合成生物学 ›› 2025, Vol. 6 ›› Issue (1): 136-156.DOI: 10.12211/2096-8280.2024-031
基因工程辅助萝卜硫苷在十字花科作物中的高效生物合成
刘晓悦, 王盼娣, 吴刚, 刘芳
- 中国农业科学院油料作物研究所,农业农村部油料作物生物学与遗传育种重点实验室,农业农村部植物生态环境安全监督检验测试中心,湖北 武汉 430062
-
收稿日期:2024-03-28修回日期:2024-05-30出版日期:2025-02-28发布日期:2025-03-12 -
通讯作者:吴刚,刘芳 -
作者简介:刘晓悦 (2000—),女,硕士研究生。研究方向为植物分子生物学与基因工程。E-mail:17860395910@139.com吴刚 (1976—),男,研究员,博士生导师。研究方向为基因工程与转基因安全评价。E-mail:wugang@caas.cn刘芳 (1979—),女,副研究员,硕士生导师。研究方向为基因工程与转基因安全评价。E-mail:liufang03@caas.cn
Efficient biosynthesis of glucoraphanin in Brassicaceae crops by genetic engineering
LIU Xiaoyue, WANG Pandi, WU Gang, LIU Fang
- Oil Crops Research Institute,Chinese Academy of Agricultural Sciences,Key Laboratory of Biology and Genetic Breeding of Oil Crops,Ministry of Agriculture and Rural Affairs,Plant Ecological Environment Safety Supervision and Testing Center,Ministry of Agriculture and Rural Affairs,Wuhan 430062,Hubei,China
-
Received:2024-03-28Revised:2024-05-30Online:2025-02-28Published:2025-03-12 -
Contact:WU Gang, LIU Fang
摘要:
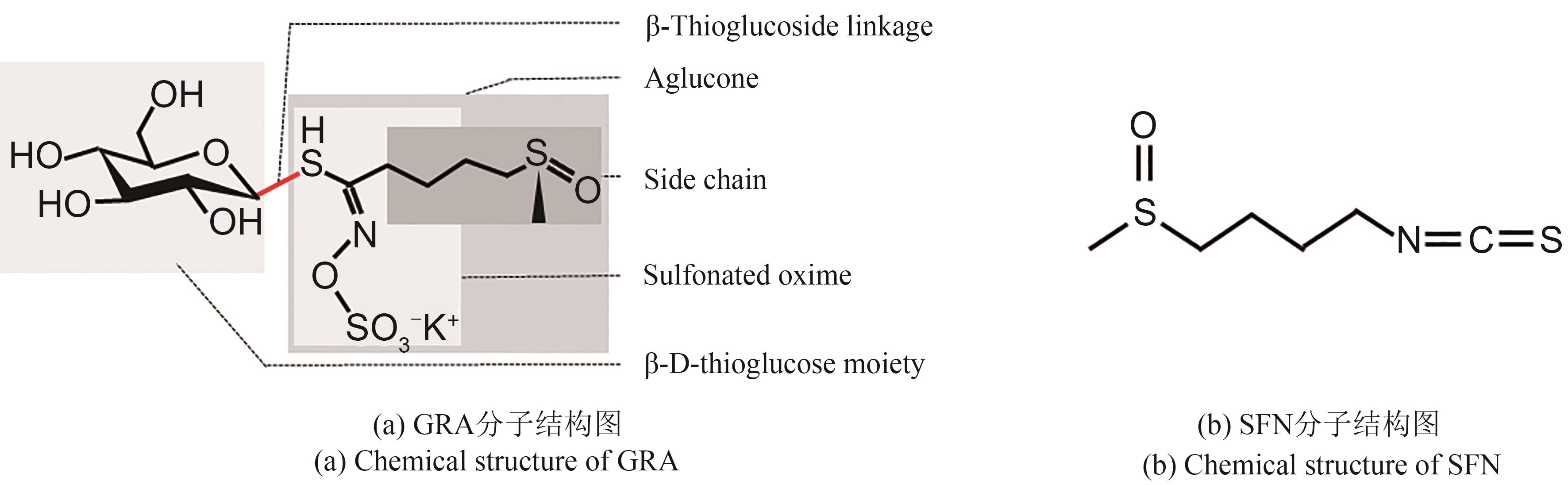
植物次级代谢物萝卜硫苷(glucoraphanin,GRA)是一种由蛋氨酸衍生的硫代葡萄糖苷(glucosinolate,GSL),性质相对稳定,其本身及水解后活性产物萝卜硫素(sulforaphane,SFN)在抵抗癌症、神经保护等方面发挥重要作用,在食品营养和科学研究中受到广泛关注。本文将综述GRA的理化性质、来源、生物学功能、合成途径以及当前生产现状,并进一步探讨未来GRA高效生物合成的潜力策略。GRA合成路径复杂,包括侧链延伸、核心结构形成以及侧链修饰三个阶段,可经植物内源黑芥子酶(myrosinase, MYR)或肠道微生物转化为具有生物活性的SFN等物质。西蓝花等十字花科作物中GRA含量较高,是当前GRA的主要来源作物,但其存在种植周期较长、产量不稳、提取率低等问题,开发经济且可再生的GRA新资源将极大地推进GRA开发应用。随着GRA生物合成及调控路径的明晰,基因工程辅助GRA的高效生物合成展现出巨大的潜力,也提示突破主流的单基因调控策略,聚合多基因多维度协同提高GRA合成的潜力。本文聚焦基因工程辅助十字花科作物高效生产GRA这一目标,系统地梳理了GRA合成各阶段的潜在候选基因并从富集部位角度指出了具高应用价值的底盘作物,以期为将来通过基因工程和分子育种技术调控植物中GRA的生物合成、实现GRA大规模可持续生产,提供一定的思路和策略。
中图分类号:
引用本文
刘晓悦, 王盼娣, 吴刚, 刘芳. 基因工程辅助萝卜硫苷在十字花科作物中的高效生物合成[J]. 合成生物学, 2025, 6(1): 136-156.
LIU Xiaoyue, WANG Pandi, WU Gang, LIU Fang. Efficient biosynthesis of glucoraphanin in Brassicaceae crops by genetic engineering[J]. Synthetic Biology Journal, 2025, 6(1): 136-156.
| 疾病 | 摄入方式 | 机制 | 效应 | 参考文献 |
|---|---|---|---|---|
| 抗癌 | ||||
| 脑癌 | 饲喂西蓝花芽提取物 | 调节Keap1/Nrf2/ARE信号通路,激活细胞抗氧化防御过程 | 抑制肿瘤生长 | [ |
| 鼻咽癌 | 添加纯品SFN | 抑制 EBV早期裂解蛋白Rta表达,阻断EBV裂解周期 | 阻断EBV在激活 | [ |
| 上调肿瘤抑制因子miRNA-124-3p表达靶向抑制STAT3信号通路表达和磷酸化 | 抑制癌增殖和转移 | [ | ||
| 肝癌 | 添加纯品SFN | 诱导 NRF2,重新连接中枢代谢调节调节氨基酸代谢支持谷胱甘肽产生,维持葡萄糖稳态 | 抗氧化 | [ |
| 肺癌 | 添加纯品SFN | 解聚微管、抑制α-微管蛋白与脂肪酸合酶、乙酰CoA羧化酶、柠檬酸裂解酶相互作用 | 抑制微管介导的线粒体自噬引起细胞凋亡 | [ |
| 降低细胞内脂肪酸以及线粒体磷酸含量 | 抑制癌细胞增殖及肿瘤干细胞自我更新 | [ | ||
| 调节Sonic Hedgehog信号通路和PHC3, 组蛋白修饰降低miR-616-5p水平 | 抑制95D和H1299非小细胞肺癌细胞转移 | [ | ||
| 胃癌 | 添加纯品SFN | 上调Bax/Bcl2蛋白以及细胞色素C、PARP-1等信号蛋白表达,促进丝裂原蛋白激酶(MAPK)JNK和 P-38的磷酸化 | 促进癌细胞凋亡 | [ |
| 下调EGFR(上皮生长因子受体),p-ERK1/2表达 | 抑制癌细胞转移 | |||
| 胰腺癌 | 添加纯品SFN | 抑制 PI3K/AKT 和 MEK/ERK 通路,激活转录因子 FOXO | 诱导细胞周期停滞 | [ |
| 诱导产生过量活性氧ROS,激活Nrf2-AMPK信号传导途径 | 抑制癌细胞生长 | [ | ||
| 结肠癌 | 添加纯品SFN | 靶向降低癌细胞HDAC3活性 | 表观修饰 | [ |
| 调节免疫细胞产生的TNFa、IL-1b和IL-6等炎症细胞因子 | 抗炎活性 | [ | ||
| 激活AMPK信号通路 | 抑制癌细胞生长 | [ | ||
| 宫颈癌 | 添加纯品SFN | 激活LATS2,阻断Rad51/MDC1修复 DNA 损伤 | 促进癌细胞凋亡 | [ |
| 前列腺癌 | 添加纯品SFN | 组蛋白H3和H4乙酰化,细胞周期停滞于S和G2/M期 | 表观修饰,抑制细胞周期 | [ |
| 神经保护 | ||||
| 帕金森病 | 添加纯品SFN | Nrf2蛋白、Nrf2mRNA和总谷胱甘肽水平的增加以及神经元组织凋亡的抑制 | Nrf2机制调节神经元与小胶质细胞 | [ |
| 阿尔兹海默病 | 添加纯品SFN | 激活Nrf2抗氧化反应元件(ARE),上调细胞对氧化应激的防御,减少神经元丢失 | 抗氧化 | [ |
| 促进小胶质细胞从促炎的M1表型向抗炎的M2表型分化,减少神经炎症 | 抗炎活性 | [ | ||
| 自闭症 | 摄入SFN | Nrf2介导的Trx1/TrxR1系统的诱导逆转中性粒细胞损伤 | 调控细胞周期 | [ |
| 脑内出血 | 饲喂SFN | SFN激活Nrf2ARE信号通路,发挥抗氧化和抗炎作用,改善脑出血后的神经功能障碍 | 抗氧化、抗炎 | [ |
| 胎儿神经保护 | 食用西蓝花芽 | SFN与酚类物质协调作用,清除自由基及金属络合作用 | 抗氧化 | [ |
| 其他健康益处 | ||||
| 心肌病 | 饮用SFN水溶液 | 通过PI3k/Akt/Nrf2信号通路去除砷代谢产生的过量自由基 | 抗氧化 | [ |
| 防止砷引起的心脏损伤、氧化应激、线粒体复合物功能障碍 | 抗氧化 | |||
| 骨质疏松 | 膳食ITC | 诱导NAD(P)H:醌氧化还原酶1的活性,抑制基质中金属蛋白酶1的产生 | 抗氧化 | [ |
| 肥胖及并发症 | 进食西蓝花等蔬菜 | 激活Nrf2或有效调节 AMP 激活蛋白激酶 | 抗氧化、抗炎 | [ |
| 皮下注射SFN | 减少机体氧化应激以及炎症生物标志物,促进脂肪组织中的巨噬细胞极化为 M2 表型 | 调控脂肪族纤维化相关的基因表达 | [ | |
| 牛皮癣 | 腹腔注射SFN | 激活 KEAP1-NRF2 通路和减弱炎症信号传导 | 强抗氧化 | [ |
表1 GRA生物学功能
Table 1 Biological functions of GRA
| 疾病 | 摄入方式 | 机制 | 效应 | 参考文献 |
|---|---|---|---|---|
| 抗癌 | ||||
| 脑癌 | 饲喂西蓝花芽提取物 | 调节Keap1/Nrf2/ARE信号通路,激活细胞抗氧化防御过程 | 抑制肿瘤生长 | [ |
| 鼻咽癌 | 添加纯品SFN | 抑制 EBV早期裂解蛋白Rta表达,阻断EBV裂解周期 | 阻断EBV在激活 | [ |
| 上调肿瘤抑制因子miRNA-124-3p表达靶向抑制STAT3信号通路表达和磷酸化 | 抑制癌增殖和转移 | [ | ||
| 肝癌 | 添加纯品SFN | 诱导 NRF2,重新连接中枢代谢调节调节氨基酸代谢支持谷胱甘肽产生,维持葡萄糖稳态 | 抗氧化 | [ |
| 肺癌 | 添加纯品SFN | 解聚微管、抑制α-微管蛋白与脂肪酸合酶、乙酰CoA羧化酶、柠檬酸裂解酶相互作用 | 抑制微管介导的线粒体自噬引起细胞凋亡 | [ |
| 降低细胞内脂肪酸以及线粒体磷酸含量 | 抑制癌细胞增殖及肿瘤干细胞自我更新 | [ | ||
| 调节Sonic Hedgehog信号通路和PHC3, 组蛋白修饰降低miR-616-5p水平 | 抑制95D和H1299非小细胞肺癌细胞转移 | [ | ||
| 胃癌 | 添加纯品SFN | 上调Bax/Bcl2蛋白以及细胞色素C、PARP-1等信号蛋白表达,促进丝裂原蛋白激酶(MAPK)JNK和 P-38的磷酸化 | 促进癌细胞凋亡 | [ |
| 下调EGFR(上皮生长因子受体),p-ERK1/2表达 | 抑制癌细胞转移 | |||
| 胰腺癌 | 添加纯品SFN | 抑制 PI3K/AKT 和 MEK/ERK 通路,激活转录因子 FOXO | 诱导细胞周期停滞 | [ |
| 诱导产生过量活性氧ROS,激活Nrf2-AMPK信号传导途径 | 抑制癌细胞生长 | [ | ||
| 结肠癌 | 添加纯品SFN | 靶向降低癌细胞HDAC3活性 | 表观修饰 | [ |
| 调节免疫细胞产生的TNFa、IL-1b和IL-6等炎症细胞因子 | 抗炎活性 | [ | ||
| 激活AMPK信号通路 | 抑制癌细胞生长 | [ | ||
| 宫颈癌 | 添加纯品SFN | 激活LATS2,阻断Rad51/MDC1修复 DNA 损伤 | 促进癌细胞凋亡 | [ |
| 前列腺癌 | 添加纯品SFN | 组蛋白H3和H4乙酰化,细胞周期停滞于S和G2/M期 | 表观修饰,抑制细胞周期 | [ |
| 神经保护 | ||||
| 帕金森病 | 添加纯品SFN | Nrf2蛋白、Nrf2mRNA和总谷胱甘肽水平的增加以及神经元组织凋亡的抑制 | Nrf2机制调节神经元与小胶质细胞 | [ |
| 阿尔兹海默病 | 添加纯品SFN | 激活Nrf2抗氧化反应元件(ARE),上调细胞对氧化应激的防御,减少神经元丢失 | 抗氧化 | [ |
| 促进小胶质细胞从促炎的M1表型向抗炎的M2表型分化,减少神经炎症 | 抗炎活性 | [ | ||
| 自闭症 | 摄入SFN | Nrf2介导的Trx1/TrxR1系统的诱导逆转中性粒细胞损伤 | 调控细胞周期 | [ |
| 脑内出血 | 饲喂SFN | SFN激活Nrf2ARE信号通路,发挥抗氧化和抗炎作用,改善脑出血后的神经功能障碍 | 抗氧化、抗炎 | [ |
| 胎儿神经保护 | 食用西蓝花芽 | SFN与酚类物质协调作用,清除自由基及金属络合作用 | 抗氧化 | [ |
| 其他健康益处 | ||||
| 心肌病 | 饮用SFN水溶液 | 通过PI3k/Akt/Nrf2信号通路去除砷代谢产生的过量自由基 | 抗氧化 | [ |
| 防止砷引起的心脏损伤、氧化应激、线粒体复合物功能障碍 | 抗氧化 | |||
| 骨质疏松 | 膳食ITC | 诱导NAD(P)H:醌氧化还原酶1的活性,抑制基质中金属蛋白酶1的产生 | 抗氧化 | [ |
| 肥胖及并发症 | 进食西蓝花等蔬菜 | 激活Nrf2或有效调节 AMP 激活蛋白激酶 | 抗氧化、抗炎 | [ |
| 皮下注射SFN | 减少机体氧化应激以及炎症生物标志物,促进脂肪组织中的巨噬细胞极化为 M2 表型 | 调控脂肪族纤维化相关的基因表达 | [ | |
| 牛皮癣 | 腹腔注射SFN | 激活 KEAP1-NRF2 通路和减弱炎症信号传导 | 强抗氧化 | [ |
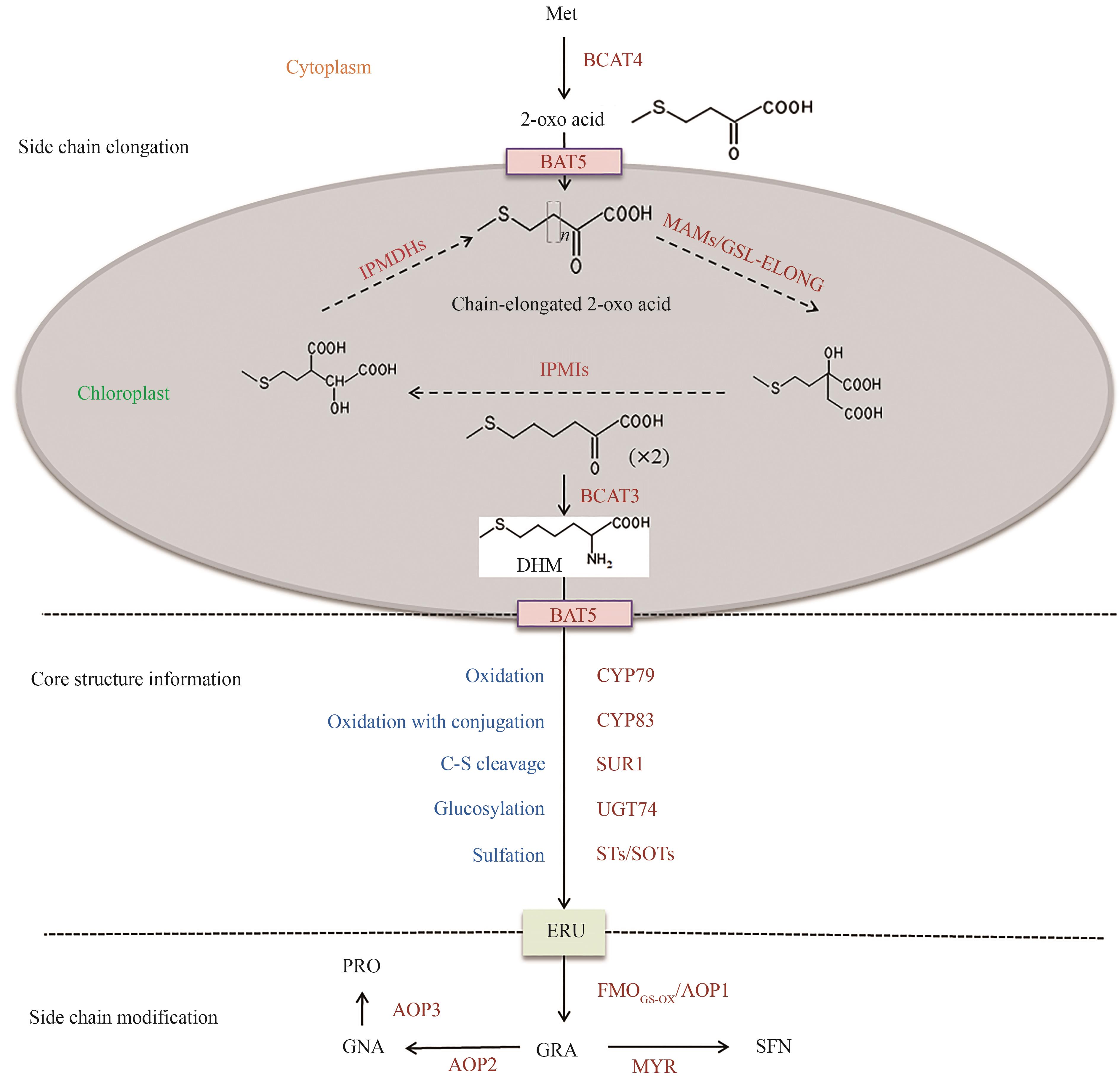
图 2 GRA从头合成完整途径(Met—蛋氨酸; BCAT4—支链氨基酸氨基转移酶4; BAT5—胆汁酸转运体5; MAMs/GSL-ELONG—硫代烷基苹果酸合成酶; IPMIs—苹果酸异丙酯异构酶; IPMDHs—苹果酸异丙酯脱氢酶; BCAT3—支链氨基酸氨基转移酶3; DHM—二高蛋氨酸; CYP79—细胞色素P450单加氧酶CYP79家族; CYP83—细胞色素P450单加氧酶CYP83家族; SUR1—C-S裂解酶; UGT74—糖基转移酶转移酶; STs/SOTs—硫基转移酶; ERU—4-甲硫基-丁基硫代葡萄糖苷; FMOGS-OX/AOP1—黄素单加氧酶; GRA—萝卜硫苷; AOP2—α-酮戊二酸依赖性双加氧酶;AOP3—α-酮戊二酸依赖性双加氧酶; GNA—3-丁烯基硫代葡萄糖苷, 葡萄糖芜菁芥素; PRO—2-羟基-3-丁烯基硫代葡萄糖苷, 甲状腺肿素原; MYR—黑芥子酶; SFN—萝卜硫素)
Fig. 2 De novo synthesis pathway of GRA(Met—methionine; BCAT4—branched-chain aminotransferase 4; BAT5—bile acid transporter 5; MAMs—methylthioalkylmalates ynthases; IPMIs—isopropylmalate isomerases; IPMDHs—isopropylmalate dehydrogenases; BCAT3—branched-chain aminotransferase 3; DHM—dihomoMet; CYP79—cytochrome P450 enzymes CYP79 family; CYP83—cytochrome P450 enzymes CYP83 family; SUR1—SUPERROOT1; UGT74—UDP-glycosyltransferase 74; STs/SOTs—sulfotransferases; ERU—glucoerucin; FMOGS-OX/AOP1—flavin-monooxygenase; GRA—glucoraphanin; AOP2—alkenyl hydroxalkyl producing 2; AOP3—alkenyl hydroxalkyl producing 3; GNA—gluconapin; PRO—progoitrin; MYR—myrosinase; SFN—sulforaphane)
| 俗名 | 拉丁名 | 种子 | 苗 (种子刚发芽) | 叶 | 芽 | 根 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 西蓝花 | Brassica oleracea var. italica | +/+++ | ++/+++ | ++/++ | +/+ | +/+ | [ |
| 萝卜 | Raphanus sativus | +/- | +/- | -/+ | +/+ | +/- | [ |
| 白萝卜 | xBrassicoraphanus | +/+++ | -/++ | +/+ | +/++ | +/++ | [ |
| 甘蓝 | Brassica oleracea var. capitata | ++/+++ | ++/+++ | ++/+ | ++/++ | +/++ | [ |
| 花椰菜 | Brassica oleracea var. botrytis | +/+ | +/+ | N/N | +/+ | +/+ | [ |
| 大白菜 | Brassica rapa ssp. pekinensis | +/+++ | -/+++ | +/+ | +/+ | +/+ | [ |
| 羽衣甘蓝 | Brassica oleracea var. acephala | +/++ | +/++ | ++/+ | +/++ | +/+ | [ |
| 芥菜 | Brassica juncea | -/- | +/++ | -/N | -/+ | -/+ | [ |
| 小白菜 | Brassica rapa ssp. chinensis | +/+ | +/+ | N/N | +/+ | +/++ | [ |
| 欧洲油菜 | Brassica napus | +/++ | +/+++ | +/++ | N/N | N/N | [ |
表2 常见十字花科作物中GRA/PRO含量
Table 2 GRA/PRO content in common Brassicaceae crops
| 俗名 | 拉丁名 | 种子 | 苗 (种子刚发芽) | 叶 | 芽 | 根 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 西蓝花 | Brassica oleracea var. italica | +/+++ | ++/+++ | ++/++ | +/+ | +/+ | [ |
| 萝卜 | Raphanus sativus | +/- | +/- | -/+ | +/+ | +/- | [ |
| 白萝卜 | xBrassicoraphanus | +/+++ | -/++ | +/+ | +/++ | +/++ | [ |
| 甘蓝 | Brassica oleracea var. capitata | ++/+++ | ++/+++ | ++/+ | ++/++ | +/++ | [ |
| 花椰菜 | Brassica oleracea var. botrytis | +/+ | +/+ | N/N | +/+ | +/+ | [ |
| 大白菜 | Brassica rapa ssp. pekinensis | +/+++ | -/+++ | +/+ | +/+ | +/+ | [ |
| 羽衣甘蓝 | Brassica oleracea var. acephala | +/++ | +/++ | ++/+ | +/++ | +/+ | [ |
| 芥菜 | Brassica juncea | -/- | +/++ | -/N | -/+ | -/+ | [ |
| 小白菜 | Brassica rapa ssp. chinensis | +/+ | +/+ | N/N | +/+ | +/++ | [ |
| 欧洲油菜 | Brassica napus | +/++ | +/+++ | +/++ | N/N | N/N | [ |
| 涉及基因 | 来源物种 | 受体物种 | 器官 | 结果/(µmol/g) | 参考文献 | |
|---|---|---|---|---|---|---|
| 传统育种 | ||||||
| 杂交育种 | MYB28 | Brassica villosa×GD33 X | Beneforté® | 花茎 花蕾 | GRA 28 DW(2.5~3倍) | [ |
| GRS1/grs1 | Brassica oleracea var. acephala ×Raphanus sativus L. | Raphanobrassica grs1 | 叶 | GRA 34.1 DW(2倍) | [ | |
| 微生物代谢工程 | ||||||
| 同工酶替代 | BCAT3 | Brassica rapa | Escherichia coli BL21(DE3) | 菌液 | GRA 2~3μg/L | [ |
| GSL-ELONG | Brassica oleracea | |||||
| IPMI(LSU1和SSU3) | Arabidopsis thaliana | |||||
| IPMDH1 | Arabidopsis thaliana | |||||
| 整合外源基因到染色体 | CYP79F1 | Brassica oleracea | Escherichia coli MG1655 | GRA 0.675μg/L | [ | |
| CYP83A1 | Brassica rapa | |||||
| EGT2 | Neurospora crassa | |||||
| UGT74B1 | Arabidopsis thaliana | |||||
| ST5c | Brassica rapa | |||||
| FMOGS-OX1 | Arabidopsis thaliana | |||||
| 植物代谢工程 | ||||||
| 过表达 | MAM1 | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶混合物 | SFN 增加1.7~3.4倍 FW | [ |
| 分子标记辅助回交育种 | braop2.2/braop2.3 | Brassica rapa “R-O-18”(ssp. trilocularis) | Brassica rapa“L58”(ssp. parachinensis) | 叶 | GRA增加18倍 DW | [ |
| 过表达AOP1 | FMOGS-OX2 | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶混合物 | SFN增加1.6~2.7倍 FW | [ |
| 抑制GRA 代谢基因 | GSL-ALK 基因家族 | Brassica napus | Brassica napus | 种子 | GRA 42.6 | [ |
| AOP2 | Brassica oleracea var. alboglabra Bailey | Brassica oleracea var. alboglabra Gailan-04 | 茎 | GRA 3.03 DW (3.09倍) | [ | |
| AOP2(GSL-ALK) | Brassica juncea | Brassica juncea | 种子 | GRA 24.1 DW | [ | |
| BoaAOP2s | Brassica oleracea var. alboglabra | Brassica oleracea var. alboglabra | 叶 | GRA 0.082-0.289 FW(11.71~41.29倍) | [ | |
| 过表达 转录因子 | BoMYB29 | Brassica oleracea Winspit | Brassica oleracea DH AG1012 | 叶 | GRA 2.542 FW | [ |
| csmyb28,csmyb29 | Camelina sativa | Camelina sativa | 种子、根 | GSL完全消失 | [ | |
| 增加MYR | Myrosianse gene | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶 混合物 | SFN 增加3.7倍 FW | [ |
| 敲除转运 蛋白 | gtr2 | Arabidopsis thaliana | Arabidopsis thaliana | 种子 | GSL下降 | [ |
| BnaA06.GTR2 | Brassica napus | Brassica napus | 种子 | GSLS下降 | [ | |
| csgtr1 csgtr2 | Camelina sativa | Camelina sativa | 种子、根 | GSLS减少,种子中减少0.85~0.88倍 | [ | |
| gtr1 gtr2 | Arabidopsis thaliana | Arabidopsis thaliana | 种子、根 | GSL种子中几乎消失, 根中显著积累 | [ | |
| Myrosinase-FMOGS-OX2-MAM1(M-F-A) | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶 混合物 | SFN增加1.8~5.5倍(FW) | [ | |
| 优化基 因组合 | BCAT3 | Arabidopsis thaliana | Nicotiana benthamiana | 叶 | 2.05±0.32(DW,4.74倍) | [ |
| dCGS | ||||||
| IPMI2 | ||||||
| Aconitase | ||||||
| CGBP | ||||||
表3 GRA生产现状
Table 3 Current status of GRA production
| 涉及基因 | 来源物种 | 受体物种 | 器官 | 结果/(µmol/g) | 参考文献 | |
|---|---|---|---|---|---|---|
| 传统育种 | ||||||
| 杂交育种 | MYB28 | Brassica villosa×GD33 X | Beneforté® | 花茎 花蕾 | GRA 28 DW(2.5~3倍) | [ |
| GRS1/grs1 | Brassica oleracea var. acephala ×Raphanus sativus L. | Raphanobrassica grs1 | 叶 | GRA 34.1 DW(2倍) | [ | |
| 微生物代谢工程 | ||||||
| 同工酶替代 | BCAT3 | Brassica rapa | Escherichia coli BL21(DE3) | 菌液 | GRA 2~3μg/L | [ |
| GSL-ELONG | Brassica oleracea | |||||
| IPMI(LSU1和SSU3) | Arabidopsis thaliana | |||||
| IPMDH1 | Arabidopsis thaliana | |||||
| 整合外源基因到染色体 | CYP79F1 | Brassica oleracea | Escherichia coli MG1655 | GRA 0.675μg/L | [ | |
| CYP83A1 | Brassica rapa | |||||
| EGT2 | Neurospora crassa | |||||
| UGT74B1 | Arabidopsis thaliana | |||||
| ST5c | Brassica rapa | |||||
| FMOGS-OX1 | Arabidopsis thaliana | |||||
| 植物代谢工程 | ||||||
| 过表达 | MAM1 | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶混合物 | SFN 增加1.7~3.4倍 FW | [ |
| 分子标记辅助回交育种 | braop2.2/braop2.3 | Brassica rapa “R-O-18”(ssp. trilocularis) | Brassica rapa“L58”(ssp. parachinensis) | 叶 | GRA增加18倍 DW | [ |
| 过表达AOP1 | FMOGS-OX2 | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶混合物 | SFN增加1.6~2.7倍 FW | [ |
| 抑制GRA 代谢基因 | GSL-ALK 基因家族 | Brassica napus | Brassica napus | 种子 | GRA 42.6 | [ |
| AOP2 | Brassica oleracea var. alboglabra Bailey | Brassica oleracea var. alboglabra Gailan-04 | 茎 | GRA 3.03 DW (3.09倍) | [ | |
| AOP2(GSL-ALK) | Brassica juncea | Brassica juncea | 种子 | GRA 24.1 DW | [ | |
| BoaAOP2s | Brassica oleracea var. alboglabra | Brassica oleracea var. alboglabra | 叶 | GRA 0.082-0.289 FW(11.71~41.29倍) | [ | |
| 过表达 转录因子 | BoMYB29 | Brassica oleracea Winspit | Brassica oleracea DH AG1012 | 叶 | GRA 2.542 FW | [ |
| csmyb28,csmyb29 | Camelina sativa | Camelina sativa | 种子、根 | GSL完全消失 | [ | |
| 增加MYR | Myrosianse gene | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶 混合物 | SFN 增加3.7倍 FW | [ |
| 敲除转运 蛋白 | gtr2 | Arabidopsis thaliana | Arabidopsis thaliana | 种子 | GSL下降 | [ |
| BnaA06.GTR2 | Brassica napus | Brassica napus | 种子 | GSLS下降 | [ | |
| csgtr1 csgtr2 | Camelina sativa | Camelina sativa | 种子、根 | GSLS减少,种子中减少0.85~0.88倍 | [ | |
| gtr1 gtr2 | Arabidopsis thaliana | Arabidopsis thaliana | 种子、根 | GSL种子中几乎消失, 根中显著积累 | [ | |
| Myrosinase-FMOGS-OX2-MAM1(M-F-A) | Brassica oleracea var. oleracea | Brassica oleracea var. oleracea | 茎叶 混合物 | SFN增加1.8~5.5倍(FW) | [ | |
| 优化基 因组合 | BCAT3 | Arabidopsis thaliana | Nicotiana benthamiana | 叶 | 2.05±0.32(DW,4.74倍) | [ |
| dCGS | ||||||
| IPMI2 | ||||||
| Aconitase | ||||||
| CGBP | ||||||
| 1 | THINH NGUYEN V P T, STEWART J, LOPEZ M, et al. Glucosinolates: natural occurrence, biosynthesis, accessibility, isolation, structures, and biological activities[J]. Molecules, 2020, 25(19): 4537. |
| 2 | BLAŽEVIĆ I, MONTAUT S, BURČUL F, et al. Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants[J]. Phytochemistry, 2020, 169: 112100. |
| 3 | ASIF ALI M, KHAN N, KALEEM N, et al. Anticancer properties of sulforaphane: current insights at the molecular level[J]. Frontiers in Oncology, 2023, 13: 1168321. |
| 4 | YAGISHITA Y, FAHEY J W, DINKOVA-KOSTOVA A T, et al. Broccoli or sulforaphane: is it the source or dose that matters?[J]. Molecules, 2019, 24(19): 3593. |
| 5 | CAO H Y, LIU R N, ZHANG J H, et al. Improving sulforaphane content in transgenic broccoli plants by overexpressing MAM1, FMOGS-OX2, and Myrosinase [J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2021, 146(3): 461-471. |
| 6 | ROSA E, HEANEY R, FENWICK G, et al. Glucosinolates in crop plants[M/OL]//JANICK J. Horticultural reviews, 1996, 18: 99-125 [2024-02-01]. . |
| 7 | MIAO H Y, ZENG W, WANG J S, et al. Improvement of glucosinolates by metabolic engineering in Brassica crops[J]. aBIOTECH, 2021, 2(3): 314-329. |
| 8 | QIN H, KING G J, BORPATRAGOHAIN P, et al. Developing multifunctional crops by engineering Brassicaceae glucosinolate pathways[J]. Plant Communications, 2023, 4(4): 100565. |
| 9 | SUT S, BOSCHIERO I, SOLANA M, et al. Supercritical CO₂ extraction of Eruca sativa using cosolvents: phytochemical composition by LC-MS analysis[J]. Molecules, 2018, 23(12): 3240. |
| 10 | ZHANG S, YING D Y, CHENG L J, et al. Sulforaphane in broccoli-based matrices: effects of heat treatment and addition of oil[J]. LWT, 2020, 128: 109443. |
| 11 | YU X, MA F, ZHANG L X, et al. Extraction and quantification of sulforaphane and indole-3-carbinol from rapeseed tissues using QuEChERS coupled with UHPLC-MS/MS[J]. Molecules, 2020, 25(9): 2149. |
| 12 | FÖRSTER N, ULRICHS C, SCHREINER M, et al. Development of a reliable extraction and quantification method for glucosinolates in Moringa oleifera [J]. Food Chemistry, 2015, 166: 456-464. |
| 13 | WU C C, CHUANG H Y, LIN C Y, et al. Inhibition of epstein-barr virus reactivation in nasopharyngeal carcinoma cells by dietary sulforaphane[J]. Molecular Carcinogenesis, 2013, 52(12): 946-958. |
| 14 | MAHN A, CASTILLO A. Potential of sulforaphane as a natural immune system enhancer: a review[J]. Molecules, 2021, 26(3): 752. |
| 15 | ELKASHTY O A, TRAN S D. Sulforaphane as a promising natural molecule for cancer prevention and treatment[J]. Current Medical Science, 2021, 41(2): 250-269. |
| 16 | OTOO R A, ALLEN A R. Sulforaphane’s multifaceted potential: from neuroprotection to anticancer action[J]. Molecules, 2023, 28(19): 6902. |
| 17 | SINGH D, ARORA R, BHATIA A, et al. Molecular targets in cancer prevention by 4-(methylthio)butyl isothiocyanate - a comprehensive review[J]. Life Sciences, 2020, 241: 117061. |
| 18 | SCHEPICI G, BRAMANTI P, MAZZON E. Efficacy of sulforaphane in neurodegenerative diseases[J]. International Journal of Molecular Sciences, 2020, 21(22): 8637. |
| 19 | GLADE M J, MEGUID M M. A Glance at… broccoli, glucoraphanin, and sulforaphane[J]. Nutrition, 2015, 31(9): 1175-1178. |
| 20 | BAENAS N, VEGA-GARCÍA A, MANJARREZ-MARMOLEJO J, et al. The preventive effects of broccoli bioactives against cancer: evidence from a validated rat glioma model[J]. Biomedecine &Pharmacotherapy, 2023, 168: 115720. |
| 21 | LI X Q, ZHAO Z L, LI M, et al. Sulforaphane promotes apoptosis, and inhibits proliferation and self-renewal of nasopharyngeal cancer cells by targeting STAT signal through miRNA-124-3p[J]. Biomedecine & Pharmacotherapy, 2018, 103: 473-481. |
| 22 | CHEN L, CHAN L S, LUNG H L, et al. Crucifera sulforaphane (SFN) inhibits the growth of nasopharyngeal carcinoma through DNA methyltransferase 1 (DNMT1)/Wnt inhibitory factor 1 (WIF1) axis[J]. Phytomedicine, 2019, 63: 153058. |
| 23 | BERNUZZI F, MAERTENS A, SAHA S, et al. Sulforaphane rewires central metabolism to support antioxidant response and achieve glucose homeostasis[J]. Redox Biology, 2023, 67: 102878. |
| 24 | YAN Y T, ZHOU Y, LI J T, et al. Sulforaphane downregulated fatty acid synthase and inhibited microtubule-mediated mitophagy leading to apoptosis[J]. Cell Death & Disease, 2021, 12(10): 917. |
| 25 | WANG F P, SUN Y W, HUANG X Y, et al. Sulforaphane inhibits self-renewal of lung cancer stem cells through the modulation of sonic Hedgehog signaling pathway and polyhomeotic homolog 3[J]. AMB Express, 2021, 11(1): 121. |
| 26 | WANG D X, ZOU Y J, ZHUANG X B, et al. Sulforaphane suppresses EMT and metastasis in human lung cancer through miR-616-5p-mediated GSK3β/β-catenin signaling pathways[J]. Acta Pharmacologica Sinica, 2017, 38(2): 241-251. |
| 27 | MONDAL A, BISWAS R, RHEE Y H, et al. Sulforaphene promotes Ba x /Bcl2, MAPK-dependent human gastric cancer AGS cells apoptosis and inhibits migration via EGFR, p-ERK1/2 down-regulation[J]. General Physiology and Biophysics, 2016, 35(1): 25-34. |
| 28 | ROY S K, SRIVASTAVA R K, SHANKAR S. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer[J]. Journal of Molecular Signaling, 2010, 5: 10. |
| 29 | CHEN X, JIANG Z D, ZHOU C C, et al. Activation of Nrf2 by sulforaphane inhibits high glucose-induced progression of pancreatic cancer via AMPK dependent signaling[J]. Cellular Physiology and Biochemistry, 2018, 50(3): 1201-1215. |
| 30 | RAJENDRAN P, KIDANE A I, YU T W, et al. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates[J]. Epigenetics, 2013, 8(6): 612-623. |
| 31 | BESSLER H, DJALDETTI M. Broccoli and human health: immunomodulatory effect of sulforaphane in a model of colon cancer[J]. International Journal of Food Sciences and Nutrition, 2018, 69(8): 946-953. |
| 32 | WANG S Y, WANG Y N, LIU X N, et al. SFN enhanced the radiosensitivity of cervical cancer cells via activating LATS2 and blocking Rad51/MDC1 recruitment to DNA damage site[J]. Cancers, 2022, 14(8): 1872. |
| 33 | RUTZ J, THALER S, MAXEINER S, et al. Sulforaphane reduces prostate cancer cell growth and proliferation in vitro by modulating the cdk-cyclin axis and expression of the CD44 variants 4, 5, and 7[J]. International Journal of Molecular Sciences, 2020, 21(22): 8724. |
| 34 | MORRONI F, SITA G, DJEMIL A, et al. Comparison of adaptive neuroprotective mechanisms of sulforaphane and its interconversion product erucin in in vitro and in vivo models of Parkinson’s disease[J]. Journal of Agricultural and Food Chemistry, 2018, 66(4): 856-865. |
| 35 | KRAFT A D, JOHNSON D A, JOHNSON J A. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult[J]. The Journal of Neuroscience, 2004, 24(5): 1101-1112. |
| 36 | TRIO P Z, FUJISAKI S, TANIGAWA S, et al. DNA microarray highlights Nrf2-mediated neuron protection targeted by wasabi-derived isothiocyanates in IMR-32 cells[J]. Gene Regulation and Systems Biology, 2016, 10: 73-83. |
| 37 | HERNANDEZ-RABAZA V, CABRERA-PASTOR A, TAORO-GONZALEZ L, et al. Neuroinflammation increases GABAergic tone and impairs cognitive and motor function in hyperammonemia by increasing GAT-3 membrane expression. Reversal by sulforaphane by promoting M2 polarization of microglia[J]. Journal of Neuroinflammation, 2016, 13(1): 83. |
| 38 | ALSHEHRI S, AHMAD S F, ALBEKAIRI N A, et al. Thioredoxin 1 and thioredoxin reductase 1 redox system is dysregulated in neutrophils of subjects with autism: in vitro effects of environmental toxicant, methylmercury[J]. Toxics, 2023, 11(9): 739. |
| 39 | IMAI T, MATSUBARA H, HARA H. Potential therapeutic effects of Nrf2 activators on intracranial hemorrhage[J]. Journal of Cerebral Blood Flow and Metabolism, 2021, 41(7): 1483-1500. |
| 40 | BOWEN-FORBES C, ARMSTRONG E, MOSES A, et al. Broccoli, kale, and radish sprouts: key phytochemical constituents and DPPH free radical scavenging activity[J]. Molecules, 2023, 28(11): 4266. |
| 41 | THANGAPANDIYAN S, HEMA T, MILTONPRABU S, et al. Sulforaphane ameliorate arsenic induced cardiotoxicity in rats: role of PI3k/Akt mediated Nrf2 signaling pathway[J]. Journal of Biochemical and Molecular Toxicology, 2024, 38(1): e23576. |
| 42 | VANDUCHOVA A, ANZENBACHER P, ANZENBACHEROVA E. Isothiocyanate from broccoli, sulforaphane, and its properties[J]. Journal of Medicinal Food, 2019, 22(2): 121-126. |
| 43 | MTHEMBU S X H, MAZIBUKO-MBEJE S E, MOETLEDIWA M T, et al. Sulforaphane: a nutraceutical against diabetes-related complications[J]. Pharmacological Research, 2023, 196: 106918. |
| 44 | ZHANG Z Z, CHEN H L, PAN C, et al. Sulforaphane reduces adipose tissue fibrosis via promoting M2 macrophages polarization in HFD fed-mice[J]. Biochimica et Biophysica Acta(BBA) - Molecular Cell Research, 2024, 1871(2): 119626. |
| 45 | MA C J, GU C D, LIAN P P, et al. Sulforaphane alleviates psoriasis by enhancing antioxidant defense through KEAP1-NRF2 pathway activation and attenuating inflammatory signaling[J]. Cell Death & Disease, 2023, 14(11): 768. |
| 46 | FAHEY J W, TALALAY P. Antioxidant functions of sulforaphane: a potent inducer of phase Ⅱ detoxication enzymes[J]. Food and Chemical Toxicology, 1999, 37(9-10): 973-979. |
| 47 | ZHANG Y, TALALAY P, CHO C G, et al. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure[J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(6): 2399-2403. |
| 48 | HANAHAN D, WEINBERG R A. Hallmarks of cancer: the next generation[J]. Cell, 2011, 144(5): 646-674. |
| 49 | ABDULL RAZIS A F, DE NICOLA G R, PAGNOTTA E, et al. 4-Methylsulfanyl-3-butenyl isothiocyanate derived from glucoraphasatin is a potent inducer of rat hepatic phase Ⅱ enzymes and a potential chemopreventive agent[J]. Archives of Toxicology, 2012, 86(2): 183-194. |
| 50 | IPPOUSHI K, TAKEUCHI A, ITO H, et al. Antioxidative effects of daikon sprout (Raphanus sativus L.) and ginger (Zingiber officinale Roscoe) in rats[J]. Food Chemistry, 2007, 102(1): 237-242. |
| 51 | JUGE N, MITHEN R F, TRAKA M. Molecular basis for chemoprevention by sulforaphane: a comprehensive review[J]. Cellular and Molecular Life Sciences, 2007, 64(9): 1105-1127. |
| 52 | RUSSO M, SPAGNUOLO C, RUSSO G L, et al. Nrf2 targeting by sulforaphane: a potential therapy for cancer treatment[J]. Critical Reviews in Food Science and Nutrition, 2018, 58(8): 1391-1405. |
| 53 | YANG H E, KANG M J, HUR G, et al. Sulforaphene suppresses adipocyte differentiation via induction of post-translational degradation of CCAAT/enhancer binding protein beta (C/EBPβ)[J]. Nutrients, 2020, 12(3): 758. |
| 54 | SIVAPALAN T, MELCHINI A, SAHA S, et al. Bioavailability of glucoraphanin and sulforaphane from high-glucoraphanin broccoli[J]. Molecular Nutrition & Food Research, 2018, 62(18): e1700911. |
| 55 | SHAPIRO T A, FAHEY J W, WADE K L, et al. Chemoprotective glucosinolates and isothiocyanates of broccoli sprouts: metabolism and excretion in humans[J]. Cancer Epidemiology, Biomarkers & Prevention, 2001, 10(5): 501-508. |
| 56 | MANGLA B, JAVED S, SULTAN M H, et al. Sulforaphane: a review of its therapeutic potentials, advances in its nanodelivery, recent patents, and clinical trials[J]. Phytotherapy Research, 2021, 35(10): 5440-5458. |
| 57 | ANGELINO D, JEFFERY E. Glucosinolate hydrolysis and bioavailability of resulting isothiocyanates: focus on glucoraphanin[J]. Journal of Functional Foods, 2014, 7: 67-76. |
| 58 | FAHEY J W, HOLTZCLAW W D, WEHAGE S L, et al. Sulforaphane bioavailability from glucoraphanin-rich broccoli: control by active endogenous myrosinase[J]. PLoS One, 2015, 10(11): e0140963. |
| 59 | CRAMER J M, TERAN-GARCIA M, JEFFERY E H. Enhancing sulforaphane absorption and excretion in healthy men through the combined consumption of fresh broccoli sprouts and a glucoraphanin-rich powder[J]. British Journal of Nutrition, 2012, 107(9): 1333-1338. |
| 60 | MAGRATH R, BANO F, MORGNER M, et al. Genetics of aliphatic glucosinolates. Ⅰ. Side chain elongation in Brassica napus and Arabidopsis thaliana [J]. Heredity, 1994, 72(3): 290-299. |
| 61 | REDOVNIKOVIĆ I R, TEXTOR S, LISNIĆ B, et al. Expression pattern of the glucosinolate side chain biosynthetic genes MAM1 and MAM3 of Arabidopsis thaliana in different organs and developmental stages[J]. Plant Physiology and Biochemistry, 2012, 53: 77-83. |
| 62 | GIGOLASHVILI T, YATUSEVICH R, ROLLWITZ I, et al. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana [J]. The Plant Cell, 2009, 21(6): 1813-1829. |
| 63 | KUMAR R, LEE S G, AUGUSTINE R, et al. Molecular basis of the evolution of methylthioalkylmalate synthase and the diversity of methionine-derived glucosinolates[J]. The Plant Cell, 2019, 31(7): 1633-1647. |
| 64 | PETERSEN A, HANSEN L G, MIRZA N, et al. Changing substrate specificity and iteration of amino acid chain elongation in glucosinolate biosynthesis through targeted mutagenesis of Arabidopsis methylthioalkylmalate synthase 1[J]. Bioscience Reports, 2019, 39(7): BSR20190446. |
| 65 | LI G, RIAZ A, GOYAL S, et al. Inheritance of three major genes involved in the synthesis of aliphatic glucosinolates in Brassica oleracea [J]. Journal of the American Society for Horticultural Science, 2001, 126(4): 427-431. |
| 66 | DOUGLAS GRUBB C, ZIPP B J, LUDWIG-MÜLLER J, et al. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis[J]. The Plant Journal, 2004, 40(6): 893-908. |
| 67 | MIKKELSEN M D, PETERSEN B L, OLSEN C E, et al. Biosynthesis and metabolic engineering of glucosinolates[J]. Amino Acids, 2002, 22(3): 279-295. |
| 68 | HANSEN C H, WITTSTOCK U, OLSEN C E, et al. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates[J]. Journal of Biological Chemistry, 2001, 276(14): 11078-11085. |
| 69 | MIKKELSEN M D, OLSEN C E, HALKIER B A. Production of the cancer-preventive glucoraphanin in tobacco[J]. Molecular Plant, 2010, 3(4): 751-759. |
| 70 | PARKIN I, MAGRATH R, KEITH D, et al. Genetics of aliphatic glucosinolates. Ⅱ. Hydroxylation of alkenyl glucosinolates in Brassica napus [J]. Heredity, 1994, 72(6): 594-598. |
| 71 | CHU M, SELTZER T F. Myxedema coma induced by ingestion of raw bok choy[J]. New England Journal of Medicine, 2010, 362(20): 1945-1946. |
| 72 | MITHEN R. Glucosinolates-biochemistry, genetics and biological activity[J]. Plant Growth Regulation, 2001, 34(1): 91-103. |
| 73 | KONG W W, LI J, YU Q Y, et al. Two novel flavin-containing monooxygenases involved in biosynthesis of aliphatic glucosinolates[J]. Frontiers in Plant Science, 2016, 7: 1292. |
| 74 | CHHAJED S, MISRA B B, TELLO N, et al. Chemodiversity of the glucosinolate-myrosinase system at the single cell type resolution[J]. Frontiers in Plant Science, 2019, 10: 618. |
| 75 | LV Q Q, LI X F, FAN B F, et al. The cellular and subcellular organization of the glucosinolate-myrosinase system against herbivores and pathogens[J]. International Journal of Molecular Sciences, 2022, 23(3): 1577. |
| 76 | SCHUSTER J, KNILL T, REICHELT M, et al. Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis [J]. The Plant Cell, 2006, 18(10): 2664-2679. |
| 77 | ANDERSEN T G, NOUR-ELDIN H H, FULLER V L, et al. Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis [J]. The Plant Cell, 2013, 25(8): 3133-3145. |
| 78 | JØRGENSEN M E, NOUR-ELDIN H H, HALKIER B A. Transport of defense compounds from source to sink: lessons learned from glucosinolates[J]. Trends in Plant Science, 2015, 20(8): 508-514. |
| 79 | NOUR-ELDIN H H, ANDERSEN T G, BUROW M, et al. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds[J]. Nature, 2012, 488(7412): 531-534. |
| 80 | BORPATRAGOHAIN P, ROSE T J, LIU L, et al. Remobilization and fate of sulphur in mustard[J]. Annals of Botany, 2019, 124(3): 471-480. |
| 81 | BHANDARI S R, JO J S, LEE J G. Comparison of glucosinolate profiles in different tissues of nine Brassica crops[J]. Molecules, 2015, 20(9): 15827-15841. |
| 82 | LIU M P, ZHANG L H, SER S L, et al. Comparative phytonutrient analysis of broccoli by-products: the potentials for broccoli by-product utilization[J]. Molecules, 2018, 23(4): 900. |
| 83 | NUGROHO A B D, HAN N, PERVITASARI A N, et al. Differential expression of major genes involved in the biosynthesis of aliphatic glucosinolates in intergeneric Baemoochae (Brassicaceae) and its parents during development[J]. Plant Molecular Biology, 2020, 102(1-2): 171-184. |
| 84 | ROBIN A H K, HOSSAIN M R, PARK J I, et al. Glucosinolate profiles in cabbage genotypes influence the preferential feeding of diamondback moth (Plutella xylostella)[J]. Frontiers in Plant Science, 2017, 8: 1244. |
| 85 | CÁMARA-MARTOS F, OBREGÓN-CANO S, MESA-PLATA O, et al. Quantification and in vitro bioaccessibility of glucosinolates and trace elements in Brassicaceae leafy vegetables[J]. Food Chemistry, 2021, 339: 127860. |
| 86 | ASSEFA A D, KIM S H, KO H C, et al. Leaf mustard (Brassica juncea) germplasm resources showed diverse characteristics in agro-morphological traits and glucosinolate levels[J]. Foods, 2023, 12(23): 4374. |
| 87 | MOCNIAK L E, ELKIN K R, DILLARD S L, et al. Building comprehensive glucosinolate profiles for Brassica varieties[J]. Talanta, 2023, 251: 123814. |
| 88 | PARK C H, KIM N S, PARK J S, et al. Effects of light-emitting diodes on the accumulation of glucosinolates and phenolic compounds in sprouting canola (Brassica napus L.)[J]. Foods, 2019, 8(2): 76. |
| 89 | VELASCO P, SOENGAS P, VILAR M, et al. Comparison of glucosinolate profiles in leaf and seed tissues of different Brassica napus crops[J]. Journal of the American Society for Horticultural Science, 2008, 133(4): 551-558. |
| 90 | WITTSTOCK U, BUROW M. Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance[J]. The Arabidopsis Book, 2010, 8: e0134. |
| 91 | SANGKRET S, PONGMALAI P, DEVAHASTIN S, et al. Enhanced production of sulforaphane by exogenous glucoraphanin hydrolysis catalyzed by myrosinase extracted from Chinese flowering cabbage (Brassica rapa var. parachinensis)[J]. Scientific Reports, 2019, 9(1): 9882. |
| 92 | WITTSTOCK U, BUROW M. Tipping the scales: specifier proteins in glucosinolate hydrolysis[J]. IUBMB Life, 2007, 59(12): 744-751. |
| 93 | WU Y F, LV C Z, ZOU L G, et al. Approaches for enhancing the stability and formation of sulforaphane[J]. Food Chemistry, 2021, 345: 128771. |
| 94 | DOSZ E B, JEFFERY E H. Modifying the processing and handling of frozen broccoli for increased sulforaphane formation[J]. Journal of Food Science, 2013, 78(9): H1459-H1463. |
| 95 | BASKAR V, GURURANI M A, YU J W, et al. Engineering glucosinolates in plants: current knowledge and potential uses[J]. Applied Biochemistry and Biotechnology, 2012, 168(6): 1694-1717. |
| 96 | BOURANIS J A, BEAVER L M, WONG C P, et al. Sulforaphane and sulforaphane-nitrile metabolism in humans following broccoli sprout consumption: inter-individual variation, association with gut microbiome composition, and differential bioactivity[J]. Molecular Nutrition & Food Research, 2024, 68(4): e2300286. |
| 97 | LI F, HULLAR M A J, BERESFORD S A A, et al. Variation of glucoraphanin metabolism in vivo and ex vivo by human gut bacteria[J]. British Journal of Nutrition, 2011, 106(3): 408-416. |
| 98 | LUANG-IN V, NARBAD A, NUENO-PALOP C, et al. The metabolism of methylsulfinylalkyl- and methylthioalkyl-glucosinolates by a selection of human gut bacteria[J]. Molecular Nutrition & Food Research, 2014, 58(4): 875-883. |
| 99 | PALOP M L, SMITHS J P, BRINK B TEN. Degradation of sinigrin by Lactobacillus agilis strain R16[J]. International Journal of Food Microbiology, 1995, 26(2): 219-229. |
| 100 | MITREITER S, GIGOLASHVILI T. Regulation of glucosinolate biosynthesis[J]. Journal of Experimental Botany, 2021, 72(1): 70-91. |
| 101 | SØNDERBY I E, HANSEN B G, BJARNHOLT N, et al. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates[J]. PLoS One, 2007, 2(12): e1322. |
| 102 | SEO M S, KIM J S. Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae[J]. Molecules, 2017, 22(9): 1549. |
| 103 | TRAKA M H, SAHA S, HUSEBY S, et al. Genetic regulation of glucoraphanin accumulation in Beneforté® broccoli[J]. New Phytologist, 2013, 198(4): 1085-1095. |
| 104 | AUGUSTINE R, MAJEE M, GERSHENZON J, et al. Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea [J]. Journal of Experimental Botany, 2013, 64(16): 4907-4921. |
| 105 | ABDULL RAZIS A F, IORI R, IOANNIDES C. The natural chemopreventive phytochemical R-sulforaphane is a far more potent inducer of the carcinogen-detoxifying enzyme systems in rat liver and lung than the S-isomer[J]. International Journal of Cancer, 2011, 128(12): 2775-2782. |
| 106 | AZIZI NASER S, AMIRI-BESHELI B, SHARIFI-MEHR S. The isolation and determination of sulforaphane from broccoli tissues by reverse phase-high performance liquid chromatography[J]. Journal of the Chinese Chemical Society, 2011, 58(7): 906-910. |
| 107 | KRÄLING K, RÖBBELEN G, THIES W, et al. Variation of seed glucosinolates in lines of Brassica napus [J]. Plant Breeding, 1990, 105(1): 33-39. |
| 108 | KUSHAD M M, BROWN A F, KURILICH A C, et al. Variation of glucosinolates in vegetable crops of Brassica oleracea [J]. Journal of Agricultural and Food Chemistry, 1999, 47(4): 1541-1548. |
| 109 | ENDO R, CHIKANO H, ITABASHI E, et al. Large insertion in radish GRS1 enhances glucoraphanin content in intergeneric hybrids, Raphanobrassica (Raphanus sativus L.×Brassica oleracea var. acephala)[J]. Frontiers in Plant Science, 2023, 14: 1132302. |
| 110 | LIU F X, YANG H, WANG L M, et al. Biosynthesis of the high-value plant secondary product benzyl isothiocyanate via functional expression of multiple heterologous enzymes in Escherichia coli [J]. ACS Synthetic Biology, 2016, 5(12): 1557-1565. |
| 111 | YANG H, LIU F X, LI Y, et al. Reconstructing biosynthetic pathway of the plant-derived cancer chemopreventive-precursor glucoraphanin in Escherichia coli [J]. ACS Synthetic Biology, 2018, 7(1): 121-131. |
| 112 | YANG H, QIN J Y, WANG X W, et al. Production of plant-derived anticancer precursor glucoraphanin in chromosomally engineered Escherichia coli [J]. Microbiological Research, 2020, 238: 126484. |
| 113 | LIU Z Y, LIANG J L, ZHENG S N, et al. Enriching glucoraphanin in Brassica rapa through replacement of BrAOP2.2/BrAOP2.3 with non-functional genes[J]. Frontiers in Plant Science, 2017, 8: 1329. |
| 114 | LIU Z, HIRANI A H, MCVETTY P B E, et al. Reducing progoitrin and enriching glucoraphanin in Braasica napus seeds through silencing of the GSL-ALK gene family[J]. Plant Molecular Biology, 2012, 79(1): 179-189. |
| 115 | QIAN H M, SUN B, MIAO H Y, et al. Variation of glucosinolates and quinone reductase activity among different varieties of Chinese kale and improvement of glucoraphanin by metabolic engineering[J]. Food Chemistry, 2015, 168: 321-326. |
| 116 | AUGUSTINE R, BISHT N C. Biofortification of oilseed Brassica juncea with the anti-cancer compound glucoraphanin by suppressing GSL-ALK gene family[J]. Scientific Reports, 2015, 5: 18005. |
| 117 | ZHENG H, HUANG W L, LI X X, et al. CRISPR/Cas9-mediated BoaAOP2s editing alters aliphatic glucosinolate side-chain metabolic flux and increases the glucoraphanin content in Chinese kale[J]. Food Research International, 2023, 170: 112995. |
| 118 | ZULUAGA D L, GRAHAM N S, KLINDER A, et al. Overexpression of the MYB29 transcription factor affects aliphatic glucosinolate synthesis in Brassica oleracea [J]. Plant Molecular Biology, 2019, 101(1-2): 65-79. |
| 119 | HÖLZL G, REZAEVA B R, KUMLEHN J, et al. Ablation of glucosinolate accumulation in the oil crop Camelina sativa by targeted mutagenesis of genes encoding the transporters GTR1 and GTR2 and regulators of biosynthesis MYB28 and MYB29[J]. Plant Biotechnology Journal, 2023, 21(1): 189-201. |
| 120 | HE Y Z, YANG Z Q, TANG M Q, et al. Enhancing canola breeding by editing a glucosinolate transporter gene lacking natural variation[J]. Plant Physiology, 2022, 188(4): 1848-1851. |
| 121 | BARNUM C R, ENDELMAN B J, ORNELAS I J, et al. Optimization of heterologous glucoraphanin production in planta [J]. ACS Synthetic Biology, 2022, 11(5): 1865-1873. |
| 122 | BARNUM C R, ENDELMAN B J, SHIH P M. Utilizing plant synthetic biology to improve human health and wellness[J]. Frontiers in Plant Science, 2021, 12: 691462. |
| 123 | MAGRATH R, MITHEN R. Maternal effects on the expression of individual aliphatic glucosinolates in seeds and seedlings of Brassica napus [J]. Plant Breeding, 1993, 111(3): 249-252. |
| 124 | BELL L, WAGSTAFF C. Glucosinolates, myrosinase hydrolysis products, and flavonols found in rocket (Eruca sativa and Diplotaxis tenuifolia)[J]. Journal of Agricultural and Food Chemistry, 2014, 62(20): 4481-4492. |
| 125 | HARUN S, ABDULLAH-ZAWAWI M R, GOH H H, et al. A comprehensive gene inventory for glucosinolate biosynthetic pathway in Arabidopsis thaliana [J]. Journal of Agricultural and Food Chemistry, 2020, 68(28): 7281-7297. |
| 126 | SØNDERBY I E, GEU-FLORES F, HALKIER B A. Biosynthesis of glucosinolates-gene discovery and beyond[J]. Trends in Plant Science, 2010, 15(5): 283-290. |
| 127 | LIU S, HUANG H B, YI X Q, et al. Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome-wide association study[J]. Plant Biotechnology Journal, 2020, 18(6): 1472-1484. |
| 128 | TANG Y S, ZHANG G R, JIANG X Y, et al. Genome-wide association study of glucosinolate metabolites (mGWAS) in Brassica napus L.[J]. Plants, 2023, 12(3): 639. |
| 129 | WEI D Y, CUI Y X, MEI J Q, et al. Genome-wide identification of loci affecting seed glucosinolate contents in Brassica napus L.[J]. Journal of Integrative Plant Biology, 2019, 61(5): 611-623. |
| 130 | BELL L, WAGSTAFF C. Enhancement of glucosinolate and isothiocyanate profiles in Brassicaceae crops: addressing challenges in breeding for cultivation, storage, and consumer-related traits[J]. Journal of Agricultural and Food Chemistry, 2017, 65(43): 9379-9403. |
| 131 | BELL L, ORUNA-CONCHA M J, DE HARO-BAILON A. Editorial: nutritional quality and nutraceutical properties of Brassicaceae (Cruciferae)[J]. Frontiers in Nutrition, 2023, 10: 1292964. |
| 132 | SHEN J J, LIU Y J, WANG X L, et al. A comprehensive review of health-benefiting components in rapeseed oil[J]. Nutrients, 2023, 15(4): 999. |
| 133 | YI G E, ROBIN A H, YANG K, et al. Identification and expression analysis of glucosinolate biosynthetic genes and estimation of glucosinolate contents in edible organs of Brassica oleracea subspecies[J]. Molecules, 2015, 20(7): 13089-13111. |
| 134 | CROCOLL C, MIRZA N, REICHELT M, et al. Optimization of engineered production of the glucoraphanin precursor dihomomethionine in Nicotiana benthamiana [J]. Frontiers in Bioengineering and Biotechnology, 2016, 4: 14. |
| 135 | ZANG Y X, KIM J H, PARK Y D, et al. Metabolic engineering of aliphatic glucosinolates in Chinese cabbage plants expressing Arabidopsis MAM1, CYP79F1, and CYP83A1[J]. BMB Reports, 2008, 41(6): 472-478. |
| 136 | CHEN S X, GLAWISCHNIG E, JØRGENSEN K, et al. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis [J]. The Plant Journal, 2003, 33(5): 923-937. |
| 137 | REINTANZ B, LEHNEN M, REICHELT M, et al. Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates[J]. The Plant Cell, 2001, 13(2): 351-367. |
| 138 | NAUR P, PETERSEN B L, MIKKELSEN M D, et al. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis [J]. Plant Physiology, 2003, 133(1): 63-72. |
| 139 | ZHANG Y Y, HUAI D X, YANG Q Y, et al. Overexpression of three glucosinolate biosynthesis genes in Brassica napus identifies enhanced resistance to Sclerotinia sclerotiorum and Botrytis cinerea [J]. PLoS One, 2015, 10(10): e0140491. |
| 140 | HANSEN B G, KLIEBENSTEIN D J, HALKIER B A. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis [J]. The Plant Journal, 2007, 50(5): 902-910. |
| 141 | YIN L, CHEN H C, CAO B H, et al. Molecular characterization of MYB28 involved in aliphatic glucosinolate biosynthesis in Chinese kale (Brassica oleracea var. alboglabra Bailey)[J]. Frontiers in Plant Science, 2017, 8: 1083. |
| 142 | ARAKI R, HASUMI A, NISHIZAWA O I, et al. Novel bioresources for studies of Brassica oleracea: identification of a kale MYB transcription factor responsible for glucosinolate production[J]. Plant Biotechnology Journal, 2013, 11(8): 1017-1027. |
| 143 | WANG L L, JIANG H, QIU Y J, et al. Biochemical characterization of a novel myrosinase rmyr from Rahnella inusitata for high-level preparation of sulforaphene and sulforaphane[J]. Journal of Agricultural and Food Chemistry, 2022, 70(7): 2303-2311. |
| 144 | YE Q W, FANG Y W, LI M J, et al. Characterization of a novel myrosinase with high activity from marine bacterium Shewanella baltica myr-37[J]. International Journal of Molecular Sciences, 2022, 23(19): 11258. |
| 145 | WANG L L, HAMOUDA H I, DONG Y Y, et al. High-level and reusable preparation of sulforaphane by yeast cells expressing myrosinase[J]. Food Chemistry, 2023, 18: 100668. |
| 146 | BORGEN B H, THANGSTAD O P, AHUJA I, et al. Removing the mustard oil bomb from seeds: transgenic ablation of myrosin cells in oilseed rape (Brassica napus) produces MINELESS seeds[J]. Journal of Experimental Botany, 2010, 61(6): 1683-1697. |
| 147 | THANGSTAD O P, GILDE B, CHADCHAWAN S, et al. Cell specific, cross-species expression of myrosinases in Brassica napus, Arabidopsis thaliana and Nicotiana tabacum [J]. Plant Molecular Biology, 2004, 54(4): 597-611. |
| 148 | MIRZA N, CROCOLL C, ERIK OLSEN C, et al. Engineering of methionine chain elongation part of glucoraphanin pathway in E. coli [J]. Metabolic Engineering, 2016, 35: 31-37. |
| 149 | GUO R F, SHEN W S, QIAN H M, et al. Jasmonic acid and glucose synergistically modulate the accumulation of glucosinolates in Arabidopsis thaliana [J]. Journal of Experimental Botany, 2013, 64(18): 5707-5719. |
| 150 | GUO Y Y, GONG C Y, CAO B E, et al. Blue light enhances health-promoting sulforaphane accumulation in broccoli (Brassica oleracea var. italica) sprouts through inhibiting salicylic acid synthesis[J]. Plants, 2023, 12(17): 3151. |
| 151 | AARABI F, KUSAJIMA M, TOHGE T, et al. Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants[J]. Science Advances, 2016, 2(10): e1601087. |
| 152 | JEON B W, OH M H, KIM H S, et al. Glucosinolate variation among organs, growth stages and seasons suggests its dominant accumulation in sexual over asexual-reproductive organs in white radish[J]. Scientia Horticulturae, 2022, 291: 110617. |
| 153 | SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2021, 71(3): 209-249. |
| 154 | WANG Y A, YAN Q J, FAN C M, et al. Overview and countermeasures of cancer burden in China[J]. Science China Life Sciences, 2023, 66(11): 2515-2526. |
| 155 | JEFFERY E H, ARAYA M. Physiological effects of broccoli consumption[J]. Phytochemistry Reviews, 2009, 8(1): 283-298. |
| 156 | GUO L J, HUANG F F, LIU M R, et al. Generational differences in food consumption among Chinese adults of different ages[J]. Nutrients, 2023, 15(20): 4451. |
| 157 | HEMATI H, HAGHIRALSADAT F, HEMATI M, et al. Design and evaluation of liposomal sulforaphane-loaded polyvinyl alcohol/polyethylene glycol (PVA/PEG) hydrogels as a novel drug delivery system for wound healing[J]. Gels, 2023, 9(9): 748. |
| 158 | ZAMBRANO V, BUSTOS R, AROZARENA Y, et al. Optimization of a microencapsulation process using oil-in-water (O/W) emulsion to increase thermal stability of sulforaphane[J]. Foods, 2023, 12(20): 3869. |
| [1] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [2] | 文志琼, 李煜真, 张金刚, 王菲菲, 马小清, 李福利. 化能驱动的产乙酸菌转化利用CO2研究进展[J]. 合成生物学, 2023, 4(6): 1178-1190. |
| [3] | 潘家豪, 潘炜松, 邱健, 谢东玲, 邹奇, 吴川. 重组胶原蛋白表达体系研究进展[J]. 合成生物学, 2023, 4(4): 808-823. |
| [4] | 郑涵奇, 吴晴, 李洪军, 顾臻. 合成生物学与纳米生物学的交叉融合及其在生物医药领域的应用[J]. 合成生物学, 2022, 3(2): 279-301. |
| [5] | 刘如欣, 杜磊, 徐晓庆, 丁金鹏, 张伟, 李盛英. 基于紫外诱变与生物合成基因簇倍增的多氧霉素高产菌株构建[J]. 合成生物学, 2020, 1(5): 609-620. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||