合成生物学 ›› 2023, Vol. 4 ›› Issue (1): 141-164.DOI: 10.12211/2096-8280.2022-050
RNA转录后代谢时空精密控制技术
刘韧玫1,2,3, 李乐诗1,2, 杨小燕1,2, 陈显军1,2, 杨弋1,2
- 1.华东理工大学光遗传学与合成生物学交叉学科研究中心,生物反应器工程国家重点实验室,上海 200237
2.华东理工大学药学院,上海市细胞代谢光遗传学技术前沿科学研究基地,上海 200237
3.华东理工大学生物工程学院,上海 200237
-
收稿日期:2022-09-13修回日期:2022-11-18出版日期:2023-02-28发布日期:2023-03-07 -
通讯作者:杨弋 -
作者简介:刘韧玫 (1989—),女,博士,讲师。研究方向为光遗传学与合成生物学(细胞代谢监测与控制技术)。E-mail:renmei2018@ecust.edu.cn杨弋 (1973—),男,教授,博士生导师。研究方向为控制与监测细胞内分子过程的合成生物技术与光遗传学前沿技术、癌症及代谢类疾病药理及药物筛选技术、蛋白质特异性标记与翻译后修饰的鉴定、细胞内原位成像、蛋白质药物生产技术等。E-mail:yiyang@ecust.edu.cn
第一联系人:刘韧玫(1989—),女,博士,讲师。研究方向为光遗传学与合成生物学(细胞代谢监测与控制技术)。 -
基金资助:国家重点研发计划(2022YFC3400100);国家自然科学基金(32121005)
Technologies for precise spatiotemporal control of post-transcriptional RNA metabolism
LIU Renmei1,2,3, LI Leshi1,2, YANG Xiaoyan1,2, CHEN Xianjun1,2, YANG Yi1,2
- 1.Optogenetics & Synthetic Biology Interdisciplinary Research Center,State Key Laboratory of Bioreactor Engineering,East China University of Science and Technology,Shanghai 200237,China
2.Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism,School of Pharmacy,East China University of Science and Technology,Shanghai 200237,China
3.School of Bioengineering,East China University of Science and Technology,Shanghai 200237,China
-
Received:2022-09-13Revised:2022-11-18Online:2023-02-28Published:2023-03-07 -
Contact:YANG Yi
摘要:
RNA种类繁多且功能多样,是细胞活动的核心分子之一。RNA代谢调控对于基因和RNA功能研究、细胞生命活动解析以及疾病治疗手段的开发都是至关重要的。为了深入研究RNA时间、空间分布以及功能机制,科学家们一直在追求可以在活细胞内对RNA分子活动进行精密控制的技术,这也是近些年生命科学领域的研究热点之一。目前基于基因编辑、转录调控等可以控制RNA转录生成的技术已较为成熟,但对于RNA转录后代谢的控制技术尚在发展与突破阶段。此前,RNA转录后代谢调控工具是通过调节RNA或基于RNA结合蛋白的RNA效应因子来实现的,但它们的时空分辨率较低,很难对RNA转录后代谢进行定时、定量和定位精密调控。光遗传学凭借其独特的高时空分辨率、非侵入性等优势已经被逐步用于发展活细胞RNA代谢时空精确控制技术。目前,基于核苷酸光化学修饰、遗传编码光响应因子的光遗传学工具已经可实现在转录或转录后水平对RNA多种代谢活动的时空精密控制,包括生成、运输、翻译、降解等。本文将介绍RNA代谢调控系统的研究进展,并聚焦于RNA转录后代谢的光遗传学调控技术,同时对其未来发展前景进行了展望。
中图分类号:
引用本文
刘韧玫, 李乐诗, 杨小燕, 陈显军, 杨弋. RNA转录后代谢时空精密控制技术[J]. 合成生物学, 2023, 4(1): 141-164.
LIU Renmei, LI Leshi, YANG Xiaoyan, CHEN Xianjun, YANG Yi. Technologies for precise spatiotemporal control of post-transcriptional RNA metabolism[J]. Synthetic Biology Journal, 2023, 4(1): 141-164.
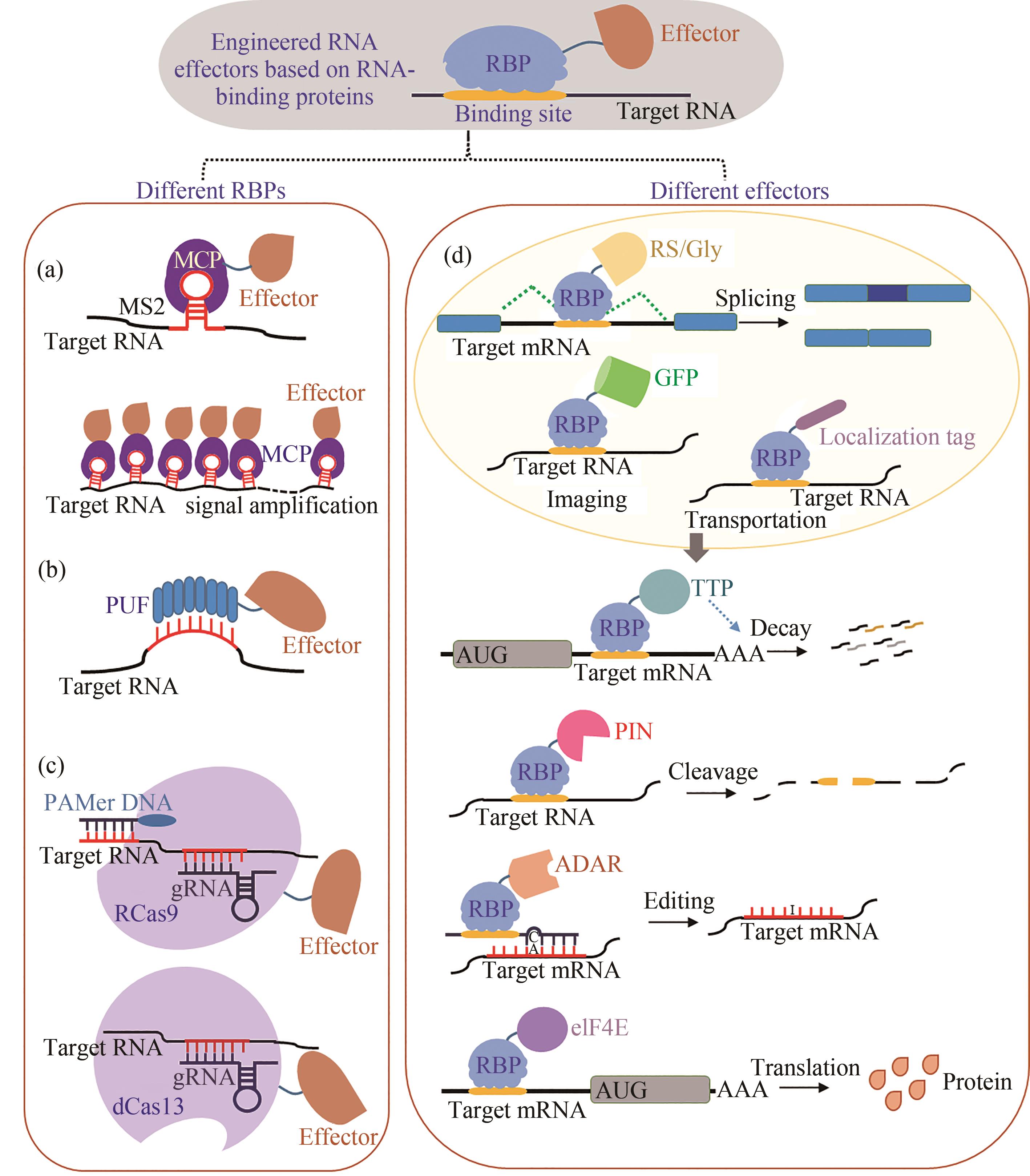
| 项目 | 基于调节RNA的RNA效应因子 | 基于RNA结合蛋白的RNA效应因子 | 基于光化学修饰的核苷酸 | 基于光笼蛋白质或其配体 | 基于光诱导蛋白异二聚化和RNA结合蛋白 | 基于光控RNA结合蛋白PAL的RNA效应因子 | 基于光控RNA结合蛋白LicV的RNA效应因子 |
|---|---|---|---|---|---|---|---|
| 工作原理 | 自带或招募内源功能蛋白或空间位阻效应 | RNA结合蛋白与不同功能结构域融合获得系列人工合成RNA效应因子 | DNA/RNA中引入光化学修饰的寡核苷酸,光照调控DNA/RNA活性 | 利用光可移除的囚笼基团对RNA结合蛋白或其配体活性进行调控 | 光诱导蛋白异二聚化来调控RNA结合蛋白的活性 | 光诱导的PAL蛋白与RNA适配体的结合 | 光诱导的LicV与RAT RNA的结合 |
| 诱导条件 | 无 | 无 | 多数为UV光,少量为可见光 | UV光 | 蓝光 | 蓝光 | 蓝光 |
| 细胞毒性 | 低 | 低 | 高 | 高 | 低 | 低 | 低 |
| 制备方法 | 遗传编码 | 遗传编码 | 体外合成 | 体外合成 | 遗传编码 | 遗传编码 | 遗传编码 |
| 或体外合成 | |||||||
| 可调性 | 难 | 难 | 适中 | 适中 | 容易 | 容易 | 容易 |
| 时间分辨率 | 低 | 低 | 适中 | 适中 | 高 | 高 | 高 |
| 空间分辨率 | 低 | 低 | 高 | 高 | 高 | 高 | 高 |
| 普适性 | 高 | 高 | 低 | 低 | 未知 | 未知 | 高 |
表1 RNA转录后代谢调控系统
| 项目 | 基于调节RNA的RNA效应因子 | 基于RNA结合蛋白的RNA效应因子 | 基于光化学修饰的核苷酸 | 基于光笼蛋白质或其配体 | 基于光诱导蛋白异二聚化和RNA结合蛋白 | 基于光控RNA结合蛋白PAL的RNA效应因子 | 基于光控RNA结合蛋白LicV的RNA效应因子 |
|---|---|---|---|---|---|---|---|
| 工作原理 | 自带或招募内源功能蛋白或空间位阻效应 | RNA结合蛋白与不同功能结构域融合获得系列人工合成RNA效应因子 | DNA/RNA中引入光化学修饰的寡核苷酸,光照调控DNA/RNA活性 | 利用光可移除的囚笼基团对RNA结合蛋白或其配体活性进行调控 | 光诱导蛋白异二聚化来调控RNA结合蛋白的活性 | 光诱导的PAL蛋白与RNA适配体的结合 | 光诱导的LicV与RAT RNA的结合 |
| 诱导条件 | 无 | 无 | 多数为UV光,少量为可见光 | UV光 | 蓝光 | 蓝光 | 蓝光 |
| 细胞毒性 | 低 | 低 | 高 | 高 | 低 | 低 | 低 |
| 制备方法 | 遗传编码 | 遗传编码 | 体外合成 | 体外合成 | 遗传编码 | 遗传编码 | 遗传编码 |
| 或体外合成 | |||||||
| 可调性 | 难 | 难 | 适中 | 适中 | 容易 | 容易 | 容易 |
| 时间分辨率 | 低 | 低 | 适中 | 适中 | 高 | 高 | 高 |
| 空间分辨率 | 低 | 低 | 高 | 高 | 高 | 高 | 高 |
| 普适性 | 高 | 高 | 低 | 低 | 未知 | 未知 | 高 |
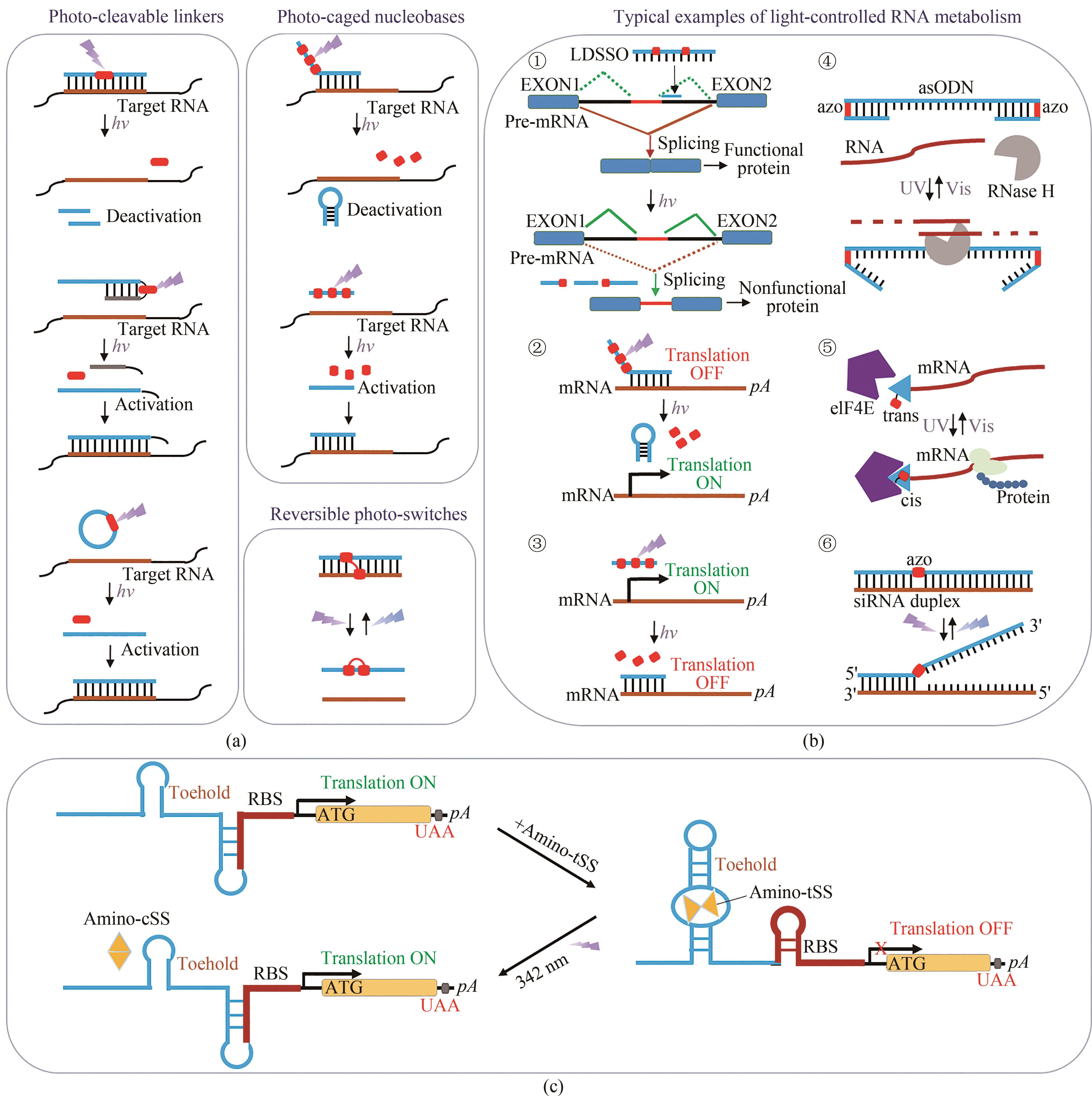
图3 基于核苷酸光化学修饰的调控技术(a)几种不同的寡核苷酸功能光调节方法。(b)利用光可转换基团邻硝基苄基(oNB)或偶氮苯(AzoB)对RNA代谢的调控:①oNB修饰的LDSSO对RNA剪接的调控;②通过核碱基笼状反义剂对基因沉默的光激活;③通过光诱导形成发夹,从而对反义剂进行光化学失活;④使用偶氮苯修饰的哑铃反义寡脱氧核苷酸进行RNA降解的光响应调节;⑤PC-cap对翻译的可逆光调节;⑥siRNAzos的光诱导失活和再激活。(c)Were-1核糖开关的翻译调节。在没有配体的情况下,核糖体结合位点(RBS)暴露并翻译荧光素酶,而在存在amino-tSS的情况下,RBS被隔离,从而Fluc不表达。342 nm光照射下,Amino-tSS切换为Amino-cSS构象,使得Were-1构象发生变化,进而重新激活RBS活性
Fig. 3 Regulation technology based on photochemical modifications of nucleotides(a) Different approaches for regulating oligonucleotide functions by light. (b) Regulation of RNA metabolism by photoswitchable o-nitrobenzyl (oNB) or azobenzene (AzoB). ① Regulation of RNA splicing by oNB-modified LDSSOs; ② Light-activation of gene silencing through nucleobase-caged antisense agents; ③ Optochemical deactivation of an antisense agent through light-induced hairpin formation; ④ Photoregulating RNA digestion using azobenzene linked dumbbell antisense oligodeoxynucleotides; ⑤ Light-mediated reversible activation of translation via PC-cap; ⑥ Photoinduced inactivation and reactivation of siRNAzos. (c) Translation regulation by the Were-1 riboswitch. In absence of the ligand, the ribosomal binding site (RBS) is exposed, and luciferase is translated, whereas in presence of amino-tSS, the RBS is sequestered, repressing the expression of Fluc. With light (342 nm) irradiation, amino-tSS is switched to amino-cSS conformation, causing theconformation change of Were-1 to activate RBS activities.
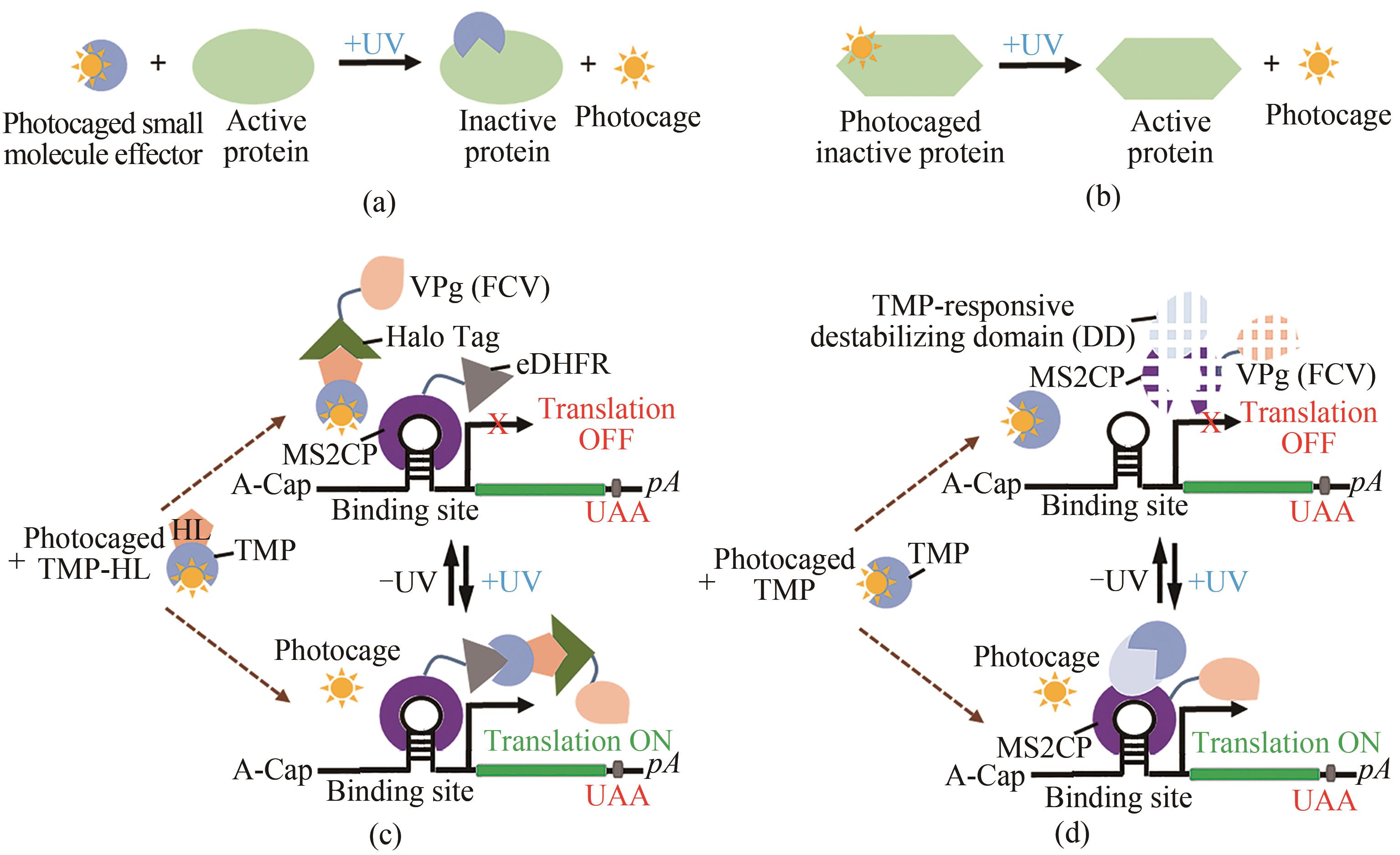
图4 基于光笼蛋白质配体的调控技术(a)“光笼”效应小分子。(b)“光笼”氨基酸。(c)基于分裂CaVT系统和光笼甲氧苄啶-HaloTag配体的光诱导翻译激活。光介导TMP-HL去除光笼,使得TMP-HL介导MS2CP-eDHFR和HaloTag-VPg(FCV)发生相互作用。由此产生的MS2CP-eDHFR-HaloTag-VPg(FCV)复合物可激活含有1xMS2(U)site1-hmAG1基序的靶modRNA的翻译。(d)基于光笼TMP与融合有响应TMP的去稳定域的CaVT(DD-CaVT)的光诱导翻译激活。光介导TMP去除光笼,使得TMP与DD-CaVT发生相互作用,从而防止DD-CaVT的快速降解。稳定的DD-CaVT与含有弱结合基序1xMS2(U) site2-hmAG1的靶modRNA结合并激活其翻译
Fig. 4 Regulatory technology based on photocage protein ligands(a) Photocaged small effector molecules. (b) Photocaged amino acids. (c) Light-inducible translational activation by split CaVT systems and photocaged trimethoprim-HaloTag ligand through light-mediated removal of the photocage from TMP-HL to enable the TMP-HL-mediated interactions between MS2CP-eDHFR and HaloTag-VPg(FCV), resulting MS2CP-eDHFR-HaloTag-VPg (FCV) complex to activate the translation of target modRNA containing a motif (1xMS2(U)site1-hmAG1). (d) Light-inducible translational activation by photocaged TMP and TMP-responsive destabilizing domain-fused CaVT (DD-CaVT) through light-mediated removal of the photocage from TMP to enable the interaction between TMP and DD-CaVT, preventing the rapid degradation of DD-CaVT for binding to a target modRNA with a weak binding motif (1xMS2(U) site2-hmAG1) and activate its translation.

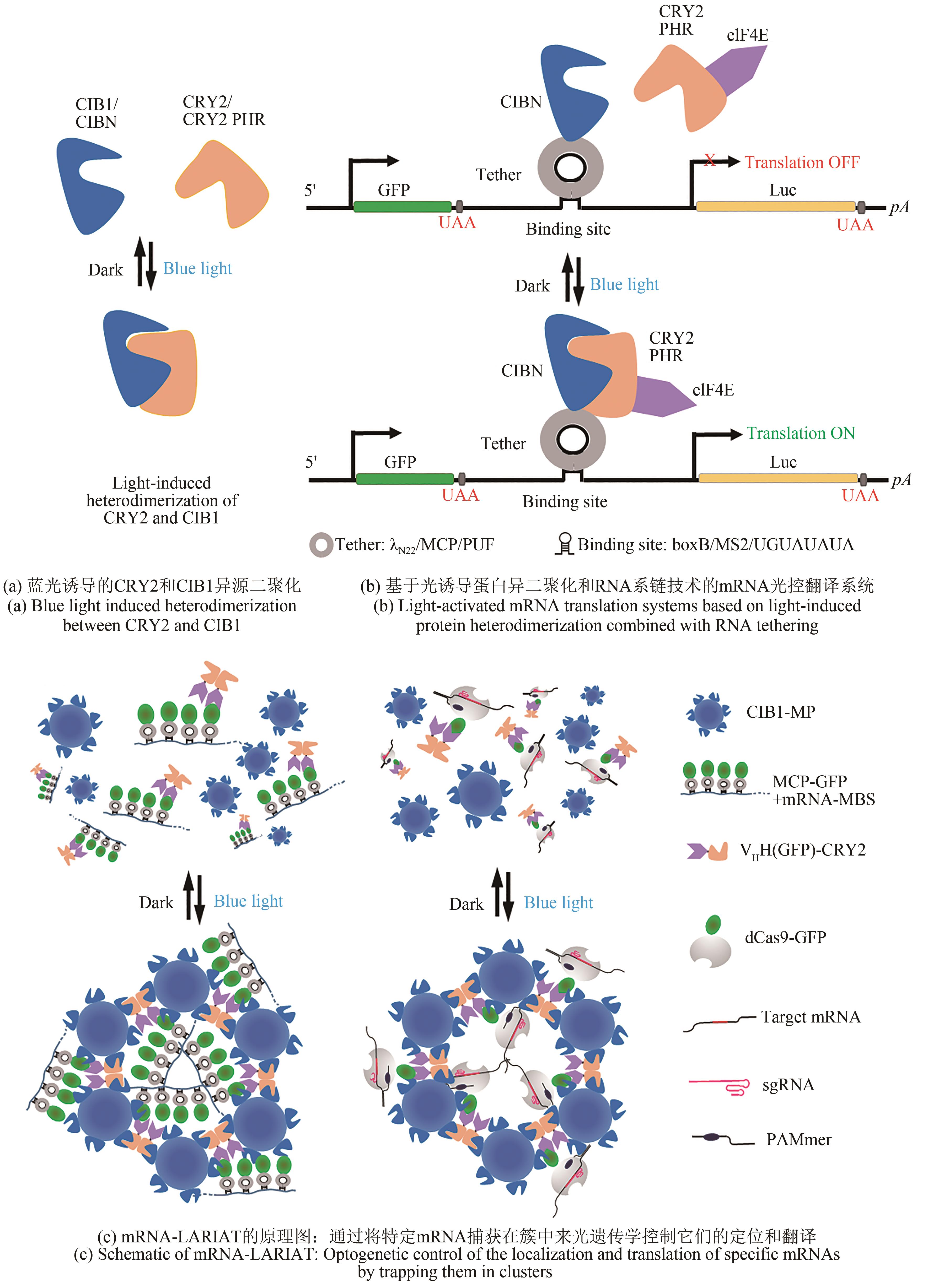
图7 基于LicV光诱导同源二聚化的RNA功能与代谢的光遗传学控制(a)蓝光可诱导LicV同源二聚化,使其与RAT短发夹RNA结合;(b)~(e)利用LicV-RAT光诱导相互作用实现RNA定位(b)、剪接(c)、翻译(d)和降解(e)的光遗传学控制;(f)LA-CRISPR系统原理;将LicV-RAT光诱导相互作用引入CRISPR-Cas系统,可实现对基因转录的光遗传学控制
Fig. 7 Optogenetic control of RNA function and metabolism based on light-induced homodimerization of LicVBlue light induced homodimerization of LicV for its binding to RAT short hairpin RNA (a). Optogenetic control of RNA through light-induced LicV-RAT interaction for localization (b), splicing (c), translation (d), and degradation (e). The LA-CRISPR system based on the introduction of the light-induced LicV-RAT interaction into the CRISPR-Cas system to enable optogenetic control of gene transcription (f)..
| 136 | MOMOTAKE A, LINDEGGER N, NIGGLI E, et al. The nitrodibenzofuran chromophore:a new caging group for ultra-efficient photolysis in living cells[J]. Nature Methods, 2006, 3(1): 35-40. |
| 137 | FURUTA T, WANG S S, DANTZKER J L, et al. Brominated 7-hydroxycoumarin-4-ylmethyls: photolabile protecting groups with biologically useful cross-sections for two photon photolysis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(4): 1193-1200. |
| 138 | FURUTA T, TAKEUCHI H, ISOZAKI M, et al. Bhc-cNMPs as either water-soluble or membrane-permeant photoreleasable cyclic nucleotides for both one- and two-photon excitation[J]. ChemBioChem, 2004, 5(8): 1119-1128. |
| 139 | NAKANISHI H, YOSHII T, KAWASAKI S, et al. Light-controllable RNA-protein devices for translational regulation of synthetic mRNAs in mammalian cells[J]. Cell Chemical Biology, 2021, 28(5): 662-674.e5. |
| 140 | LIU H T, YU X H, LI K W, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis [J]. Science, 2008, 322(5907): 1535-1539. |
| 141 | CAO J C, ARHA M, SUDRIK C, et al. Light-inducible activation of target mRNA translation in mammalian cells[J]. Chemical Communications (Cambridge, England), 2013, 49(75): 8338-8340. |
| 142 | CAO J C, ARHA M, SUDRIK C, et al. Bidirectional regulation of mRNA translation in mammalian cells by using PUF domains[J]. Angewandte Chemie International Edition, 2014, 53(19): 4900-4904. |
| 143 | KIM N Y, LEE S, YU J, et al. Optogenetic control of mRNA localization and translation in live cells[J]. Nature Cell Biology, 2020, 22(3): 341-352. |
| 144 | WEBER A M, KAISER J, ZIEGLER T, et al. A blue light receptor that mediates RNA binding and translational regulation[J]. Nature Chemical Biology, 2019, 15(11): 1085-1092. |
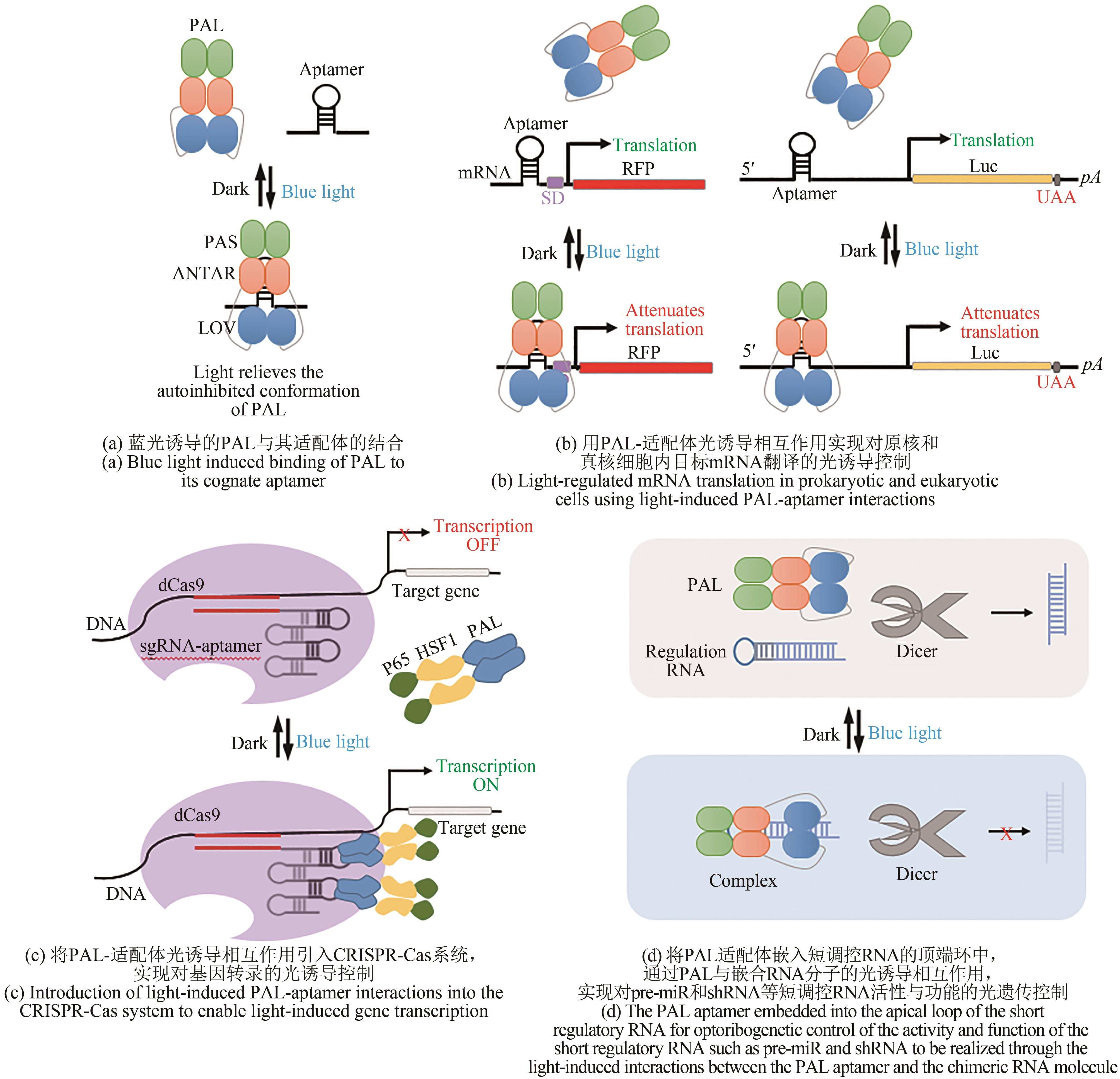
| 145 | RENZL C, KAKOTI A, MAYER G. Aptamer-mediated reversible transactivation of gene expression by light[J]. Angewandte Chemie International Edition, 2020, 59(50): 22414-22418. |
| 146 | PILSL S, MORGAN C, CHOUKEIFE M, et al. Optoribogenetic control of regulatory RNA molecules[J]. Nature Communications, 2020, 11: 4825. |
| 147 | LIU R M, YANG J, YAO J, et al. Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins[J]. Nature Biotechnology, 2022, 40(5): 779-786. |
| 148 | HÜBNER S, DECLERCK N, DIETHMAIER C, et al. Prevention of cross-talk in conserved regulatory systems: Identification of specificity determinants in RNA-binding anti-termination proteins of the BglG family[J]. Nucleic Acids Research, 2011, 39(10): 4360-4372. |
| 1 | CECH T R, STEITZ J A. The noncoding RNA revolution-trashing old rules to forge new ones[J]. Cell, 2014, 157(1): 77-94. |
| 2 | BHATTI G K, KHULLAR N, SIDHU I S, et al. Emerging role of non-coding RNA in health and disease[J]. Metabolic Brain Disease, 2021, 36(6): 1119-1134. |
| 3 | ESTELLER M. Non-coding RNAs in human disease[J]. Nature Reviews Genetics, 2011, 12(12): 861-874. |
| 4 | MONTES M, SANFORD B L, COMISKEY D F, et al. RNA splicing and disease: animal models to therapies[J]. Trends in Genetics: TIG, 2019, 35(1): 68-87. |
| 5 | BATTICH N, STOEGER T, PELKMANS L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution[J]. Nature Methods, 2013, 10(11): 1127-1133. |
| 6 | WANG K C, CHANG H Y. Molecular mechanisms of long noncoding RNAs[J]. Molecular Cell, 2011, 43(6): 904-914. |
| 7 | BUXBAUM A R, HAIMOVICH G, SINGER R H. In the right place at the right time: visualizing and understanding mRNA localization[J]. Nature Reviews Molecular Cell Biology, 2015, 16(2): 95-109. |
| 8 | Method of the year 2011[J]. Nature Methods, 2012, 9(1): 1. |
| 9 | KHAN S H. Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application[J]. Molecular Therapy Nucleic Acids, 2019, 16: 326-334. |
| 10 | DE MENA L, RIZK P, RINCON-LIMAS D E. Bringing light to transcription: the optogenetics repertoire[J]. Frontiers in Genetics, 2018, 9: 518. |
| 11 | TAN P, HE L, HUANG Y, et al. Optophysiology: iIlluminating cell physiology with optogenetics[J]. Physiological Reviews, 2022, 102(3): 1263-1325. |
| 12 | XU X S, QI L S. A CRISPR-dCas toolbox for genetic engineering and synthetic biology[J]. Journal of Molecular Biology, 2019, 431(1): 34-47. |
| 13 | KONERMANN S, BRIGHAM M D, TREVINO A, et al. Optical control of mammalian endogenous transcription and epigenetic states[J]. Nature, 2013, 500(7463): 472-476. |
| 149 | DECLERCK N, VINCENT F, HOH F, et al. RNA recognition by transcriptional antiterminators of the BglG/SacY family: Functional and structural comparison of the CAT domain from SacY and LicT[J]. Journal of Molecular Biology, 1999, 294(2): 389-402. |
| 150 | ZOLTOWSKI B D, SCHWERDTFEGER C, WIDOM J, et al. Conformational switching in the fungal light sensor Vivid[J]. Science, 2007, 316(5827): 1054-1057. |
| 151 | SHARMA E, STERNE-WEILER T, O'HANLON D, et al. Global mapping of human RNA-RNA interactions[J]. Molecular Cell, 2016, 62(4): 618-626. |
| 152 | WAN R X, BAI R, SHI Y G. Molecular choreography of pre-mRNA splicing by the spliceosome[J]. Current Opinion in Structural Biology, 2019, 59: 124-133. |
| 153 | DOUDNA J A, RATH V L. Structure and function of the eukaryotic ribosome: The next frontier[J]. Cell, 2002, 109(2): 153-156. |
| 154 | HENTZE M W, CASTELLO A, SCHWARZL T, et al. A brave new world of RNA-binding proteins[J]. Nature Reviews Molecular Cell Biology, 2018, 19(5): 327-341. |
| 155 | OPLÄNDER C, HIDDING S, WERNERS F B, et al. Effects of blue light irradiation on human dermal fibroblasts[J]. Journal of Photochemistry and Photobiology B: Biology, 2011, 103(2): 118-125. |
| 156 | NGUYEN N T, HUANG K, ZENG H X, et al. Nano-optogenetic engineering of CAR T cells for precision immunotherapy with enhanced safety[J]. Nature Nanotechnology, 2021, 16(12): 1424-1434. |
| 157 | LI T, CHEN X J, QIAN Y J, et al. A synthetic BRET-based optogenetic device for pulsatile transgene expression enabling glucose homeostasis in mice[J]. Nature Communications, 2021, 12: 615. |
| 158 | MÜLLER K, ENGESSER R, METZGER S, et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells[J]. Nucleic Acids Research, 2013, 41(7): e77. |
| 159 | ASH C, DUBEC M, DONNE K, et al. Effect of wavelength and beam width on penetration in light-tissue interaction using computational methods[J]. Lasers in Medical Science, 2017, 32(8): 1909-1918. |
| 14 | WARYAH C B, MOSES C, AROOJ M, et al. Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing[J]. Methods in Molecular Biology, 2018, 1767: 19-63. |
| 15 | LO C L, CHOUDHURY S R, IRUDAYARAJ J, et al. Epigenetic editing of Ascl1 gene in neural stem cells by optogenetics[J]. Scientific Reports, 2017, 7: 42047. |
| 16 | CORBETT A H. Post-transcriptional regulation of gene expression and human disease[J]. Current Opinion in Cell Biology, 2018, 52: 96-104. |
| 17 | ZACCARA S, RIES R J, JAFFREY S R. Reading, writing and erasing mRNA methylation[J]. Nature Reviews Molecular Cell Biology, 2019, 20(10): 608-624. |
| 18 | GERSTBERGER S, HAFNER M, TUSCHL T. A census of human RNA-binding proteins[J]. Nature Reviews Genetics, 2014, 15(12): 829-845. |
| 19 | MORRIS K V, MATTICK J S. The rise of regulatory RNA[J]. Nature Reviews Genetics, 2014, 15(6): 423-437. |
| 20 | IWASAKI Y W, SIOMI M C, SIOMI H. PIWI-interacting RNA: its biogenesis and functions[J]. Annual Review of Biochemistry, 2015, 84: 405-433. |
| 21 | PANNI S, LOVERING R C, PORRAS P, et al. Non-coding RNA regulatory networks[J]. Biochimica et Biophysica Acta Gene Regulatory Mechanisms, 2020, 1863(6): 194417. |
| 22 | GOODALL G J, WICKRAMASINGHE V O. RNA in cancer[J]. Nature Reviews Cancer, 2021, 21(1): 22-36. |
| 23 | JONKHOUT N, TRAN J, SMITH M A, et al. The RNA modification landscape in human disease[J]. RNA, 2017, 23(12): 1754-1769. |
| 24 | HUANG Y, ZHANG J L, YU X L, et al. Molecular functions of small regulatory noncoding RNA[J]. Biochemistry Biokhimiia, 2013, 78(3): 221-230. |
| 25 | GUO S, KEMPHUES K J. Par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed[J]. Cell, 1995, 81(4): 611-620. |
| 26 | ZHAO J H, GUO H S. RNA silencing: from discovery and elucidation to application and perspectives[J]. Journal of Integrative Plant Biology, 2022, 64(2): 476-498. |
| 27 | GHILDIYAL M, ZAMORE P D. Small silencing RNAs: an expanding universe[J]. Nature Reviews Genetics, 2009, 10(2): 94-108. |
| 28 | DANG Y K, YANG Q Y, XUE Z H, et al. RNA interference in fungi: Pathways, functions, and applications[J]. Eukaryotic Cell, 2011, 10(9): 1148-1155. |
| 29 | MORAN Y H, AGRON M, PRAHER D, et al. The evolutionary origin of plant and animal microRNAs[J]. Nature Ecology & Evolution, 2017, 1: 27. |
| 30 | OPHINNI Y, PALATINI U, HAYASHI Y, et al. piRNA-guided CRISPR-like immunity in eukaryotes[J]. Trends in Immunology, 2019, 40(11): 998-1010. |
| 31 | LIU H C, RAUCH S, DICKINSON B C. Programmable technologies to manipulate gene expression at the RNA level[J]. Current Opinion in Chemical Biology, 2021, 64: 27-37. |
| 32 | MERKLE T, MERZ S, REAUTSCHNIG P, et al. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides[J]. Nature Biotechnology, 2019, 37(2): 133-138. |
| 33 | QU L, YI Z Y, ZHU S Y, et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs[J]. Nature Biotechnology, 2019, 37(9): 1059-1069. |
| 34 | CROOKE S T, WITZTUM J L, BENNETT C F, et al. RNA-targeted therapeutics[J]. Cell Metabolism, 2018, 27(4): 714-739. |
| 35 | KOLE R, KRIEG A M. Exon skipping therapy for Duchenne muscular dystrophy[J]. Advanced Drug Delivery Reviews, 2015, 87(4): 104-107. |
| 36 | ZHONG G C, WANG H M, HE W H, et al. A reversible RNA on-switch that controls gene expression of AAV-delivered therapeutics in vivo [J]. Nature Biotechnology, 2020, 38(2): 169-175. |
| 37 | ZHAO E M, MAO A S, DE PUIG H, et al. RNA-responsive elements for eukaryotic translational control[J]. Nature Biotechnology, 2022, 40(4): 539-545. |
| 38 | HOSEINPOOR R, KAZEMI B, RAJABIBAZL M, et al. Improving the expression of anti-IL-2Rα monoclonal antibody in the CHO cells through optimization of the expression vector and translation efficiency[J]. Journal of Biotechnology, 2020, 324: 112-120. |
| 39 | BON C, LUFFARELLI R, RUSSO R, et al. SINEUP non-coding RNAs rescue defective frataxin expression and activity in a cellular model of Friedreich's Ataxia[J]. Nucleic Acids Research, 2019, 47(20): 10728-10743. |
| 40 | TOKI N, TAKAHASHI H, SHARMA H, et al. SINEUP long non-coding RNA acts via PTBP1 and HNRNPK to promote translational initiation assemblies[J]. Nucleic Acids Research, 2020, 48(20): 11626-11644. |
| 41 | SCHEITL C P M, GHAEM MAGHAMI M, LENZ A K, et al. Site-specific RNA methylation by a methyltransferase ribozyme[J]. Nature, 2020, 587(7835): 663-667. |
| 42 | AGRAWAL N, DASARADHI P V N, MOHMMED A, et al. RNA interference: biology, mechanism, and applications[J]. Microbiology and Molecular Biology Reviews: MMBR, 2003, 67(4): 657-685. |
| 43 | KELLEHER A D, CORTEZ-JUGO C, CAVALIERI F, et al. RNAi therapeutics: an antiviral strategy for human infections[J]. Current Opinion in Pharmacology, 2020, 54: 121-129. |
| 44 | CORBETT K S, EDWARDS D K, LEIST S R, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness[J]. Nature, 2020, 586(7830): 567-571. |
| 45 | MULLIGAN M J, LYKE K E, KITCHIN N, et al. Publisher Correction: Phase Ⅰ/Ⅱ study of COVID-19 RNA vaccine BNT162b1 in adults[J]. Nature, 2021, 590(7844): E26. |
| 46 | REES H A, LIU D R. Base editing: precision chemistry on the genome and transcriptome of living cells[J]. Nature Reviews Genetics, 2018, 19(12): 770-788. |
| 47 | ROBERTS T C, LANGER R, WOOD M J A. Advances in oligonucleotide drug delivery[J]. Nature Reviews Drug Discovery, 2020, 19(10): 673-694. |
| 48 | PICHON X, WILSON L A, STONELEY M, et al. RNA binding protein/RNA element interactions and the control of translation[J]. Current Protein & Peptide Science, 2012, 13(4): 294-304. |
| 49 | ABIL Z, DENARD C A, ZHAO H M. Modular assembly of designer PUF proteins for specific post-transcriptional regulation of endogenous RNA[J]. Journal of Biological Engineering, 2014, 8(1): 7. |
| 50 | QUERIDO E, Chartrand P. Using fluorescent proteins to study mRNA trafficking in living cells[J]. Methods in Cell Biology, 2008, 85: 273-292. |
| 51 | BERTRAND E, CHARTRAND P, SCHAEFER M, et al. Localization of ASH1 mRNA particles in living yeast[J]. Molecular Cell, 1998, 2(4): 437-445. |
| 52 | MA H H, TU L C, NASERI A, et al. CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging[J]. Nature Methods, 2018, 15(11): 928-931. |
| 53 | QIN P W, PARLAK M, KUSCU C, et al. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9[J]. Nature Communications, 2017, 8: 14725. |
| 54 | FUJII S, SMALL I. The evolution of RNA editing and pentatricopeptide repeat genes[J]. New Phytologist, 2011, 191(1): 37-47. |
| 55 | AZAD M T A, BHAKTA S, TSUKAHARA T. Site-directed RNA editing by adenosine deaminase acting on RNA for correction of the genetic code in gene therapy[J]. Gene Therapy, 2017, 24(12): 779-786. |
| 56 | SHAO J W, WANG M Y, YU G L, et al. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(29): E6722-E6730. |
| 57 | ZALATAN J G, LEE M E, ALMEIDA R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds[J]. Cell, 2015, 160(1/2): 339-350. |
| 58 | CZAPLINSKI K. Techniques for single-molecule mRNA imaging in living cells[J]. Advances in Experimental Medicine and Biology, 2017, 978: 425-441. |
| 59 | CHAO J A, PATSKOVSKY Y, ALMO S C, et al. Structural basis for the coevolution of a viral RNA-protein complex[J]. Nature Structural & Molecular Biology, 2008, 15(1): 103-105. |
| 60 | MA H H, TU L C, NASERI A, et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow[J]. Nature Biotechnology, 2016, 34(5): 528-530. |
| 61 | SHAO S P, ZHANG W W, HU H, et al. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system[J]. Nucleic Acids Research, 2016, 44(9): e86. |
| 62 | KARAGIANNIS P, FUJITA Y, SAITO H. RNA-based gene circuits for cell regulation[J]. Proceedings of the Japan Academy Series B, Physical and Biological Sciences, 2016, 92(9): 412-422. |
| 63 | KOPNICZKY M B, MOORE S J, FREEMONT P S. Multilevel regulation and translational switches in synthetic biology[J]. IEEE Transactions on Biomedical Circuits and Systems, 2015, 9(4): 485-496. |
| 64 | LILLEY D M J. The K-turn motif in riboswitches and other RNA species[J]. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2014, 1839(10): 995-1004. |
| 65 | SAITO H, KOBAYASHI T, HARA T, et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch[J]. Nature Chemical Biology, 2010, 6(1): 71-78. |
| 66 | STAPLETON J A, ENDO K, FUJITA Y, et al. Feedback control of protein expression in mammalian cells by tunable synthetic translational inhibition[J]. ACS Synthetic Biology, 2012, 1(3): 83-88. |
| 67 | ZHAO Y Y, MAO M W, ZHANG W J, et al. Expanding RNA binding specificity and affinity of engineered PUF domains[J]. Nucleic Acids Research, 2018, 46(9): 4771-4782. |
| 68 | JAMIESON A C, MILLER J C, PABO C O. Drug discovery with engineered zinc-finger proteins[J]. Nature Reviews Drug Discovery, 2003, 2(5): 361-368. |
| 69 | MACKAY J P, FONT J, SEGAL D J. The prospects for designer single-stranded RNA-binding proteins[J]. Nature Structural & Molecular Biology, 2011, 18(3): 256-261. |
| 70 | FERRÉ-D'AMARÉ A R, DOUDNA J A. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module[J]. Journal of Molecular Biology, 2000, 295(3): 541-556. |
| 71 | SHAV-TAL Y, DARZACQ X, SHENOY S M, et al. Dynamics of single mRNPs in nuclei of living cells[J]. Science, 2004, 304(5678): 1797-1800. |
| 72 | FERRÉ-D'AMARÉ A R. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs[J]. Methods, 2010, 52(2): 159-167. |
| 73 | GOLDING I PAULSSON J, ZAWILSKI S M, et al. Real-time kinetics of gene activity in individual bacteria[J]. Cell, 2005, 123(6): 1025-1036. |
| 74 | VASUDEVAN S, STEITZ J A. AU-rich-element-mediated upregulation of translation by FXR1 and argonaute 2[J]. Cell, 2007, 128(6): 1105-1118. |
| 75 | WANG Y, CHEONG C G, TANAKA HALL T M, et al. Engineering splicing factors with designed specificities[J]. Nature Methods, 2009, 6(11): 825-830. |
| 76 | NELLES D A, FANG M Y, O'CONNELL M R, et al. Programmable RNA tracking in live cells with CRISPR/Cas9[J]. Cell, 2016, 165(2): 488-496. |
| 77 | BATRA R, NELLES D A, PIRIE E, et al. Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9[J]. Cell, 2017, 170(5): 899-912.e10. |
| 78 | KERYER-BIBENS C, BARREAU C, OSBORNE H B. Tethering of proteins to RNAs by bacteriophage proteins[J]. Biology of the Cell, 2008, 100(2): 125-138. |
| 79 | RAUCH S, HE C, DICKINSON B C. Targeted m6A reader proteins to study epitranscriptomic regulation of single RNAs[J]. Journal of the American Chemical Society, 2018, 140(38): 11974-11981. |
| 80 | COX D B T, GOOTENBERG J S, ABUDAYYEH O O, et al. RNA editing with CRISPR-cas13[J]. Science, 2017, 358(6366): 1019-1027. |
| 81 | GOLDFLESS S J, BELMONT B J, DE PAZ A M, et al. Direct and specific chemical control of eukaryotic translation with a synthetic RNA-protein interaction[J]. Nucleic Acids Research, 2012, 40(9): e64. |
| 82 | DU M, JILLETTE N, ZHU J J, et al. CRISPR artificial splicing factors[J]. Nature Communications, 2020, 11(1): 2973. |
| 83 | PASTRANA E. Optogenetics: controlling cell function with light[J]. Nature Methods, 2011, 8(1): 24-25. |
| 84 | DEISSEROTH K. Optogenetics[J]. Nature Methods, 2011, 8(1): 26-29. |
| 85 | LIU X, TONEGAWA S. Optogenetics 3.0[J]. Cell, 2010, 141(1): 22-24. |
| 86 | NOWAK V A, PEREIRA E A C, GREEN A L, et al. Optogenetics—shining light on neurosurgical conditions[J]. British Journal of Neurosurgery, 2010, 24(6): 618-624. |
| 87 | TISCHER D, WEINER O D. Illuminating cell signalling with optogenetic tools[J]. Nature Reviews Molecular Cell Biology, 2014, 15(8): 551-558 |
| 88 | REPINA N A, ROSENBLOOM A, MUKHERJEE A, et al. At light speed: Advances in optogenetic systems for regulating cell signaling and behavior[J]. Annual Review of Chemical and Biomolecular Engineering, 2017, 8: 13-39. |
| 89 | SOMUNCU Ö S, BERNS H M, SANCHEZ J G. New pioneers of optogenetics in neuroscience[J]. Advances in Experimental Medicine and Biology, 2020, 1288: 47-60. |
| 90 | REDCHUK T A, KARASEV M M, VERKHUSHA P V, et al. Optogenetic regulation of endogenous proteins[J]. Nature Communications, 2020, 11: 605. |
| 91 | GOGLIA A G, TOETTCHER J E. A bright future: optogenetics to dissect the spatiotemporal control of cell behavior[J]. Current Opinion in Chemical Biology, 2019, 48: 106-113. |
| 92 | TAVAKOLI A, MIN J H. Photochemical modifications for DNA/RNA oligonucleotides[J]. RSC Advances, 2022, 12(11): 6484-6507. |
| 93 | LIU Q Y, DEITERS A. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach[J]. Accounts of Chemical Research, 2014, 47(1): 45-55. |
| 94 | DUSSY A, MEYER C, QUENNET E, et al. New light-sensitive nucleosides for caged DNA strand breaks[J]. ChemBioChem, 2002, 3(1): 54-60. |
| 95 | TANG X J, SU M, YU L L, et al. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides[J]. Nucleic Acids Research, 2010, 38(11): 3848-3855. |
| 96 | GOVAN J M, UPRETY R, THOMAS M, et al. Cellular delivery and photochemical activation of antisense agents through a nucleobase caging strategy[J]. ACS Chemical Biology, 2013, 8(10): 2272-2282. |
| 97 | MCCONNELL E M, COZMA I, MOU Q B, et al. Biosensing with DNAzymes[J]. Chemical Society Reviews, 2021, 50(16): 8954-8994. |
| 98 | HWANG K, WU P W, KIM T, et al. Photocaged DNAzymes as a general method for sensing metal ions in living cells[J]. Angewandte Chemie International Edition, 2014, 53(50): 13798-13802. |
| 99 | YOUNG D D, GOVAN J M, LIVELY M O, et al. Photochemical regulation of restriction endonuclease activity[J]. ChemBioChem, 2009, 10(10): 1612-1616. |
| 100 | TANG X J, MAEGAWA S, WEINBERG E S, et al. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids[J]. Journal of the American Chemical Society, 2007, 129(36): 11000-11001. |
| 101 | KIMURA Y, SHU Z M, ITO M, et al. Intracellular build-up RNAi with single-strand circular RNAs as siRNA precursors[J]. Chemical Communications (Cambridge, England), 2020, 56(3): 466-469. |
| 102 | WU L, PEI F, ZHANG J H, et al. Synthesis of site-specifically phosphate-caged siRNAs and evaluation of their RNAi activity and stability[J]. Chemistry, 2014, 20(38): 12114-12122. |
| 103 | GOVAN J M, YOUNG D D, LUSIC H, et al. Optochemical control of RNA interference in mammalian cells[J]. Nucleic Acids Research, 2013, 41(22): 10518-10528. |
| 104 | KALA A, JAIN P K, KARUNAKARAN D, et al. The synthesis of tetra-modified RNA for the multidimensional control of gene expression via light-activated RNA interference[J]. Nature Protocols, 2014, 9(1): 11-20. |
| 105 | CARLSON-STEVERMER J, KELSO R, KADINA A, et al. CRISPRoff enables spatio-temporal control of CRISPR editing[J]. Nature Communications, 2020, 11: 5041. |
| 106 | ZOU R S, LIU Y, WU B, et al. Cas9 deactivation with photocleavable guide RNAs[J]. Molecular Cell, 2021, 81(7): 1553-1565.e8. |
| 107 | LIU Y, ZOU R S, HE S X, et al. Very fast CRISPR on demand[J]. Science, 2020, 368(6496): 1265-1269. |
| 108 | HEMPHILL J, LIU Q Y, UPRETY R, et al. Conditional control of alternative splicing through light-triggered splice-switching oligonucleotides[J]. Journal of the American Chemical Society, 2015, 137(10): 3656-3662. |
| 109 | GOVAN J M, YOUNG D D, LIVELY M O, et al. Optically triggered immune response through photocaged oligonucleotides[J]. Tetrahedron Letters, 2015, 56(23): 3639-3642. |
| 110 | YOUNG D D, LIVELY M O, DEITERS A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells[J]. Journal of the American Chemical Society, 2010, 132(17): 6183-6193. |
| 111 | LUBBE A S, SZYMANSKI W, FERINGA B L. Recent developments in reversible photoregulation of oligonucleotide structure and function[J]. Chemical Society Reviews, 2017, 46(4): 1052-1079. |
| 112 | WU L, HE Y, TANG X J, et al. Photoregulating RNA digestion using azobenzene linked dumbbell antisense oligodeoxynucleotides[J]. Bioconjugate Chemistry, 2015, 26(6): 1070-1079. |
| 113 | OGASAWARA S. Duration control of protein expression in vivo by light-mediated reversible activation of translation[J]. ACS Chemical Biology, 2017, 12(2): 351-356. |
| 114 | HAMMILL M L, ISLAM G, DESAULNIERS J P. Controlling gene-silencing with azobenzene-containing siRNAs (siRNAzos)[J]. Current Protocols in Nucleic Acid Chemistry, 2020, 83(1): e119. |
| 115 | RESENDIZ M J E, SCHÖN A, FREIRE E, et al. Photochemical control of RNA structure by disrupting π-stacking[J]. Journal of the American Chemical Society, 2012, 134(30): 12478-12481. |
| 116 | ANHÄUSER L, KLÖCKER N, MUTTACH F, et al. A benzophenone-based photocaging strategy for the N7 position of guanosine[J]. Angewandte Chemie International Edition, 2020, 59(8): 3161-3165. |
| 117 | JAKUBOVSKA J, TAURAITĖ D, MEŠKYS R. A versatile method for the UVA-induced cross-linking of acetophenone- or benzophenone-functionalized DNA[J]. Scientific Reports, 2018, 8: 16484. |
| 118 | ANDO H, FURUTA T, TSIEN R Y, et al. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos[J]. Nature Genetics, 2001, 28(4): 317-325. |
| 119 | SAKAMOTO T, SHIGENO A, OHTAKI Y, et al. Photo-regulation of constitutive gene expression in living cells by using ultrafast photo-cross-linking oligonucleotides[J]. Biomaterials Science, 2014, 2(9): 1154-1157. |
| 120 | CHOUDHARY A, VANICHKINA D P, ENDER C, et al. Identification of miR-29b targets using 3-cyanovinylcarbazole containing mimics[J]. RNA, 2018, 24(4): 597-608. |
| 121 | WATANABE Y, FUJIMOTO K. Complete photochemical regulation of 8-17 DNAzyme activity by using reversible DNA photo-crosslinking[J]. ChemBioChem, 2020, 21(22): 3244-3248. |
| 122 | OGASAWARA S, MAEDA M. Photoresponsive 5′-cap for the reversible photoregulation of gene expression[J]. Bioorganic & Medicinal Chemistry Letters, 2011, 21(18): 5457-5459. |
| 123 | OGASAWARA S. Control of cellular function by reversible photoregulation of translation[J]. ChemBioChem, 2014, 15(18): 2652-2655. |
| 124 | ROTSTAN K A, ABDELSAYED M M, PASSALACQUA L F, et al. Regulation of mRNA translation by a photoriboswitch[J]. eLife, 2020, 9: e51737. |
| 125 | GONZAGA E R. Role of UV light in photodamage, skin aging, and skin cancer: importance of photoprotection[J]. American Journal of Clinical Dermatology, 2009, 10(S1): 19-24. |
| 126 | YOU M X, JAFFREY S R. Designing optogenetically controlled RNA for regulating biological systems[J]. Annals of the New York Academy of Sciences, 2015, 1352(1): 13-19. |
| 127 | HAMMILL M L, ISLAM G, DESAULNIERS J P. Synthesis, derivatization and photochemical control of ortho-functionalized tetrachlorinated azobenzene-modified siRNAs[J]. ChemBioChem, 2020, 21(16): 2367-2372. |
| 128 | YOUNG D D, DEITERS A. Photochemical control of biological processes[J]. Organic & Biomolecular Chemistry, 2007, 5(7): 999-1005. |
| 129 | MAYER G, HECKEL A. Biologically active molecules with a "light switch"[J]. Angewandte Chemie International Edition, 2006, 45(30): 4900-4921. |
| 130 | LEE H M, LARSON D R, LAWRENCE D S. Illuminating the chemistry of life: Design, synthesis, and applications of "caged" and related photoresponsive compounds[J]. ACS Chemical Biology, 2009, 4(6): 409-427. |
| 131 | DEITERS A. Principles and applications of the photochemical control of cellular processes[J]. ChemBioChem, 2010, 11(1): 47-53. |
| 132 | DEITERS A. Light activation as a method of regulating and studying gene expression[J]. Current Opinion in Chemical Biology, 2009, 13(5/6): 678-686. |
| 133 | RIGGSBEE C W, DEITERS A. Recent advances in the photochemical control of protein function[J]. Trends in Biotechnology, 2010, 28(9): 468-475. |
| 134 | ADAMS S R, TSIEN R Y. Controlling cell chemistry with caged compounds[J]. Annual Review of Physiology, 1993, 55: 755-784. |
| 135 | ZHU Y, PAVLOS C M, TOSCANO J P, et al. 8-Bromo-7-hydroxyquinoline as a photoremovable protecting group for physiological use: mechanism and scope[J]. Journal of the American Chemical Society, 2006, 128(13): 4267-4276. |
| [1] | 郑益坤, 郑婕, 胡国鹏. 光遗传学工具在学习记忆中的应用研究[J]. 合成生物学, 2025, 6(1): 87-104. |
| [2] | 赵静宇, 张健, 祁庆生, 王倩. 基于细菌双组分系统的生物传感器的研究进展[J]. 合成生物学, 2024, 5(1): 38-52. |
| [3] | 于袁欢, 周阳, 王欣怡, 孔德强, 叶海峰. 光遗传学照进生物医学研究进展[J]. 合成生物学, 2023, 4(1): 102-140. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||