• 研究论文 •
遗传开关的活细胞定向进化平台的建立及应用
周玉洁1,2, 易啸1,2
- 1.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,定量合成生物学全国重点实验室,广东 深圳 518055
2.中国科学院大学,北京 100049
-
收稿日期:2025-03-24修回日期:2025-04-23出版日期:2025-04-26 -
通讯作者:易啸 -
作者简介:周玉洁 (2000—),女,硕士研究生。研究方向为合成生物学。E-mail:yj.zhou@siat.ac.cn易啸 (1986—),男,研究员,博士,博士生导师。研究方向为合成生物学,定向进化。E-mail:xiao.yi@siat.ac.cn
Engineering an in vivo directed evolution system for developing genetic switches
ZHOU Yujie1,2, YI Xiao1,2
- 1.Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen Institute of Synthetic Biology,Key Laboratory of Quantitative Synthetic Biology,Shenzhen 518055,Guangdong,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2025-03-24Revised:2025-04-23Online:2025-04-26 -
Contact:YI Xiao
摘要:
在合成生物学中,转录因子作为遗传开关,可通过模块化组合策略构建复杂的调控网络。现有工程化的基因元件种类有限,因此建立高效的遗传开关定向进化平台对于基因线路设计和代谢通路优化具有重要价值。本研究整合TADR活细胞定向进化系统和基于半乳糖磷酸激酶GalK及绿色荧光蛋白GFP的双重正负筛选系统,构建了优化遗传开关的实验平台。通过转录因子TetR的进化实验验证了该平台的有效性,并成功将转录因子AcuR从阻遏模式反转为激活模式(AcuR-OFF)。AcuR-OFF蛋白的开关性能与野生型类似。相较于常用的易错PCR技术,TADR构建突变文库成本更为低廉、突变规模多样性更高。双重正负筛选系统极大地降低了假阳性率,在目标种群丰度低的极端情况下仍然具有高效筛选能力。该实验平台有望成为遗传开关开发与优化的有力工具。
中图分类号:
引用本文
周玉洁, 易啸. 遗传开关的活细胞定向进化平台的建立及应用[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-023.
ZHOU Yujie, YI Xiao. Engineering an in vivo directed evolution system for developing genetic switches[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-023.
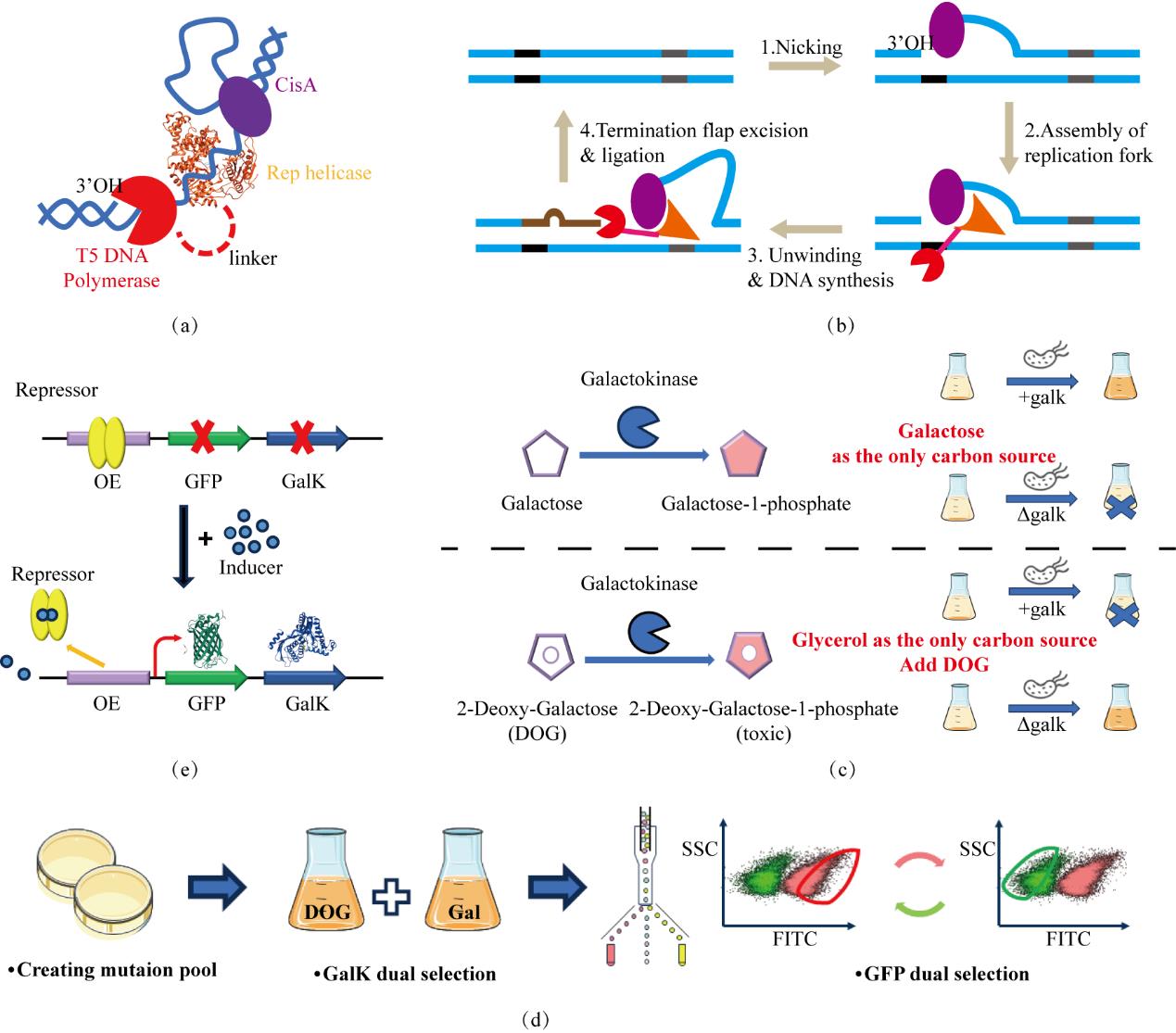
图1 搭建定向进化实验平台(a)TADR系统的组成元件;(b)TADR系统引入突变的过程,黑色和灰色片段分别为起始和终止序列;(c)GalK正负筛选标记的工作原理;(d)TADR-GalK-GFP实验平台的工作流程;(c)以阻遏蛋白为例,筛选系统GFP-GalK的表达受阻遏蛋白调控,通过改变诱导条件实现正向和负向筛选
Fig. 1 Construction of the directed evolution experimental platform(a) Components of the TADR system; (b) The process of introducing mutations by the TADR system, the black and gray segments represent the initiation and termination; (c) Taking the repressor protein as an example, in the screening system, the expression of GFP-GalK is regulated by the repressor protein, and positive and negative screening are achieved by changing the induction conditions; (d) The working principle of the positive and negative screening markers of GalK; (e) The working process of the TADR-GalK-GFP experimental platform
RBS名称 RBS name | RBS序列 RBS sequence |
|---|---|
| RBS1 | TCGAGGT |
| RBS2 | TCCTGGT |
| RBS4 | AGGAGGT |
| RBS5 | GAGGAGG |
| RBS28 | CTCGTGA |
| RBS166 | TCCTGGA |
| RBS1036 | TCCAGGA |
表1 实验使用的RBS序列
Table 1 The RBS sequence used in the experiment
RBS名称 RBS name | RBS序列 RBS sequence |
|---|---|
| RBS1 | TCGAGGT |
| RBS2 | TCCTGGT |
| RBS4 | AGGAGGT |
| RBS5 | GAGGAGG |
| RBS28 | CTCGTGA |
| RBS166 | TCCTGGA |
| RBS1036 | TCCAGGA |
启动子-RBS组合 Promoter-RBS groups | 启动子预测强度 Promoter strength | RBS预测强度 RBS strength |
|---|---|---|
| Ptac-RBS2 | 16000 | 500 |
| Pbla-RBS1 | 2000 | 3000 |
| Pbla-RBS2 | 2000 | 500 |
| Ptac-RBS1 | 16000 | 3000 |
表 2 多个启动子及RBS序列预测强度
Table 2 Prediction strength of multiple promoter and RBS sequences
启动子-RBS组合 Promoter-RBS groups | 启动子预测强度 Promoter strength | RBS预测强度 RBS strength |
|---|---|---|
| Ptac-RBS2 | 16000 | 500 |
| Pbla-RBS1 | 2000 | 3000 |
| Pbla-RBS2 | 2000 | 500 |
| Ptac-RBS1 | 16000 | 3000 |
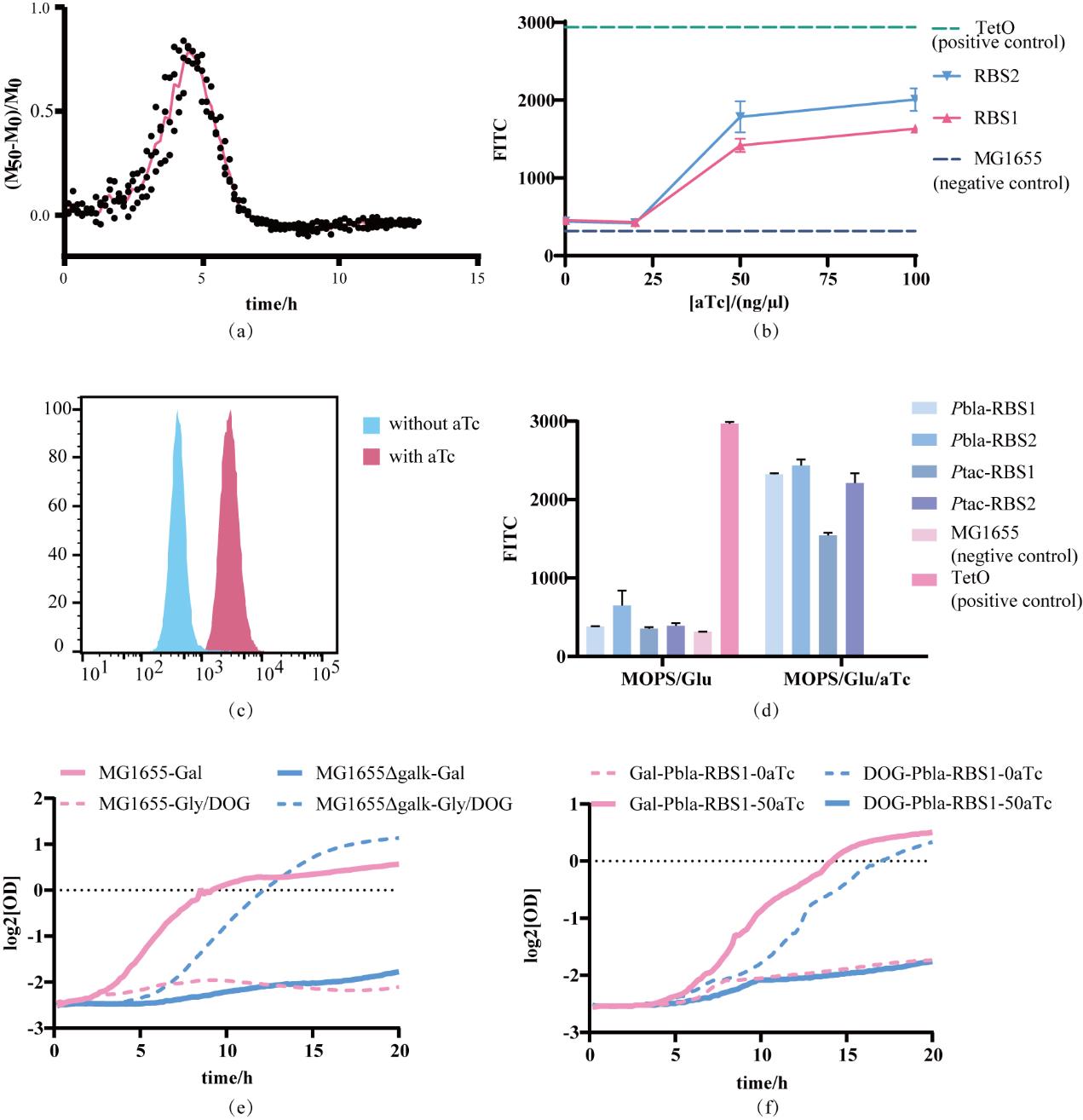
图2 TetR进化实验平台搭建(a)酶标仪测定的荧光强度与吸光值计算可得群体平均荧光强度,记为M,比较诱导前后M的差值以反应阻遏蛋白的调控状态;(b)TetR对aTc诱导剂的响应曲线,诱导剂浓度分别为0、20、50、100 ng/µl,虚线表示两组对照,TetO组为PL-tetO-GFP底盘细胞(GFP为组成型表达),MG1655组不含GFP,两组分别代表了该系统中荧光表达强度的上限及下限;(c)诱导浓度为50 ng/µl时,诱导前后的群体荧光分布;(d)不同表达强度的TetR蛋白诱导前后的荧光强度变化,以及与对照组荧光强度的比较;(e)使用MG1655(含galk)与MG1655Δgalk 两种菌株验证GalK的正负筛选性能;(f)Pbla-RBS1-TetR对GalK正负筛的响应
Fig. 2 Construction of the TetR evolution experimental platform(a) The average fluorescence intensity of the population, denoted as M, can be calculated from the fluorescence intensity and absorbance value measured by the microplate reader. Compare the difference of M before and after induction to reflect the regulatory state of the repressor protein; (b) The response curve of TetR to the aTc inducer, with the inducer concentrations being 0, 20, 50, and 100 ng/µl respectively. The dashed lines represent the two control groups. The TetO group consists of PL-tetO-GFP chassis cells, in which GFP is expressed constitutively. The MG1655 group, by contrast, lacks GFP. These two groups represent the upper and lower limits of fluorescence expression intensity within this system, respectively; (c) When the induction concentration is 50 ng/µl, the population fluorescence distribution before and after induction; (d) The variations in fluorescence intensity before and after the induction of TetR proteins with different expression levels, as well as the comparison of these fluorescence intensities with those of the control groups; (e) Use two strains, MG1655 (containing galk) and MG1655Δgalk, to verify the positive and negative screening performance of GalK; (f) The response of Pbla-RBS1-TetR to the positive and negative screening of GalK
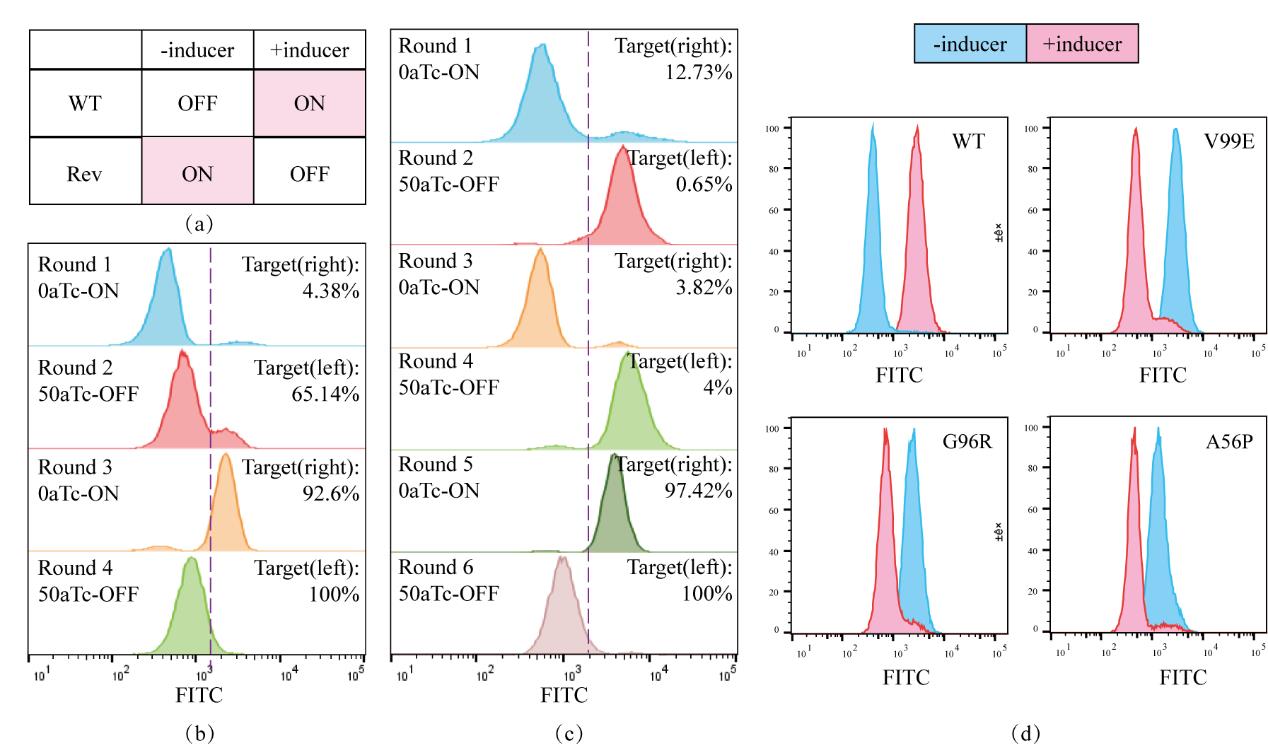
图3 筛选TetR-OFF系统(a)TetR野生型与反转表型对诱导剂的响应(b)使用GalK-GFP双正负筛的筛选过程;(c)仅使用GFP作为筛选标记的筛选过程;(d)对野生型及TetR-OFF突变子的调控性能表征,蓝色为没有诱导剂时的群体荧光分布,红色为50 ng/µl aTc 诱导后的群体荧光分布
Fig. 3 Screening of the TetR-OFF system(a) The response of the wild-type TetR and the reversed phenotype to the inducer. (b) The screening process using the GalK-GFP dual positive and negative screening. (c) The screening process using only GFP as the screening marker. (d) Characterization of the regulatory performance of the wild-type and TetR-OFF mutants. The blue represents the population fluorescence distribution without the inducer, and the red represents the population fluorescence distribution after induction with 50 ng/µl aTc.
突变工具 Mutation Tool | 组别 Groups | 位点突变 Mutation sites | ||||
|---|---|---|---|---|---|---|
| WT | N18Y | V20D | I22T | L60S | Others | |
| 易错PCR | 1 | 18N | 20V | 22I | 60L | R28H, T202A |
| 2 | 18N | 20V | 22I | |||
| TADR | 3 | 18N | 20V | 22I | Y93C, D95E | |
| 4 | 22I | |||||
表3 revTetR进化为TetR-ON的突变位点
Table 3 The mutation sites where revTetR evolved into TetR-ON.
突变工具 Mutation Tool | 组别 Groups | 位点突变 Mutation sites | ||||
|---|---|---|---|---|---|---|
| WT | N18Y | V20D | I22T | L60S | Others | |
| 易错PCR | 1 | 18N | 20V | 22I | 60L | R28H, T202A |
| 2 | 18N | 20V | 22I | |||
| TADR | 3 | 18N | 20V | 22I | Y93C, D95E | |
| 4 | 22I | |||||
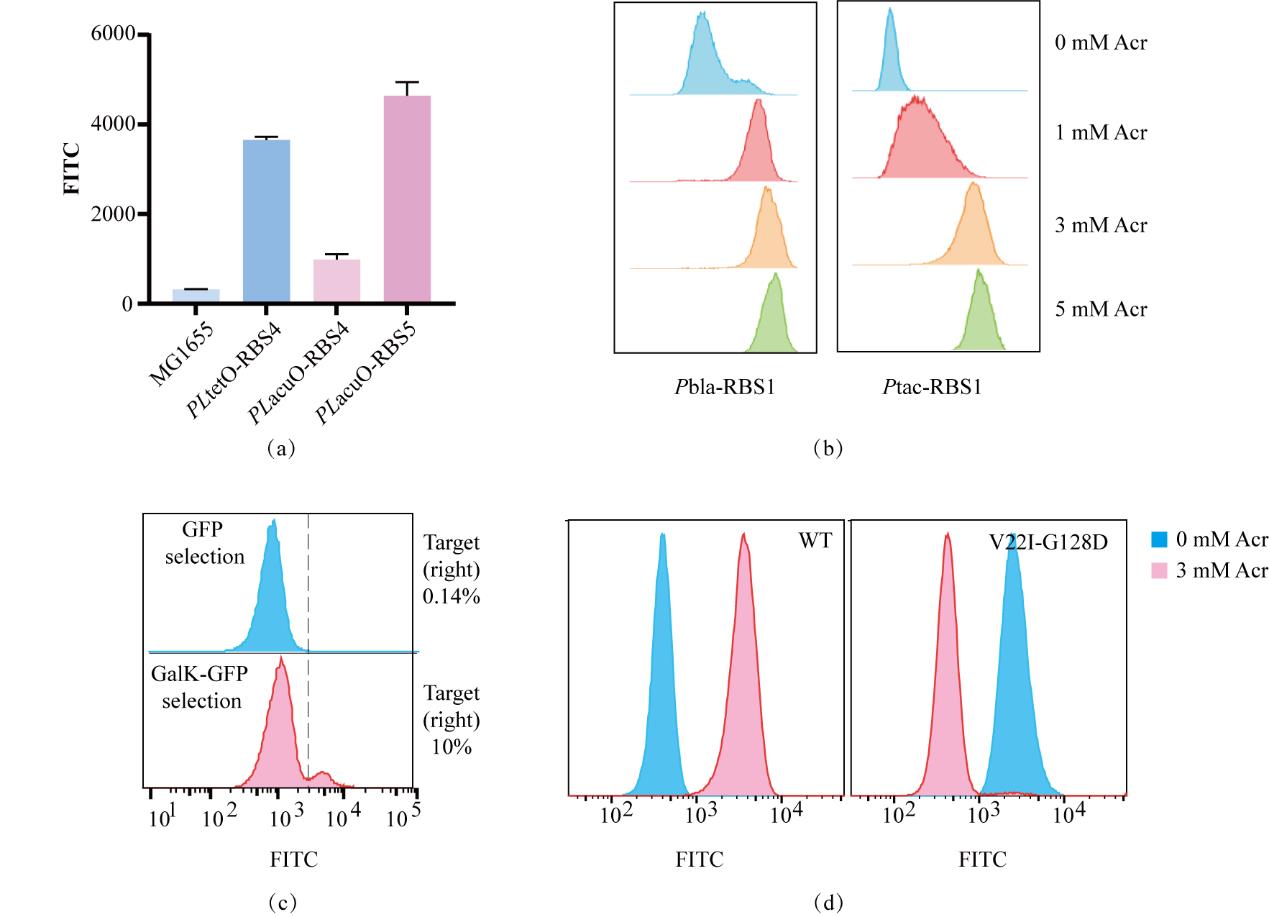
图4 搭建实验平台开发AcuR-OFF(a)以MG1655为荧光负对照,TetO组为荧光正对照,优化AcuO调控系统;(b)分别对两组不同表达强度的TetR测定诱导浓度梯度下的荧光群体分布,以选择最佳诱导条件;(c)GalK筛选前后,没有诱导剂时目标种群比例的变化,蓝色表示GalK筛选前群体荧光分布,红色表示GalK筛选后群体荧光分布;(d)对野生型及AcuR-OFF突变子的调控性能表征,蓝色为没有诱导剂时的群体荧光分布,红色为3 mM Acr诱导后的群体荧光分布
Fig. 4 Construction of the experimental platform for the development of AcuR-OFF.(a) Using MG1655 as the fluorescence negative control and the TetO group as the fluorescence positive control, the AcuO regulatory system was optimized. (b) The fluorescence population distributions of two groups of TetR with different expression intensities were measured under the induction concentration gradients respectively to select the optimal induction conditions. (c) The changes in the proportion of the target population in the absence of the inducer before and after GalK screening. The blue color represents the population fluorescence distribution before GalK screening, and the red color represents the population fluorescence distribution after GalK screening. (d) Characterization of the regulatory performance of the wild type and the AcuR-OFF mutant. The blue color represents the population fluorescence distribution in the absence of the inducer, and the red color represents the population fluorescence distribution after induction with 3 mM Acr.
| 1 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 2 | CAMPS M, NAUKKARINEN J, JOHNSON B P, et al. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(17): 9727-9732. |
| 3 | CROOK N, ABATEMARCO J, SUN J, et al. In vivo continuous evolution of genes and pathways in yeast[J]. Nature Communications, 2016, 7: 13051. |
| 4 | ENGLISH J G, OLSEN R H J, LANSU K, et al. VEGAS as a platform for facile directed evolution in mammalian cells[J]. Cell, 2019, 178(3): 748-761.e17. |
| 5 | ESVELT K M, CARLSON J C, LIU D R. A system for the continuous directed evolution of biomolecules[J]. Nature, 2011, 472(7344): 499-503. |
| 6 | FAURE G, SAITO M, WILKINSON M E, et al. TIGR-Tas: a family of modular RNA-guided DNA-targeting systems in prokaryotes and their viruses[J]. Science, 2025: eadv9789. |
| 7 | HALPERIN S O, TOU C J, WONG E B, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560(7717): 248-252. |
| 8 | HESS G T, FRÉSARD L, HAN K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells[J]. Nature Methods, 2016, 13(12): 1036-1042. |
| 9 | MOORE C L, PAPA L J 3 RD, SHOULDERS M D. A processive protein Chimera introduces mutations across defined DNA regions in vivo [J]. Journal of the American Chemical Society, 2018, 140(37): 11560-11564. |
| 10 | NYERGES Á, CSÖRGŐ B, DRASKOVITS G, et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(25): E5726-E5735. |
| 11 | RAVIKUMAR A, ARZUMANYAN G A, OBADI M K A, et al. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds[J]. Cell, 2018, 175(7): 1946-1957.e13. |
| 12 | SIMON A J, MORROW B R, ELLINGTON A D. Retroelement-based genome editing and evolution[J]. ACS Synthetic Biology, 2018, 7(11): 2600-2611. |
| 13 | TIAN R Z, ZHAO R Z, GUO H Y, et al. Engineered bacterial orthogonal DNA replication system for continuous evolution[J]. Nature Chemical Biology, 2023, 19(12): 1504-1512. |
| 14 | YI X, KAZLAUSKAS R, TRAVISANO M. Evolutionary innovation using EDGE, a system for localized elevated mutagenesis[J]. PLoS One, 2020, 15(4): e0232330. |
| 15 | YI X, KHEY J, KAZLAUSKAS R J, et al. Plasmid hypermutation using a targeted artificial DNA replisome[J]. Science Advances, 2021, 7(29): eabg8712. |
| 16 | WILSON D S, KEEFE A D. Random mutagenesis by PCR[J]. Current Protocols in Molecular Biology, 2001, Chapter 8: Unit8.3. |
| 17 | MCCULLUM E O, WILLIAMS B A R, ZHANG J L, et al. Random mutagenesis by error-prone PCR[M/OL]//BRAMAN J. Methods in molecular biology: in vitro mutagenesis protocols. third edition. Totowa, NJ: Humana Press, 2010, 634: 103-109. (2010-03-19)[2024-02-01]. . |
| 18 | IKE K, ARASAWA Y, KOIZUMI S, et al. Evolutionary design of choline-inducible and-repressible T7-based induction systems[J]. ACS Synthetic Biology, 2015, 4(12): 1352-1360. |
| 19 | RKENES T P, LAMARK T, STRØM A R. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation[J]. Journal of Bacteriology, 1996, 178(6): 1663-1670. |
| 20 | LI X T, THOMASON L C, SAWITZKE J A, et al. Positive and negative selection using the TetA-sacB cassette: recombineering and P1 transduction in Escherichia coli [J]. Nucleic Acids Research, 2013, 41(22): e204. |
| 21 | RYU Y S, CHANDRAN S P, KIM K, et al. Oligo- and dsDNA-mediated genome editing using a tetA dual selection system in Escherichia coli [J]. PLoS One, 2017, 12(7): e0181501. |
| 22 | STAVROPOULOS T A, STRATHDEE C A. Synergy between tetA and rpsL provides high-stringency positive and negative selection in bacterial artificial chromosome vectors[J]. Genomics, 2001, 72(1): 99-104. |
| 23 | BISWAS K, STAUFFER S, SHARAN S K. Using recombineering to generate point mutations: galK-based positive-negative selection method[M/OL]// PECCOUD J. Methods in molecular biology: gene synthesis. Totowa, NJ: Humana Press, 2012, 852: 121-131. (2012-01-01)[2024-02-01]. . |
| 24 | POELWIJK F J, DE VOS M G J, TANS S J. Tradeoffs and optimality in the evolution of gene regulation[J]. Cell, 2011, 146(3): 462-470. |
| 25 | SAEKI K, TOMINAGA M, KAWAI-NOMA S, et al. Rapid diversification of BetI-based transcriptional switches for the control of biosynthetic pathways and genetic circuits[J]. ACS Synthetic Biology, 2016, 5(11): 1201-1210. |
| 26 | WARMING S, COSTANTINO N, COURT D L, et al. Simple and highly efficient BAC recombineering using galK selection[J]. Nucleic Acids Research, 2005, 33(4): e36. |
| 27 | DABIRIAN Y, GONÇALVES TEIXEIRA P, NIELSEN J, et al. FadR-based biosensor-assisted screening for genes enhancing fatty acyl-CoA pools in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2019, 8(8): 1788-1800. |
| 28 | XIAO Y, BOWEN C H, LIU D, et al. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis[J]. Nature Chemical Biology, 2016, 12(5): 339-344. |
| 29 | XU X H, LI X L, LIU Y F, et al. Pyruvate-responsive genetic circuits for dynamic control of central metabolism[J]. Nature Chemical Biology, 2020, 16(11): 1261-1268. |
| 30 | DAS A T, TENENBAUM L, BERKHOUT B. Tet-on systems for doxycycline-inducible gene expression[J]. Current Gene Therapy, 2016, 16(3): 156-167. |
| 31 | GOSSEN M, BUJARD H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters[J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(12): 5547-5551. |
| 32 | RAMOS J L, MARTÍNEZ-BUENO M, MOLINA-HENARES A J, et al. The TetR family of transcriptional repressors[J]. Microbiology and Molecular Biology Reviews, 2005, 69(2): 326-356. |
| 33 | CUTHBERTSON L, NODWELL J R. The TetR family of regulators[J]. Microbiology and Molecular Biology Reviews, 2013, 77(3): 440-475. |
| 34 | BHUKYA H, ANAND R. TetR regulators: a structural and functional perspective[J]. Journal of the Indian Institute of Science, 2017, 97(2): 245-259. |
| 35 | BERENS C, HILLEN W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes[J]. European Journal of Biochemistry, 2003, 270(15): 3109-3121. |
| 36 | BERTRAM R, HILLEN W. The application of Tet repressor in prokaryotic gene regulation and expression[J]. Microbial Biotechnology, 2008, 1(1): 2-16. |
| 37 | MOL A A, GROHER F, SCHREIBER B, et al. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module[J]. Nucleic Acids Research, 2019, 47(20): e132. |
| 38 | HE S F, ZHANG Z W, LU W Y. Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae [J]. Journal of Industrial Microbiology & Biotechnology, 2023, 50(1): kuac029. |
| 39 | SCHOLZ O, HENßLER E M, BAIL J, et al. Activity reversal of Tet repressor caused by single amino acid exchanges[J]. Molecular Microbiology, 2004, 53(3): 777-789. |
| 40 | SULLIVAN M J, CURSON A R J, SHEARER N, et al. Unusual regulation of a leaderless operon involved in the catabolism of dimethylsulfoniopropionate in Rhodobacter sphaeroides [J]. PLoS One, 2011, 6(1): e15972. |
| 41 | HARMON D E, RUIZ C. The multidrug efflux regulator AcrR of Escherichia coli responds to exogenous and endogenous ligands to regulate efflux and detoxification[J]. mSphere, 2022, 7(6): e00474-22. |
| 42 | GRKOVIC S, BROWN M H, SKURRAY R A. Transcriptional regulation of multidrug efflux pumps in bacteria[J]. Seminars in Cell & Developmental Biology, 2001, 12(3): 225-237. |
| 43 | ROGERS J K, GUZMAN C D, TAYLOR N D, et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites[J]. Nucleic Acids Research, 2015, 43(15): 7648-7660. |
| 44 | JONES K A, SNODGRASS H M, BELSARE K, et al. Phage-assisted continuous evolution and selection of enzymes for chemical synthesis[J]. ACS Central Science, 2021, 7(9): 1581-1590. |
| 45 | ROGERS J K, CHURCH G M. Genetically encoded sensors enable real-time observation of metabolite production[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(9): 2388-2393. |
| [1] | 杨璐, 吴楠, 白茸茸, 董维亮, 周杰, 姜岷. 基因回路型全细胞微生物传感器的设计、优化与应用[J]. 合成生物学, 2022, 3(6): 1061-1080. |
| [2] | 郭亮, 高聪, 柳亚迪, 陈修来, 刘立明. 大肠杆菌生产饲用氨基酸的研究进展[J]. 合成生物学, 2021, 2(6): 964-981. |
| [3] | 魏笑莲, 钱智玲, 陈巧巧, 于洪巍. 遗传改造微生物制造食品和饲料的监管要求及欧盟授权案例分析[J]. 合成生物学, 2021, 2(1): 121-133. |
| [4] | 储攀, 朱静雯, 黄文琦, 刘陈立, 傅雄飞. 底盘-回路耦合:合成基因回路设计新挑战[J]. 合成生物学, 2021, 2(1): 91-105. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
