9 in DNA target association and dissociation[J]. EMBO Reports, 2020, 21(10): e50184. |
| 28 | KIM E J, KOO T Y, PARK S W, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni [J]. Nature Communications, 2017, 8: 14500. |
| 29 | MA D C, PENG S G, HUANG W R, et al. Rational design of mini-Cas9 for transcriptional activation[J]. ACS Synthetic Biology, 2018, 7(4): 978-985. |
| 30 | EDRAKI A, MIR A, IBRAHEIM R, et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing[J]. Molecular Cell, 2019, 73(4): 714-726.e4. |
| 31 | HU Z Y, WANG S, ZHANG C D, et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope[J]. PLoS Biology, 2020, 18(3): e3000686. |
| 32 | GAO N, ZHANG C D, HU Z Y, et al. Characterization of Brevibacillus laterosporus Cas9 (BlatCas9) for mammalian genome editing[J]. Frontiers in Cell and Developmental Biology, 2020, 8: 583164. |
| 33 | SHAMS A, HIGGINS S A, FELLMANN C, et al. Comprehensive deletion landscape of CRISPR-Cas9 identifies minimal RNA-guided DNA-binding modules[J]. Nature Communications, 2021, 12(1): 5664. |
| 34 | WANG S, MAO H L, HOU L H, et al. Compact SchCas9 recognizes the simple NNGR PAM[J]. Advanced Science, 2022, 9(4): e2104789. |
| 35 | WEI J J, HOU L H, LIU J T, et al. Closely related type Ⅱ-C Cas9 orthologs recognize diverse PAMs[J]. eLife, 2022, 11: e77825. |
| 36 | SCHULER G, HU C Y, KE A L. Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9[J]. Science, 2022, 376(6600): 1476-1481. |
| 37 | KATO K, OKAZAKI S, KANNAN S, et al. Structure of the IscB-ωRNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9[J]. Nature Communications, 2022, 13(1): 6719. |
| 38 | ALTAE-TRAN H, KANNAN S, DEMIRCIOGLU F E, et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases[J]. Science, 2021, 374(6563): 57-65. |
| 39 | DELTCHEVA E, CHYLINSKI K, SHARMA C M, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase Ⅲ[J]. Nature, 2011, 471(7340): 602-607. |
| 40 | MALI P, YANG L H, ESVELT K M, et al. RNA-guided human genome engineering via Cas9[J]. Science, 2013, 339(6121): 823-826. |
| 41 | ZHANG Y P, WANG J, WANG Z B, et al. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae [J]. Nature Communications, 2019, 10(1): 1053. |
| 42 | MEFFERD A L, KORNEPATI A V R, BOGERD H P, et al. Expression of CRISPR/Cas single guide RNAs using small tRNA promoters[J]. RNA, 2015, 21(9): 1683-1689. |
| 43 | FONFARA I, RICHTER H, BRATOVIČ M, et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA[J]. Nature, 2016, 532(7600): 517-521. |
| 44 | ZETSCHE B, GOOTENBERG J S, ABUDAYYEH O O, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771. |
| 45 | DONG D, REN K, QIU X L, et al. The crystal structure of Cpf1 in complex with CRISPR RNA[J]. Nature, 2016, 532(7600): 522-526. |
| 46 | GAO P, YANG H, RAJASHANKAR K R, et al. Type Ⅴ CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition[J]. Cell Research, 2016, 26(8): 901-913. |
| 47 | BURSTEIN D, HARRINGTON L B, STRUTT S C, et al. New CRISPR–Cas systems from uncultivated microbes[J]. Nature, 2017, 542(7640): 237-241. |
| 48 | LIU J J, ORLOVA N, OAKES B L, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors[J]. Nature, 2019, 566(7743): 218-223. |
| 1 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 2 | MAKAROVA K S, KOONIN E V. Annotation and classification of CRISPR-Cas systems[J]. Methods in Molecular Biology, 2015, 1311: 47-75. |
| 3 | LI J H, TANG L N, LI T X, et al. Tandem Cas13a/crRNA-mediated CRISPR-FET biosensor: a one-for-all check station for virus without amplification[J]. ACS Sensors, 2022, 7(9): 2680-2690. |
| 4 | DOETSCHMAN T, GEORGIEVA T. Gene editing with CRISPR/Cas9 RNA-directed nuclease[J]. Circulation Research, 2017, 120(5): 876-894. |
| 5 | SAFARI F, AFARID M, RASTEGARI B, et al. CRISPR systems: novel approaches for detection and combating COVID-19[J]. Virus Research, 2021, 294: 198282. |
| 6 | RAMACHANDRAN A, SANTIAGO J G. CRISPR enzyme kinetics for molecular diagnostics[J]. Analytical Chemistry, 2021, 93(20): 7456-7464. |
| 7 | FRANGOUL H, ALTSHULER D, CAPPELLINI M D, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia[J]. New England Journal of Medicine, 2021, 384(3): 252-260. |
| 8 | FASCHING C L, SERVELLITA V, MCKAY B, et al. COVID-19 variant detection with a high-fidelity CRISPR-Cas12 enzyme[J]. Journal of Clinical Microbiology, 2022, 60(7): e00261-22. |
| 9 | GOOTENBERG J S, ABUDAYYEH O O, LEE J W, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2[J]. Science, 2017, 356(6336): 438-442. |
| 10 | GUPTA D, BHATTACHARJEE O, MANDAL D, et al. CRISPR-Cas9 system: a new-fangled dawn in gene editing[J]. Life Sciences, 2019, 232: 116636. |
| 11 | CHEW W L, TABEBORDBAR M, CHENG J K W, et al. A multifunctional AAV-CRISPR-Cas9 and its host response[J]. Nature Methods, 2016, 13(10): 868-874. |
| 12 | MINGOZZI F, HIGH K A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges[J]. Nature Reviews Genetics, 2011, 12(5): 341-355. |
| 49 | TU M J, LIN L, CHENG Y L, et al. A ‘new lease of life’: FnCpf1 possesses DNA cleavage activity for genome editing in human cells[J]. Nucleic Acids Research, 2017, 45(19): 11295-11304. |
| 50 | TENG F, CUI T T, FENG G H, et al. Repurposing CRISPR-Cas12b for mammalian genome engineering[J]. Cell Discovery, 2018, 4: 63. |
| 51 | YAN W X, HUNNEWELL P, ALFONSE L E, et al. Functionally diverse type Ⅴ CRISPR-Cas systems[J]. Science, 2019, 363(6422): 88-91. |
| 52 | HUANG C J, ADLER B A, DOUDNA J A. A naturally DNase-free CRISPR-Cas12c enzyme silences gene expression[J]. Molecular Cell, 2022, 82(11): 2148-2160.e4. |
| 53 | WANG Y, QI T, LIU J T, et al. A highly specific CRISPR-Cas12j nuclease enables allele-specific genome editing[J]. Science Advances, 2023, 9(6): eabo6405. |
| 54 | PAUSCH P, AL-SHAYEB B, BISOM-RAPP E, et al. CRISPR-Casφ from huge phages is a hypercompact genome editor[J]. Science, 2020, 369(6501): 333-337. |
| 55 | XU X S, CHEMPARATHY A, ZENG L P, et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing[J]. Molecular Cell, 2021, 81(20): 4333-4345.e4. |
| 56 | HARRINGTON L B, BURSTEIN D, CHEN J S, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes[J]. Science, 2018, 362(6416): 839-842. |
| 57 | KARVELIS T, BIGELYTE G, YOUNG J K, et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage[J]. Nucleic Acids Research, 2020, 48(9): 5016-5023. |
| 58 | TAKEDA S N, NAKAGAWA R, OKAZAKI S, et al. Structure of the miniature type Ⅴ-F CRISPR-Cas effector enzyme[J]. Molecular Cell, 2021, 81(3): 558-570.e3. |
| 59 | ANZALONE A V, KOBLAN L W, LIU D R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nature Biotechnology, 2020, 38(7): 824-844. |
| 60 | KIM D Y, LEE J M, MOON S B, et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus[J]. Nature Biotechnology, 2022, 40(1): 94-102. |
| 13 | ZINCARELLI C, SOLTYS S, RENGO G, et al. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection[J]. Molecular Therapy, 2008, 16(6): 1073-1080. |
| 14 | KOCH L. CRISPR systems go mini[J]. Nature Reviews Genetics, 2021, 22(11): 690. |
| 15 | SHEN B, ZHANG W S, ZHANG J, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects[J]. Nature Methods, 2014, 11(4): 399-402. |
| 16 | CHEN S A, LEE B, LEE A Y F, et al. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes[J]. Journal of Biological Chemistry, 2016, 291(28): 14457-14467. |
| 17 | NIOLA F, DAGNÆS-HANSEN F, FRÖDIN M. In vivo editing of the adult mouse liver using CRISPR/Cas9 and hydrodynamic tail vein injection[J]. Methods in Molecular Biology, 2019, 1961: 329-341. |
| 18 | MURUGAN K, BABU K, SUNDARESAN R, et al. The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit[J]. Molecular Cell, 2017, 68(1): 15-25. |
| 19 | TSUI T K M, LI H. Structure principles of CRISPR-Cas surveillance and effector complexes[J]. Annual Review of Biophysics, 2015, 44: 229-255. |
| 20 | ANDERS C, NIEWOEHNER O, DUERST A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease[J]. Nature, 2014, 513: 569-573. |
| 21 | STERNBERG S H, LAFRANCE B, KAPLAN M, et al. Conformational control of DNA target cleavage by CRISPR-Cas9[J]. Nature, 2015, 527: 110-113. |
| 22 | NISHIMASU H, RAN F A, HSU P D, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA[J]. Cell, 2014, 156(5): 935-949. |
| 23 | ISHINO Y, KRUPOVIC M, FORTERRE P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology[J]. Journal of Bacteriology, 2018, 200(7): e00580-17. |
| 24 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 61 | WU Z W, ZHANG Y F, YU H P, et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease[J]. Nature Chemical Biology, 2021, 17(11): 1132-1138. |
| 62 | HUANG H X, LV W Q, LI J H, et al. Comparison of DNA targeting CRISPR editors in human cells[J]. Cell & Bioscience, 2023, 13(1): 11. |
| 63 | KARVELIS T, DRUTEIKA G, BIGELYTE G, et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease[J]. Nature, 2021, 599(7886): 692-696. |
| 64 | XIANG G H, LI Y Q, SUN J, et al. Evolutionary mining and functional characterization of TnpB nucleases identify efficient miniature genome editors[J]. Nature Biotechnology, 2024, 42:745-757. |
| 65 | CHEN W Z, MA J C, WU Z W, et al. Cas12n nucleases, early evolutionary intermediates of type Ⅴ CRISPR, comprise a distinct family of miniature genome editors[J]. Molecular Cell, 2023, 83(15): 2768-2780.e6. |
| 66 | SHMAKOV S, SMARGON A, SCOTT D, et al. Diversity and evolution of class 2 CRISPR-Cas systems[J]. Nature Reviews Microbiology, 2017, 15(3): 169-182. |
| 67 | SASNAUSKAS G, TAMULAITIENE G, DRUTEIKA G, et al. TnpB structure reveals minimal functional core of Cas12 nuclease family[J]. Nature, 2023, 616(7956): 384-389. |
| 68 | SENTHILNATHAN R, ILANGOVAN I, KUNALE M, et al. An update on CRISPR-Cas12 as a versatile tool in genome editing[J]. Molecular Biology Reports, 2023, 50(3): 2865-2881. |
| 69 | ABUDAYYEH O O, GOOTENBERG J S, ESSLETZBICHLER P, et al. RNA targeting with CRISPR-Cas13[J]. Nature, 2017, 550(7675): 280-284. |
| 70 | KONERMANN S, LOTFY P, BRIDEAU N J, et al. Transcriptome engineering with RNA-targeting type Ⅵ-D CRISPR effectors[J]. Cell, 2018, 173(3): 665-676.e14. |
| 71 | HU Y P, CHEN Y C, XU J, et al. Metagenomic discovery of novel CRISPR-Cas13 systems[J]. Cell Discovery, 2022, 8(1): 107. |
| 72 | LIU L, LI X Y, WANG J Y, et al. Two distant catalytic sites are responsible for C2c2 RNase activities[J]. Cell, 2017, 168(1-2): 121-134.e12. |
| 73 | WU S M, TIAN P F, TAN T W. CRISPR-Cas13 technology portfolio and alliance with other genetic tools[J]. Biotechnology Advances, 2022, 61: 108047. |
| 74 | SHMAKOV S, ABUDAYYEH O O, MAKAROVA K S, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems[J]. Molecular Cell, 2015, 60(3): 385-397. |
| 75 | SMARGON A A, COX D B T, PYZOCHA N K, et al. Cas13b is a type Ⅵ-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28[J]. Molecular Cell, 2017, 65(4): 618-630.e7. |
| 76 | YAN W X, CHONG S R, ZHANG H B, et al. Cas13d is a compact RNA-targeting type Ⅵ CRISPR effector positively modulated by a WYL-domain-containing accessory protein[J]. Molecular Cell, 2018, 70(2): 327-339.e5. |
| 77 | XU C L, ZHOU Y S, XIAO Q Q, et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes[J]. Nature Methods, 2021, 18(5): 499-506. |
| 78 | KANNAN S, ALTAE-TRAN H, JIN X, et al. Compact RNA editors with small Cas13 proteins[J]. Nature Biotechnology, 2022, 40(2): 194-197. |
| 79 | TANG T, HAN Y L, WANG Y R, et al. Programmable system of Cas13-mediated RNA modification and its biological and biomedical applications[J]. Frontiers in Cell and Developmental Biology, 2021, 9: 677587. |
| 80 | WANG F, WANG L R, ZOU X, et al. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing[J]. Biotechnology Advances, 2019, 37(5): 708-729. |
| 81 | ZHAO F Y, ZHANG T, SUN X D, et al. A strategy for Cas13 miniaturization based on the structure and AlphaFold[J]. Nature Communications, 2023, 14(1): 5545. |
| 82 | BANDARU S, TSUJI M H, SHIMIZU Y, et al. Structure-based design of gRNA for Cas13[J]. Scientific Reports, 2020, 10(1): 11610. |
| 83 | XIAO Q Q, XU Z J, XUE Y Y, et al. Rescue of autosomal dominant hearing loss by in vivo delivery of mini dCas13X-derived RNA base editor[J]. Science Translational Medicine, 2022, 14(654): eabn0449. |
| 84 | GAO S Q, WANG Y, QI T, et al. Genome editing with natural and engineered CjCas9 orthologs[J]. Molecular Therapy, 2023, 31(4): 1177-1187. |
| 85 | ZHOU F X, YU X R, GAN R, et al. CRISPRimmunity: an interactive web server for CRISPR-associated Important Molecular events and Modulators Used in geNome edIting Tool identifYing[J]. Nucleic Acids Research, 2023, 51(W1): W93-W107. |
| 86 | AWAN M J A, AMIN I, MANSOOR S. Mini CRISPR-Cas12f1: a new genome editing tool[J]. Trends in Plant Science, 2022, 27(2): 110-112. |
| 87 | CHO E Y, RYU J Y, LEE H A R, et al. Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes[J]. Journal of Nanobiotechnology, 2019, 17(1): 19. |
| [1] | 杜瑶, 高宏丹, 刘家坤, 刘孝荣, 邢志浩, 张涛, 马东礼. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216. |
| [2] | 马孟丹, 刘宇辰. 合成生物学在疾病信息记录与实时监测中的应用潜力[J]. 合成生物学, 2023, 4(2): 301-317. |
| [3] | 刘倩, 李金根, 张晨阳, 李芳雅, 田朝光. 工业丝状真菌基因组编辑技术研究进展[J]. 合成生物学, 2021, 2(2): 256-273. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||