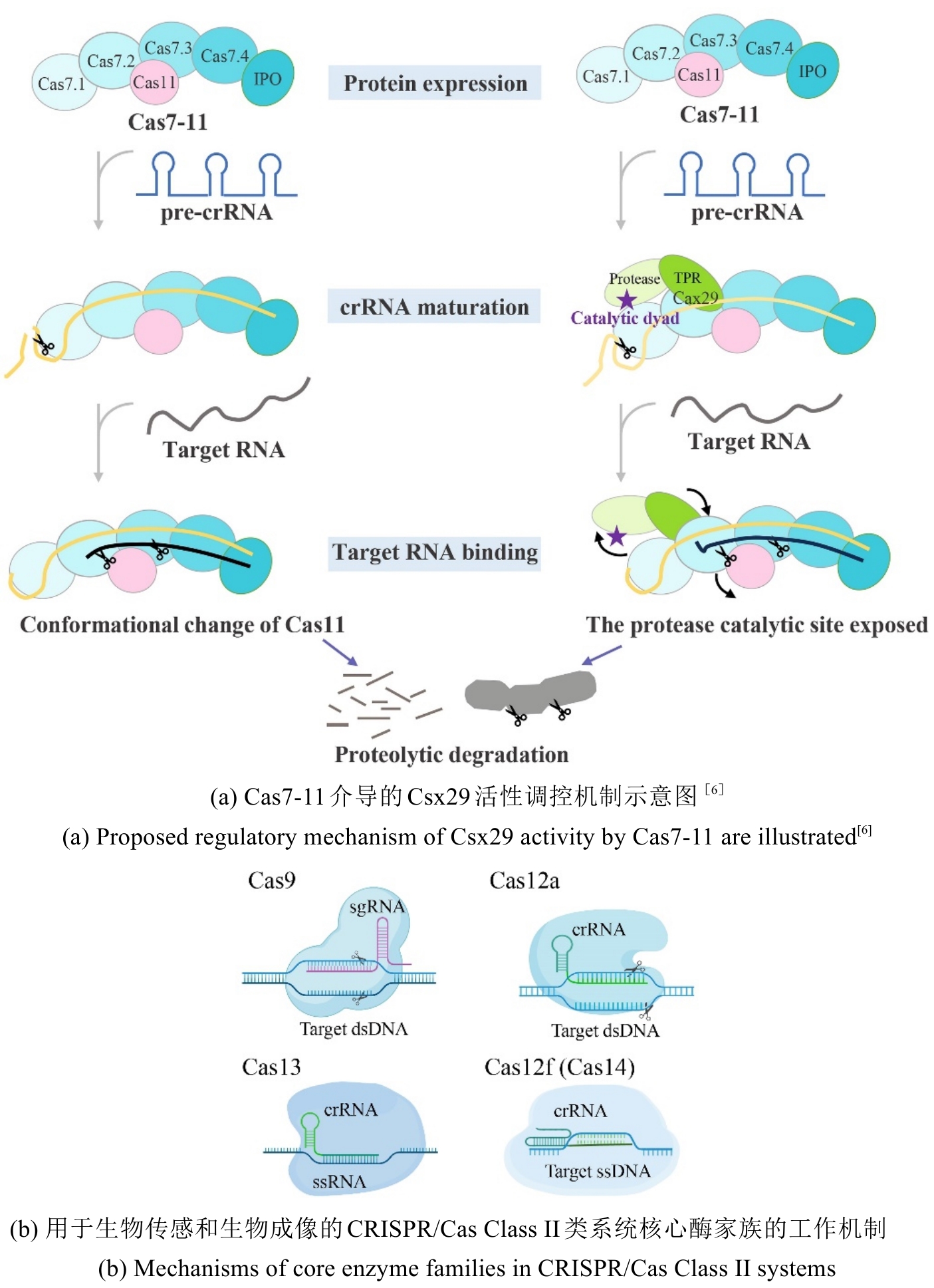
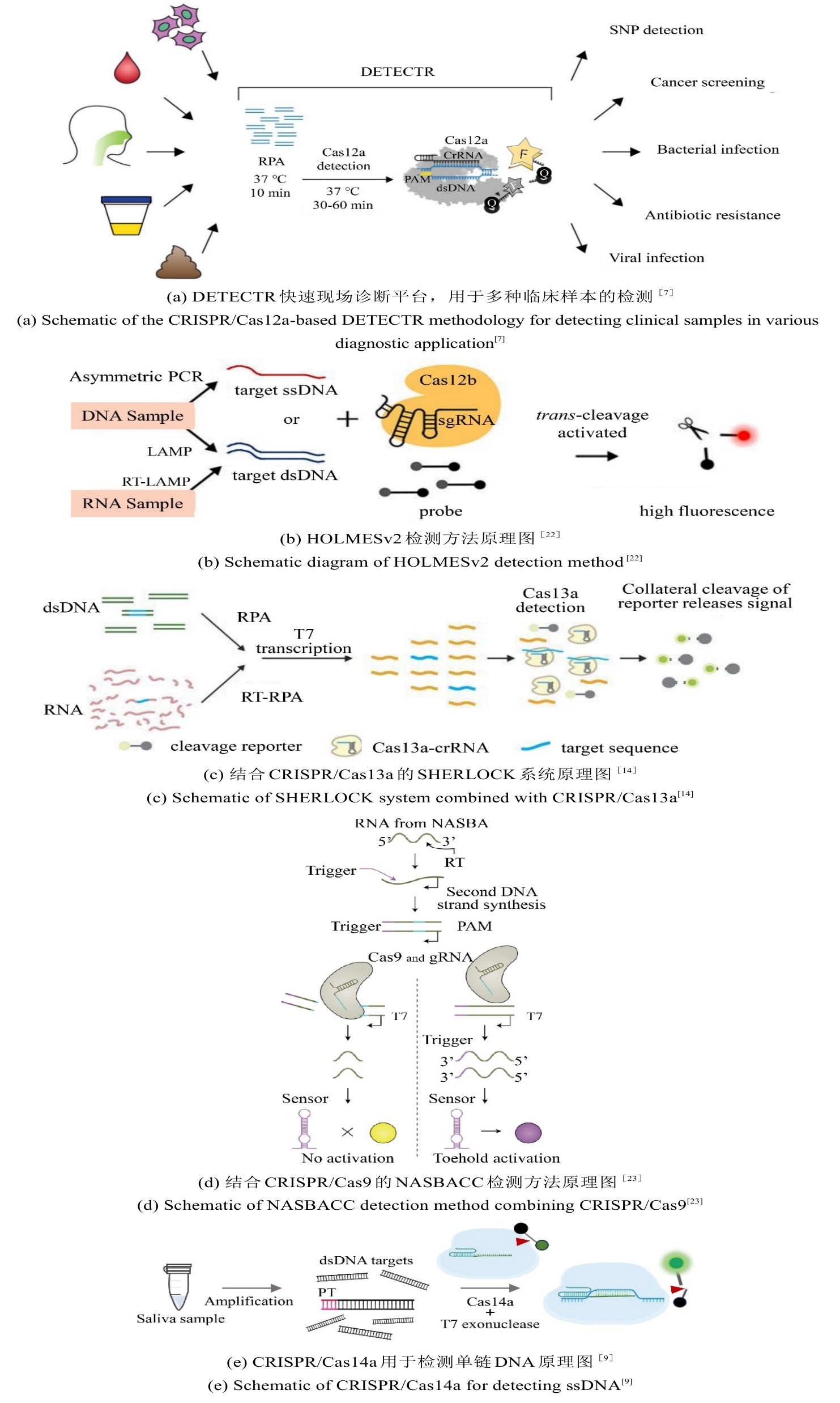
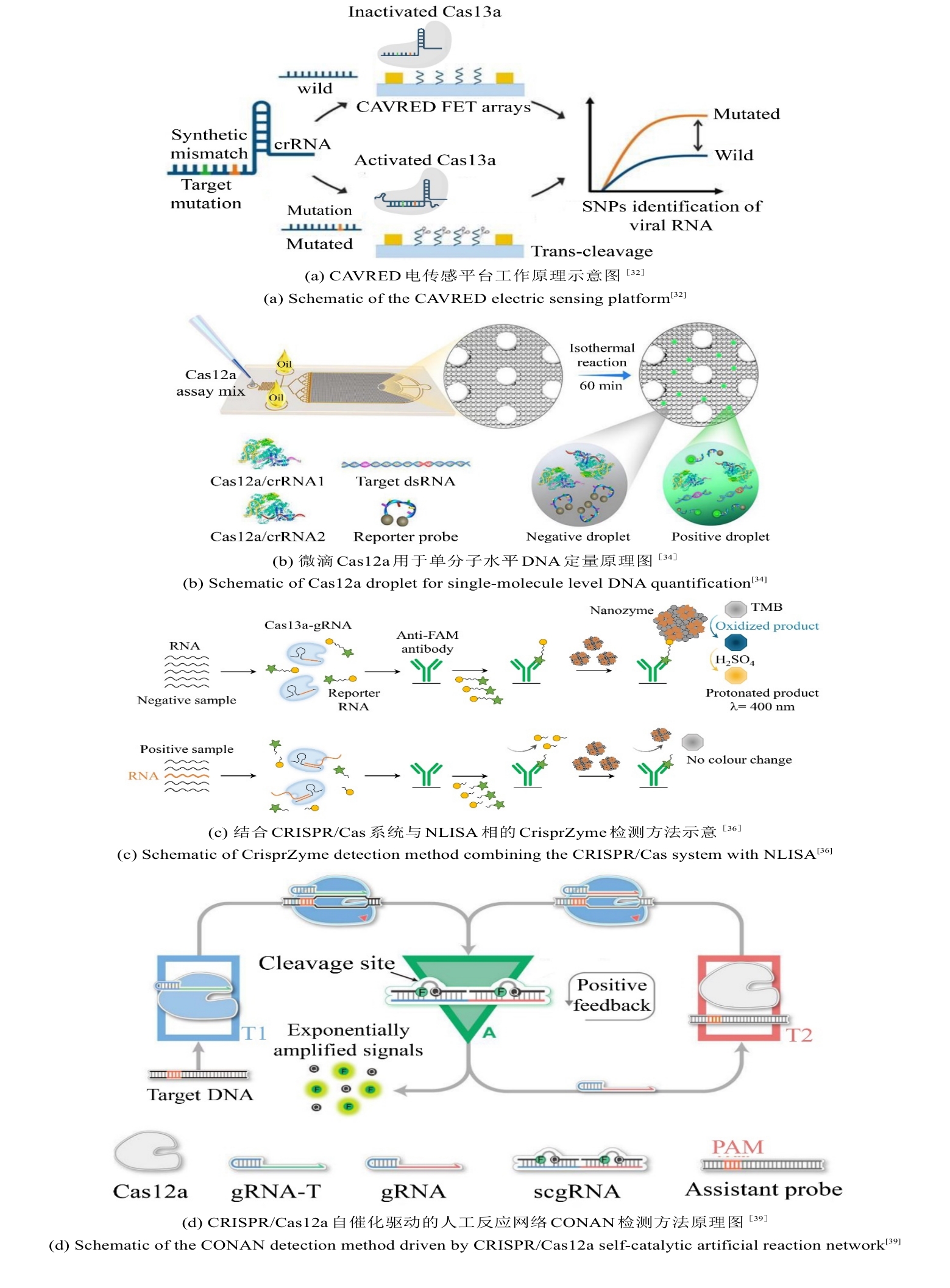
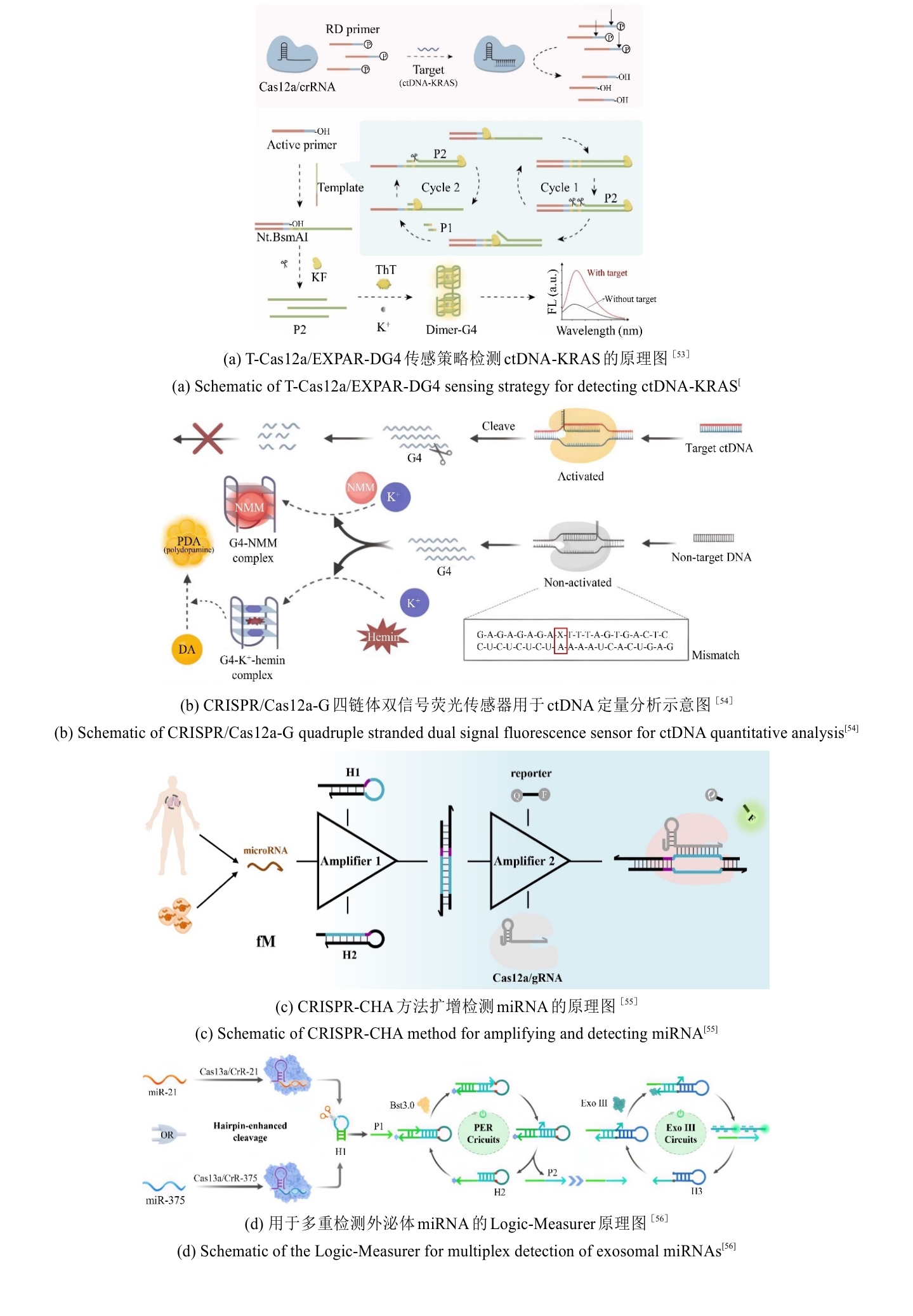
• 特约评述 •
CRISPR/Cas系统在分子诊断领域的应用研究进展
王珂1,2, 陈文慧1, 雷春阳1, 聂舟1
- 1.湖南大学化学生物传感全国重点实验室,化学化工学院,湖南 长沙 410012
2.空军军医大学药学系,陕西 西安 710032
-
收稿日期:2025-05-07修回日期:2025-06-18出版日期:2025-06-18 -
通讯作者:聂舟 -
作者简介:王珂 (1995—),女,博士,讲师。研究方向为基于CRISPR/Cas系统的可编程分子识别与信号放大机制,开发工程化高灵敏、高特异的生物传感平台,用于环境监测及疾病诊断等生命科学场景中的创新应用。E-mail:wangke123@fmmu.edu.cn聂舟 (1982—),男,博士,教授,博士生导师。研究方向:围绕功能核酸理性设计及其生物传感与成像、细胞信号通路调控应用等方面开展了深入研究,系统建立了基于物理/化学信号响应型核酸工具调控细胞受体信号通路的细胞功能重编程技术,以及RNA结构靶向活细胞/活体成像技术。E-mail:niezhou@hnu.edu.cn -
基金资助:国家自然科学基金(22034002)
Advances in the Application of CRISPR/Cas Systems in Molecular Diagnostics
WANG Ke1,2, CHEN Wenhui1, LEI Chunyang1, NIE Zhou1
- 1.State Key Laboratory of Chemo and Biosensing,College of Chemistry and Chemical Engineering,Hunan University,Changsha 410012,Hunan,China
2.Department of Pharmaceutical Analysis,School of Pharmacy,The Fourth Military Medical University,Xi’an 710032,Shaanxi,China
-
Received:2025-05-07Revised:2025-06-18Online:2025-06-18 -
Contact:NIE Zhou
摘要:
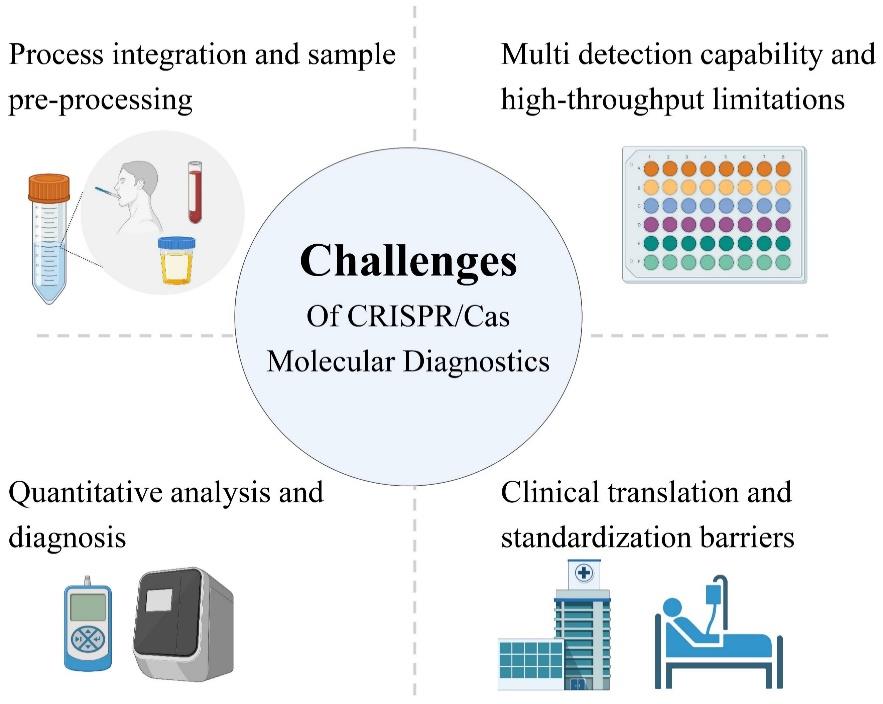
CRISPR/Cas系统因其高特异性、可编程性和便捷性,已成为分子诊断领域的重要工具。本文综述了CRISPR/Cas系统的技术原理、诊断平台优化及其在精准医学中的应用进展。首先,概述了CRISPR/Cas系统的作用机制与分类,并重点讨论了CRISPR诊断技术的创新优化策略,包括基于核酸预扩增(如SHERLOCK(specific high-sensitivity enzymatic reporter unlocking)、DETECTR(DNA endonuclease targeted CRISPR trans reporter))和免扩增的检测方法。其次,探讨了CRISPR/Cas技术在感染性疾病(病原体筛查、耐药性检测)、肿瘤分子分型(癌症早筛、遗传变异分析)及非核酸标志物检测中的临床应用。最后,本文展望了该技术的未来发展方向,包括微型化设备开发、高通量智能化诊断体系构等,并分析了其在临床转化中面临的关键挑战(如灵敏度标准化、成本控制等)。通过总结目前研究,本文旨在为CRISPR/Cas技术在分子诊断领域的进一步优化和医学应用提供理论参考。
中图分类号:
引用本文
王珂, 陈文慧, 雷春阳, 聂舟. CRISPR/Cas系统在分子诊断领域的应用研究进展[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-042.
WANG Ke, CHEN Wenhui, LEI Chunyang, NIE Zhou. Advances in the Application of CRISPR/Cas Systems in Molecular Diagnostics[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-042.
| Cas蛋白 | 大类 | 型 | 靶标 | 反式切割活性/底物 | PAM/PFS序列 |
|---|---|---|---|---|---|
| Cas9[ | Class II系统 | Type II | dsDNA | 无 | 3′ GC-rich PAM |
Cas7-11[ (多亚基复合物) | Class I系统 | Type III | ssRNA | 无 | --- |
| Cas12a (Cpf1)[ | Class II系统 | Type V | dsDNA/ssDNA | 有/ssDNA | 5′ AT-rich PAM |
| Cas12b[ | Class II系统 | Type V | dsDNA/ssDNA | 有/ssDNA | 5′ AT-rich PAM |
| Cas12f (Cas14)[ | Class II系统 | Type V | dsDNA/ssDNA | 有/ssDNA | 5′ T-rich PAM |
| Cas13[ | Class II系统 | Type VI | ssRNA | 有/ssRNA | 3′ non-G-PFS |
Cas3蛋白 (多亚基复合物)[ | Class I系统 | Type I | dsDNA | 有/ssDNA | AAG |
表1 CRISPR效应蛋白的相关特征
Table 1 The relation of CRISPR/Cas system effector proteins
| Cas蛋白 | 大类 | 型 | 靶标 | 反式切割活性/底物 | PAM/PFS序列 |
|---|---|---|---|---|---|
| Cas9[ | Class II系统 | Type II | dsDNA | 无 | 3′ GC-rich PAM |
Cas7-11[ (多亚基复合物) | Class I系统 | Type III | ssRNA | 无 | --- |
| Cas12a (Cpf1)[ | Class II系统 | Type V | dsDNA/ssDNA | 有/ssDNA | 5′ AT-rich PAM |
| Cas12b[ | Class II系统 | Type V | dsDNA/ssDNA | 有/ssDNA | 5′ AT-rich PAM |
| Cas12f (Cas14)[ | Class II系统 | Type V | dsDNA/ssDNA | 有/ssDNA | 5′ T-rich PAM |
| Cas13[ | Class II系统 | Type VI | ssRNA | 有/ssRNA | 3′ non-G-PFS |
Cas3蛋白 (多亚基复合物)[ | Class I系统 | Type I | dsDNA | 有/ssDNA | AAG |
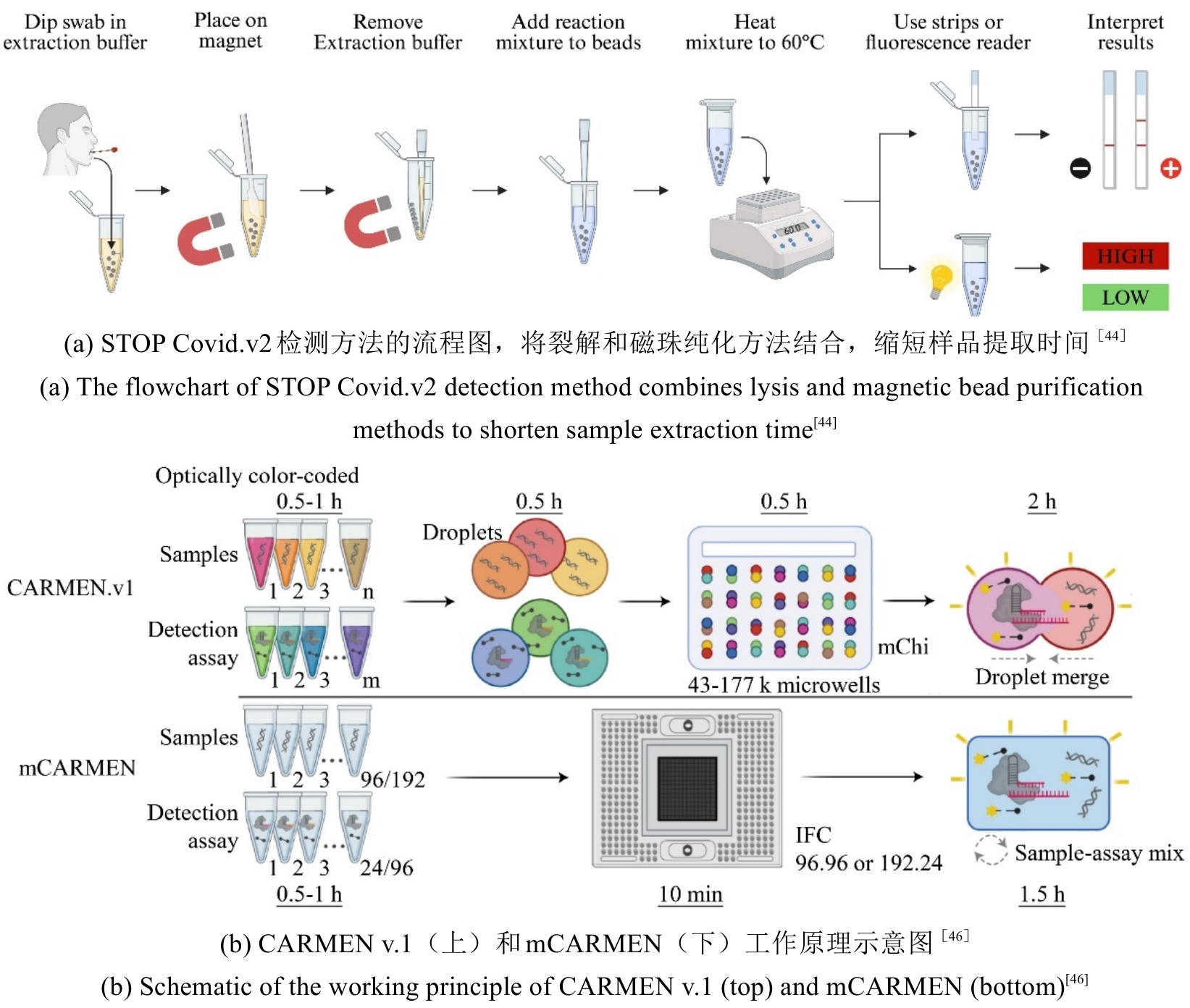
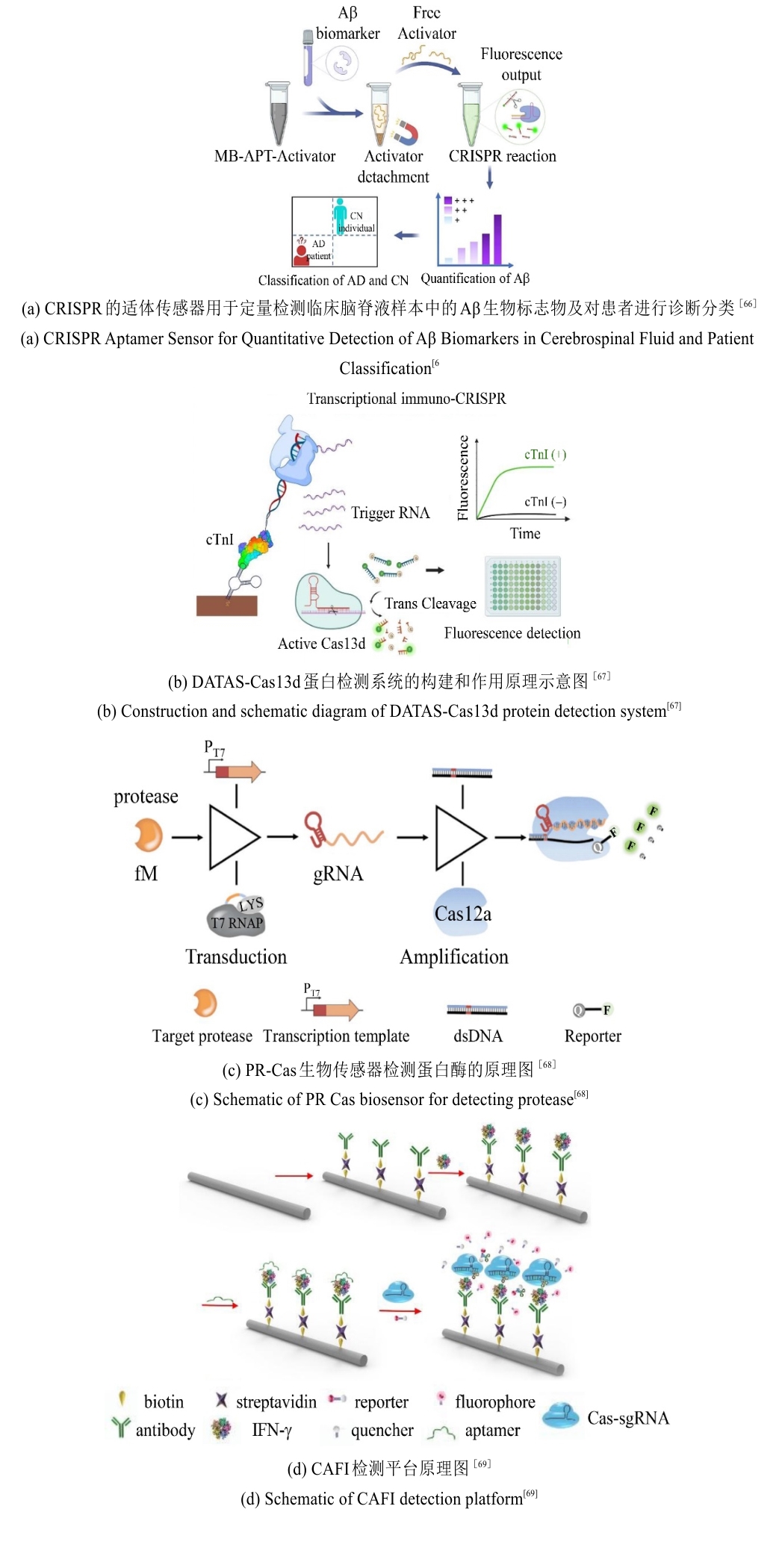
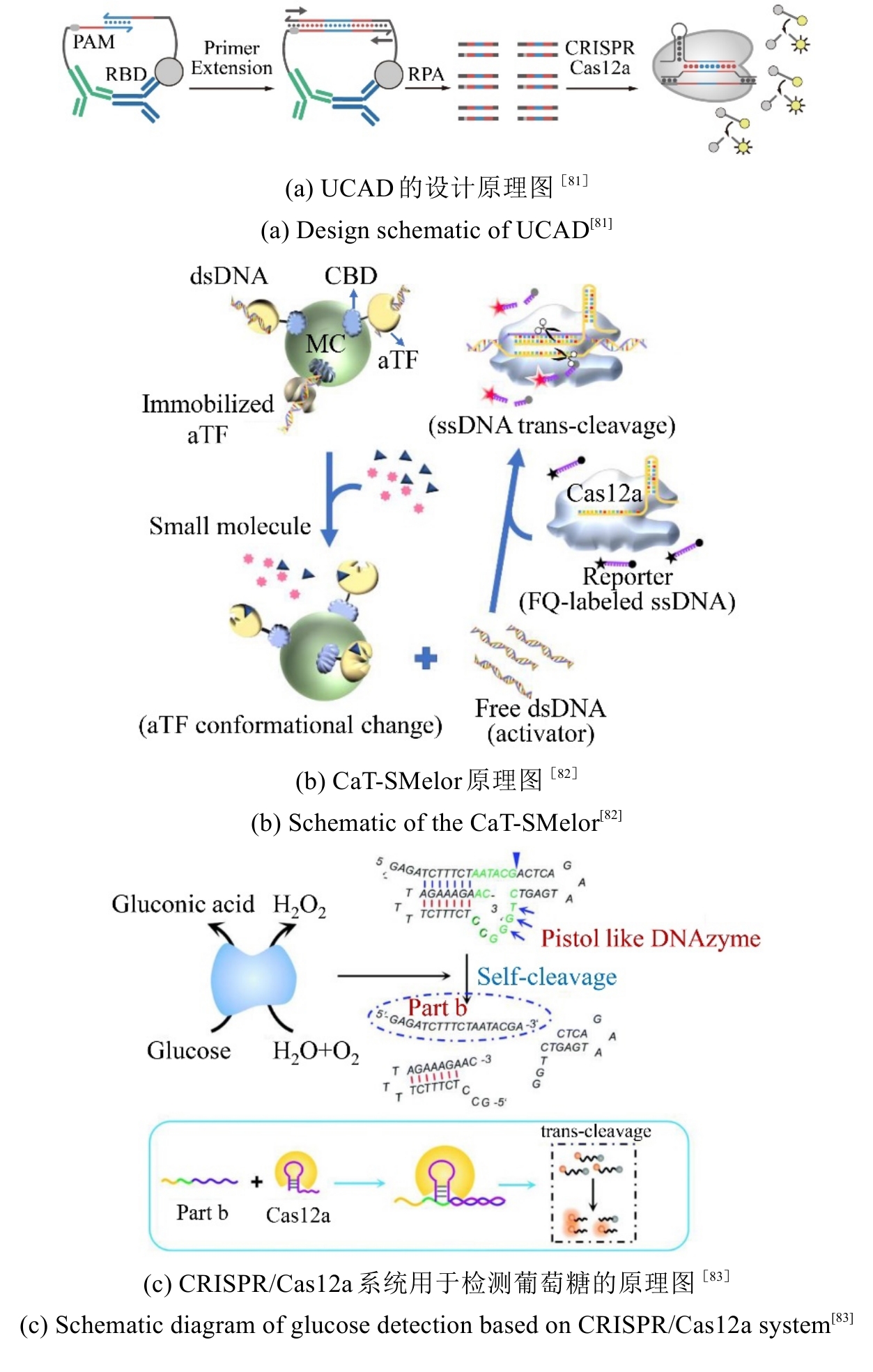
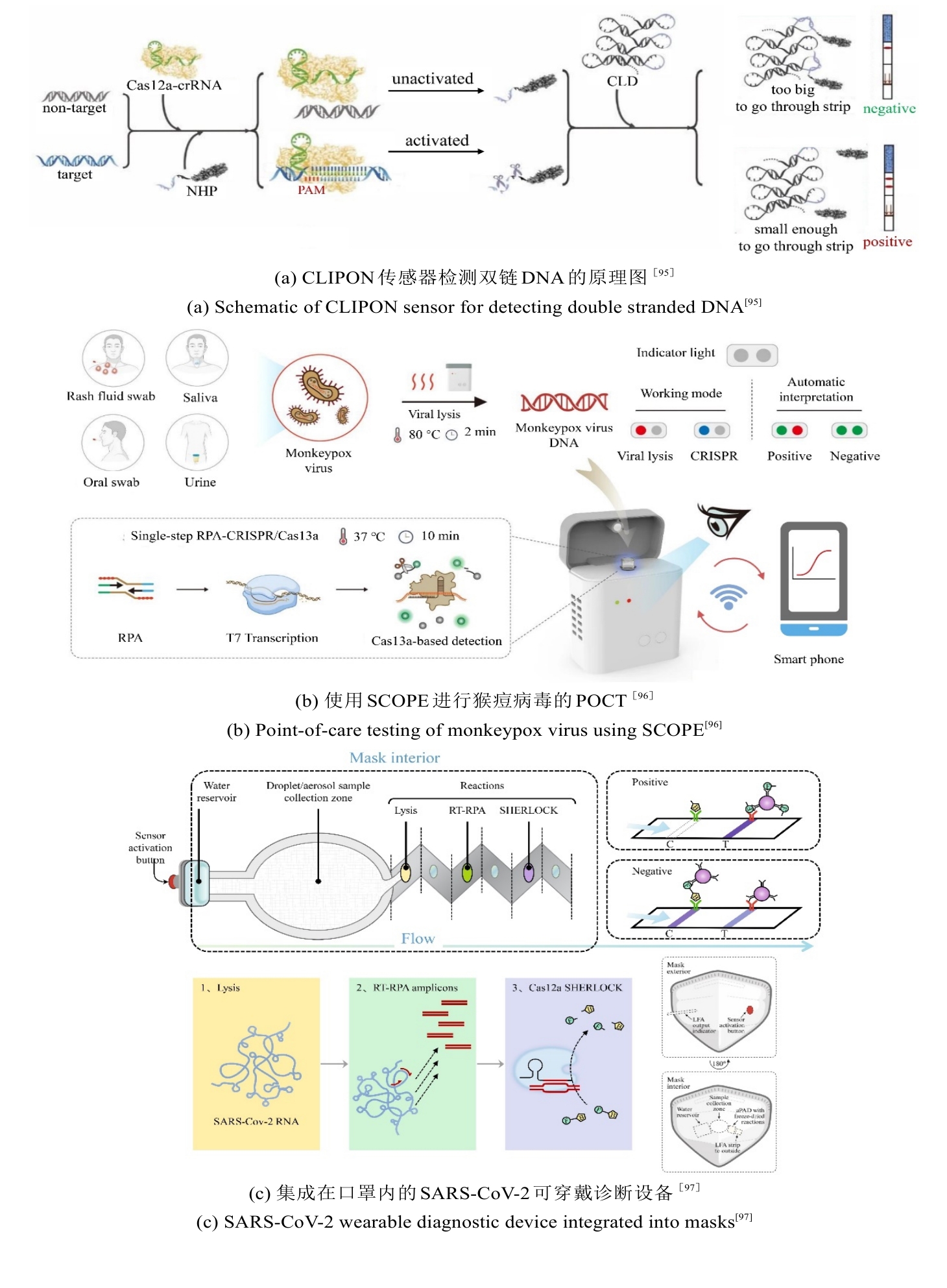
图4 CRISPR/Cas系统用于感染性疾病精准诊断的检测方法原理图
Fig. 4 Schematic diagram of detection methods using the CRISPR/Cas system for precise diagnosis of infectious diseases
| 1 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants [J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 2 | ABUDAYYEH O O, GOOTENBERG J S, ESSLETZBICHLER P, et al. RNA targeting with CRISPR-Cas13 [J]. Nature, 2017, 550(7675): 280-284. |
| 3 | ZHOU W, HU L, YING L, et al. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection [J]. Nature Communications, 2018, 9(1): 5012. |
| 4 | HU C, VAN BELJOUW S P B, NAM K H, et al. Craspase is a CRISPR RNA-guided, RNA-activated protease [J]. Science, 2022, 377(6612): 1278-1285. |
| 5 | STRECKER J, DEMIRCIOGLU F E, LI D, et al. RNA-activated protein cleavage with a CRISPR-associated endopeptidase [J]. Science, 2022, 378(6622): 874-881. |
| 6 | YU G, WANG X, ZHANG Y, et al. Structure and function of a bacterial type III-E CRISPR–Cas7-11 complex [J]. Nature Microbiology, 2022, 7(12): 2078-2088. |
| 7 | CHEN J S, MA E, HARRINGTON L B, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity [J]. Science, 2018, 360(6387): 436-439. |
| 8 | TONG X, ZHANG K, HAN Y, et al. Fast and sensitive CRISPR detection by minimized interference of target amplification [J]. Nature Chemical Biology, 2024, 20(7): 885-893. |
| 9 | HARRINGTON L B, BURSTEIN D, CHEN J S, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes [J]. Science, 2018, 362(6416): 839-842. |
| 10 | KARVELIS T, BIGELYTE G, YOUNG J K, et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage [J]. Nucleic Acids Research, 2020, 48(9): 5016-5023. |
| 11 | WU Z, ZHANG Y, YU H, et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease [J]. Nature Chemical Biology, 2021, 17(11): 1132-1138. |
| 12 | WANG Y, WANG Y, PAN D, et al. Guide RNA engineering enables efficient CRISPR editing with a miniature Syntrophomonas palmitatica Cas12f1 nuclease [J]. Cell Reports, 2022, 40(13):111418. |
| 13 | WANG Y, TANG N, JI Q. Systematic trans-Activity Comparison of Several Reported Cas12f Nucleases [J]. Chinese Journal of Chemistry, 2025, 43(12): 1339-1347. |
| 14 | GOOTENBERG J S, ABUDAYYEH O O, LEE J W, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2 [J]. Science, 2017, 356(6336): 438-442. |
| 15 | YOSHIMI K, TAKESHITA K, YAMAYOSHI S, et al. CRISPR-Cas3-based diagnostics for SARS-CoV-2 and influenza virus [J]. iScience, 2022, 25(2): 103830. |
| 16 | CHEN J, CHEN Y, HUANG L, et al. Trans-nuclease activity of Cas9 activated by DNA or RNA target binding [J]. Nature Biotechnology, 2025, 43(4): 558-568. |
| 17 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity [J]. Science, 2012, 337(6096): 816-821. |
| 18 | ZHAN T, RINDTORFF N, BETGE J, et al. CRISPR/Cas9 for cancer research and therapy [J], Seminars in Cancer Biology, 2019, 55: 106-119. |
| 19 | GOOTENBERG J S, ABUDAYYEH O O, KELLNER M J, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6 [J]. Science, 2018, 360(6387): 439-444. |
| 20 | KNOTT G J, EAST-SELETSKY A, COFSKY J C, et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme [J]. Nature Structural & Molecular Biology, 2017, 24(10): 825-833. |
| 21 | WANG B, ZHANG T, YIN J, et al. Structural basis for self-cleavage prevention by tag: anti-tag pairing complementarity in type VI Cas13 CRISPR systems [J]. Molecular Cell, 2021, 81(5): 1100-1115. e5. |
| 22 | LI L, LI S, WU N, et al. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation [J]. ACS Synthetic Biology, 2019, 8(10): 2228-2237. |
| 23 | PARDEE K, GREEN A A, TAKAHASHI M K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components [J]. Cell, 2016, 165(5): 1255-1266. |
| 24 | LI H, XIE Y, CHEN F, et al. Amplification-free CRISPR/Cas detection technology: challenges, strategies, and perspectives [J]. Chemical Society Reviews, 2023, 52(1): 361-382. |
| 25 | MYHRVOLD C, FREIJE C A, GOOTENBERG J S, et al. Field-deployable viral diagnostics using CRISPR-Cas13 [J]. Science, 2018, 360(6387): 444-448. |
| 26 | ZAGHLOUL H, EL-SHAHAT M J W J O H. Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis [J]. World Journal of Hepatology, 2014, 6(12): 916-922. |
| 27 | WAN Y, LI S, XU W, et al. Terminal chemical modifications of crRNAs enable improvement in the performance of CRISPR-Cas for point-of-care nucleic acid detection [J]. Analytical Chemistry, 2024, 96(41): 16346-16354. |
| 28 | YANG H, EREMEEVA E, ABRAMOV M, et al. CRISPR-Cas9 recognition of enzymatically synthesized base-modified nucleic acids [J]. Nucleic Acids Research, 2023, 51(4): 1501-1511. |
| 29 | HU M, QIU Z, BI Z, et al. Photocontrolled crRNA activation enables robust CRISPR-Cas12a diagnostics [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(26): e2202034119. |
| 30 | ROSSETTI M, MERLO R, BAGHERI N, et al. Enhancement of CRISPR/Cas12a trans-cleavage activity using hairpin DNA reporters [J]. Nucleic Acids Research, 2022, 50(14): 8377-8391. |
| 31 | HU M, LIU R, QIU Z, et al. Light‐start CRISPR‐Cas12a reaction with caged crRNA enables rapid and sensitive nucleic acid detection [J]. Angewandte Chemie International Edition, 2023, 62, e202300663. |
| 32 | CHEN D, HUANG W, ZHANG Y, et al. CRISPR-mediated profiling of viral RNA at single‐nucleotide resolution [J]. Angewandte Chemie International Edition, 2023, 62(30): e202304298. |
| 33 | TIAN T, SHU B, JIANG Y, et al. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics [J]. ACS Nano, 2020, 15(1): 1167-1178. |
| 34 | YUE H, SHU B, TIAN T, et al. Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level [J]. Nano Letters, 2021, 21(11): 4643-4653. |
| 35 | SHINODA H, IIDA T, MAKINO A, et al. Automated amplification-free digital RNA detection platform for rapid and sensitive SARS-CoV-2 diagnosis [J]. Communications Biology, 2022, 5(1): 473. |
| 36 | BROTO M, KAMINSKI M M, ADRIANUS C, et al. Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs [J]. Nature Nanotechnology, 2022, 17(10): 1120-1126. |
| 37 | LIU P, LIN Y, ZHUO X, et al. Universal crRNA Acylation Strategy for Robust Photo-Initiated One-Pot CRISPR-Cas12a Nucleic Acid Diagnostics [J]. Angewandte Chemie International Edition, 2024, 63(23): e202401486. |
| 38 | CHEN Y, XU X, WANG J, et al. Photoactivatable CRISPR/Cas12a Strategy for One-Pot DETECTR Molecular Diagnosis [J]. Analytical Chemistry, 2022, 94(27): 9724-9731. |
| 39 | SHI K, XIE S, TIAN R, et al. A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics [J]. Science Advances, 2021, 7(5): eabc7802 |
| 40 | SUN K, PU L, CHEN C, et al. An autocatalytic CRISPR-Cas amplification effect propelled by the LNA-modified split activators for DNA sensing [J]. Nucleic Acids Research, 2024, 52(7): e39-e39. |
| 41 | LIM J, VAN A B, KOPROWSKI K, et al. Amplification-free, OR-gated CRISPR-Cascade reaction for pathogen detection in blood samples [J]. Proceedings of the National Academy of Sciences of the United States of America, 2025, 122(11): e2420166122. |
| 42 | TANG Y, GAO L, FENG W, et al. The CRISPR-Cas toolbox for analytical and diagnostic assay development [J]. Chemical Society Reviews, 2021, 50(21): 11844-11869. |
| 43 | BROUGHTON J P, DENG X, YU G, et al. CRISPR-Cas12-based detection of SARS-CoV-2 [J]. Nature Biotechnology, 2020, 38(7): 870-874. |
| 44 | JOUNG J, LADHA A, SAITO M, et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing [J]. New England Journal of Medicine, 2020, 383(15): 1492-1494. |
| 45 | JOUNG J, LADHA A, SAITO M, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics [J]. MedRxiv, 2020. |
| 46 | WELCH N L, ZHU M, HUA C, et al. Multiplexed CRISPR-based microfluidic platform for clinical testing of respiratory viruses and identification of SARS-CoV-2 variants [J]. Nature Medicine, 2022, 28(5): 1083-1094. |
| 47 | YAN M-Y, ZHENG D, LI S-S, et al. Application of combined CRISPR screening for genetic and chemical-genetic interaction profiling in Mycobacterium tuberculosis[J]. Science Advances, 2022, 8(47): eadd5907. |
| 48 | CHEN W, LUO H, ZENG L, et al. A suite of PCR-LwCas13a assays for detection and genotyping of Treponema pallidum in clinical samples [J]. Nature Communications, 2022, 13(1): 4671. |
| 49 | ACKERMAN C M, MYHRVOLD C, THAKKU S G, et al. Massively multiplexed nucleic acid detection with Cas13 [J]. Nature, 2020, 582(7811): 277-282. |
| 50 | JAHR S, HENTZE H, ENGLISCH S, et al. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells1 [J]. Cancer Research, 2001, 61(4): 1659-1665. |
| 51 | UNDERHILL H R, KITZMAN J O, HELLWIG S, et al. Fragment Length of Circulating Tumor DNA [J]. PLoS Genetics, 2016, 12(7): e1006162. |
| 52 | MERKER J D, OXNARD G R, COMPTON C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review [J]. Archives of Pathology & Laboratory Medicine, 2018, 142(10): 1242-1253. |
| 53 | DONG J, LI X, DENG L, et al. CRISPR/Cas12a cleavage-mediated isothermal amplification lights up the dimeric G-quadruplex signal unit for ultrasensitive and label-free detection of circulating tumor DNA [J]. Sensors and Actuators B: Chemical, 2024, 404: 135292. |
| 54 | ZHAO X, LIU W, QU R, et al. Target-activated CRISPR/Cas12a recognize multifunctional G-quadruplex and dual fluorescent indicators enable rapid non-extraction analysis of circulating tumor DNA in breast cancer [J]. Sensors and Actuators B: Chemical, 2025, 430: 137372. |
| 55 | PENG S, TAN Z, CHEN S, et al. Integrating CRISPR-Cas12a with a DNA circuit as a generic sensing platform for amplified detection of microRNA [J]. Chemical Science, 2020, 11(28): 7362-7368. |
| 56 | XIE Z, ZHAO S, DENG R, et al. Logic-Measurer: A Multienzyme-Assisted Ultrasensitive Circuit for Logical Detection of Exosomal MicroRNAs [J]. ACS Nano, 2025, 19(12): 12222-12236. |
| 57 | KIM V N. MicroRNA biogenesis: coordinated cropping and dicing [J]. Nature Reviews Molecular Cell Biology, 2005, 6(5): 376-385. |
| 58 | CALURA E, FRUSCIO R, PARACCHINI L, et al. miRNA Landscape in Stage I Epithelial Ovarian Cancer Defines the Histotype Specificities [J]. Clinical Cancer Research, 2013, 19(15): 4114-4123. |
| 59 | ZHENG H, ZHANG L, ZHAO Y, et al. Plasma miRNAs as Diagnostic and Prognostic Biomarkers for Ovarian Cancer [J]. PLoS One, 2013, 8(11): e77853. |
| 60 | MILANEZ-ALMEIDA P, MARTINS A J, GERMAIN R N, et al. Cancer prognosis with shallow tumor RNA sequencing [J]. Nature Medicine, 2020, 26(2): 188-192. |
| 61 | TIAN B, MINERO GABRIEL ANTONIO S, FOCK J, et al. CRISPR-Cas12a based internal negative control for nonspecific products of exponential rolling circle amplification [J]. Nucleic Acids Research, 2020, 48(5): e30-e30. |
| 62 | XING S, LU Z, HUANG Q, et al. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification [J]. Theranostics, 2020, 10(22): 10262-10273. |
| 63 | ZHANG J, GUAN M, MA C, et al. Highly Effective Detection of Exosomal miRNAs in Plasma Using Liposome-Mediated Transfection CRISPR/Cas13a [J]. ACS Sensors, 2023, 8(2): 565-575. |
| 64 | YANG Q, DONG M-J, XU J, et al. CRISPR/RNA Aptamer System Activated by an AND Logic Gate for Biomarker-Driven Theranostics [J]. Journal of the American Chemical Society, 2025, 147(1): 169-180. |
| 65 | YAN H, WEN Y, TIAN Z, et al. A one-pot isothermal Cas12-based assay for the sensitive detection of microRNAs [J]. Nature Biomedical Engineering, 2023, 7(12): 1583-1601. |
| 66 | JIA Z, MAGHAYDAH Y, ZDANYS K, et al. CRISPR-Powered Aptasensor for Diagnostics of Alzheimer's Disease [J]. ACS Sensors, 2024, 9(1): 398-405. |
| 67 | FENG Z-Y, LIU R, LI X, et al. Harnessing the CRISPR-Cas13d System for Protein Detection by Dual-Aptamer-Based Transcription Amplification [J]. Chemistry-A European Journal, 2023, 29(10): e202202693. |
| 68 | LIU F, CHEN R, SONG W, et al. Modular Combination of Proteolysis-Responsive Transcription and Spherical Nucleic Acids for Smartphone-Based Colorimetric Detection of Protease Biomarkers [J]. Analytical Chemistry, 2021, 93(7): 3517-3525. |
| 69 | DENG F, LI Y, QIAO L, et al. A CRISPR/Cas12a-assisted on-fibre immunosensor for ultrasensitive small protein detection in complex biological samples [J]. Analytica Chimica Acta, 2022, 1192: 339351. |
| 70 | AINSWORTH M, ANDERSSON M, AUCKLAND K, et al. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison [J]. The Lancet Infectious diseases, 2020, 20(12): 1390-1400. |
| 71 | GEURTSVANKESSEL C H, OKBA N M A, IGLOI Z, et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment [J]. Nature Communications, 2020, 11(1): 3436. |
| 72 | RHOADS D D, CHERIAN S S, ROMAN K, et al. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA Emergency Use Authorization Methods for the Detection of SARS-CoV-2 from Nasopharyngeal and Nasal Swabs from Individuals Diagnosed with COVID-19 [J]. Journal of Clinical Microbiology, 2020, 58(8): e00760-20. |
| 73 | WHITMAN J D, HIATT J, MOWERY C T, et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance [J]. Nature Biotechnology, 2020, 38(10): 1174-1183. |
| 74 | ELLEDGE S K, ZHOU X X, BYRNES J R, et al. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection [J]. Nature Biotechnology, 2021, 39(8): 928-935. |
| 75 | YAO Z, DRECUN L, ABOUALIZADEH F, et al. A homogeneous split-luciferase assay for rapid and sensitive detection of anti-SARS CoV-2 antibodies [J]. Nature Communications, 2021, 12(1): 1806. |
| 76 | LONG Q-X, LIU B-Z, DENG H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19 [J]. Nature Medicine, 2020, 26(6): 845-848. |
| 77 | AGNOLON V, CONTATO A, MENEGHELLO A, et al. ELISA assay employing epitope-specific monoclonal antibodies to quantify circulating HER2 with potential application in monitoring cancer patients undergoing therapy with trastuzumab [J]. Scientific Reports, 2020, 10(1): 3016. |
| 78 | ZHANG W, RONG-HUI D, BEI L, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes [J]. Emerging Microbes & Infections, 2020, 9(1): 386-389. |
| 79 | ACHARYA A P, NAFISI P M, GARDNER A, et al. A fluorescent peroxidase probe increases the sensitivity of commercial ELISAs by two orders of magnitude [J]. Chemical Communications, 2013, 49(88): 10379-10381. |
| 80 | CARTER Q L, DOTZLAF J, SWEARINGEN C, et al. Development and characterization of a novel ELISA based assay for the quantitation of sub-nanomolar levels of neoepitope exposed NITEGE-containing aggrecan fragments [J]. Journal of Immunological Methods, 2007, 328(1): 162-168. |
| 81 | TANG Y, SONG T, GAO L, et al. A CRISPR-based ultrasensitive assay detects attomolar concentrations of SARS-CoV-2 antibodies in clinical samples [J]. Nature Communications, 2022, 13(1): 4667. |
| 82 | LIANG M, LI Z, WANG W, et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules [J]. Nature Communications, 2019, 10(1): 3672. |
| 83 | ZHANG C, YAO H, MA Q, et al. Ultrasensitive glucose detection from tears and saliva through integrating a glucose oxidase-coupled DNAzyme and CRISPR-Cas12a [J]. Analyst, 2021, 146(21): 6576-6581. |
| 84 | WHEATLEY M S, WANG Q, WEI W, et al. Cas12a-Based Diagnostics for Potato Purple Top Disease Complex Associated with Infection by ‘Candidatus Phytoplasma trifolii’-Related Strains [J]. Plant Disease, 2022, 106(8): 2039-2045. |
| 85 | AMAN R, MAHAS A, MARSIC T, et al. Efficient, rapid, and sensitive detection of plant RNA viruses with one-pot RT-RPA-CRISPR/Cas12a assay [J]. Frontiers in Microbiology, 2020, 11: 610872. |
| 86 | MARQUéS M-C, SáNCHEZ-VICENTE J, RUIZ R, et al. Diagnostics of Infections Produced by the Plant Viruses TMV, TEV, and PVX with CRISPR-Cas12 and CRISPR-Cas13 [J]. ACS Synthetic Biology, 2022, 11(7): 2384-2393. |
| 87 | MAHAS A, HASSAN N, AMAN R, et al. LAMP-Coupled CRISPR-Cas12a Module for Rapid and Sensitive Detection of Plant DNA Viruses [J]. Viruses, 2021, 13(3): 466. |
| 88 | RAMACHANDRAN V, WEILAND J J, BOLTON M D J F I M. CRISPR-based isothermal next-generation diagnostic method for virus detection in sugarbeet [J]. Frontiers in Microbiology, 2021, 12: 679994. |
| 89 | KANG H, PENG Y, HUA K, et al. Rapid Detection of Wheat Blast Pathogen Magnaporthe oryzae Triticum Pathotype Using Genome-Specific Primers and Cas12a-mediated Technology [J]. Engineering, 2021, 7(9): 1326-1335. |
| 90 | GONG X-Y, WANG Z-H, BASHIR M, et al. Recent application of CRISPR/Cas in plant disease detection [J]. TrAC Trends in Analytical Chemistry, 2025, 189: 118251. |
| 91 | LIU G. Advancing CRISPR/Cas Biosensing with Integrated Devices [J]. ACS Sensors, 2025, 10(2): 575-576. |
| 92 | LI X, WANG T, LIU X, et al. Advances of engineered microfluidic biosensors via CRISPR/Cas in bacteria and virus monitoring [J]. Chemical Engineering Journal, 2024, 491: 152038. |
| 93 | LI Z, UNO N, DING X, et al. Bioinspired CRISPR-Mediated Cascade Reaction Biosensor for Molecular Detection of HIV Using a Glucose Meter [J]. ACS Nano, 2023, 17(4): 3966-3975. |
| 94 | GE H, FENG J, HUANG L, et al. Development of a highly sensitive, high-throughput and automated CRISPR-based device for the contamination-free pathogen detection [J]. Biosensors and Bioelectronics, 2025, 278: 117323. |
| 95 | TANG Y, QI L, LIU Y, et al. CLIPON: a CRISPR-enabled strategy that turns commercial pregnancy test strips into general Point-of-Need test devices [J]. Angewandte Chemie International Edition, 2022, 61, e202115907. |
| 96 | WANG Y, CHEN H, LIN K, et al. Ultrasensitive single-step CRISPR detection of monkeypox virus in minutes with a vest-pocket diagnostic device [J]. Nature Communications., 2024, 15(1): 3279. |
| 97 | NGUYEN P Q, SOENKSEN L R, DONGHIA N M, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection [J]. Nature Biotechnology, 2021, 39(11): 1366-1374. |
| 98 | JI M, XIA Y, LOO J, et al. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform [J]. RSC Advances, 2020, 10(56): 34088-34098. |
| 99 | LIN Z, ZOU Z, PU Z, et al. Application of microfluidic technologies on COVID-19 diagnosis and drug discovery [J]. Acta Pharmaceutica Sinica B, 2023, 13(7): 2877-2896. |
| 100 | CHEN Y, ZONG N, YE F, et al. Dual-CRISPR/Cas12a-Assisted RT-RAA for Ultrasensitive SARS-CoV-2 Detection on Automated Centrifugal Microfluidics [J]. Analytical Chemistry, 2022, 94(27): 9603-9609. |
| 101 | LIM J, AHN J W, MAENG I, et al. TwinDemic detection: A non-enzymatic signal amplification system for on-site detection of multiple respiratory viruses [J]. Sensors and Actuators B: Chemical, 2025, 424: 136933. |
| 102 | NGUYEN H Q, NGUYEN V D, PHAN V M, et al. Development of a self-contained microfluidic chip and an internet-of-things-based point-of-care device for automated identification of respiratory viruses [J]. Lab on a Chip, 2024, 24(9): 2485-2496. |
| 103 | NGUYEN P Q M, WANG M, MARIA N ANN, et al. Modular micro-PCR system for the onsite rapid diagnosis of COVID-19 [J]. Microsystems & Nanoengineering, 2022, 8(1): 82. |
| 104 | SHAN X, GONG F, YANG Y, et al. Nucleic Acid Amplification-Free Digital Detection Method for SARS-CoV-2 RNA Based on Droplet Microfluidics and CRISPR-Cas13a [J]. Analytical Chemistry, 2023, 95(45): 16489-16495. |
| 105 | PENG R, LU Z, LIU M, et al. RT-RPA-assisted CRISPR/Cas12a for rapid and multiplex detection of respiratory infectious viruses based on centrifugal microfluidics [J]. Sensors and Actuators B: Chemical, 2024, 399: 134838. |
| 106 | CHEN J, YANG D, JI D, et al. A Fully Automated Point-of-Care Device Using Organic Electrochemical Transistor-Enhanced CRISPR/Cas12a for Amplification-Free Nucleic Acid Detection [J]. Advanced Functional Materials, 2025: 2420701. |
| 107 | XU J, SUO W, GOULEV Y, et al. Handheld Microfluidic Filtration Platform Enables Rapid, Low-Cost, and Robust Self-Testing of SARS-CoV-2 Virus [J]. Small, 2021, 17(52): e2104009. |
| 108 | WANG D, WANG X, YE F, et al. An Integrated Amplification-Free Digital CRISPR/Cas-Assisted Assay for Single Molecule Detection of RNA [J]. ACS Nano, 2023, 17(8): 7250-7256. |
| 109 | YIN B, WAN X, SOHAN A S M M F, et al. Microfluidics-Based POCT for SARS-CoV-2 Diagnostics [J]. Micromachines, 2022, 13(8): 1238. |
| 110 | LI Z, DING X, YIN K, et al. Instrument-free, CRISPR-based diagnostics of SARS-CoV-2 using self-contained microfluidic system [J]. Biosensors and Bioelectronics, 2022, 199: 113865. |
| 111 | FU Q, TU Y, CHENG L, et al. A fully-enclosed prototype ‘pen’ for rapid detection of SARS-CoV-2 based on RT-RPA with dipstick assay at point-of-care testing [J]. Sensors and Actuators B: Chemical, 2023, 383: 133531. |
| 112 | XIAO Y, ZHOU M, LIU C, et al. Fully integrated and automated centrifugal microfluidic chip for point-of-care multiplexed molecular diagnostics [J]. Biosensors and Bioelectronics, 2024, 255: 116240. |
| 113 | LI P, XIONG H, YANG B, et al. Recent progress in CRISPR-based microfluidic assays and applications [J]. TrAC Trends in Analytical Chemistry, 2022, 157: 116812. |
| 114 | CUI J Q, LIU F X, PARK H, et al. Droplet digital recombinase polymerase amplification (ddRPA) reaction unlocking via picoinjection [J]. Biosensors and Bioelectronics, 2022, 202: 114019. |
| 115 | LIU F X, CUI J Q, PARK H, et al. Isothermal Background-Free Nucleic Acid Quantification by a One-Pot Cas13a Assay Using Droplet Microfluidics [J]. Analytical Chemistry, 2022, 94(15): 5883-5892. |
| 116 | ZHANG L, WANG H, YANG S, et al. High-Throughput and Integrated CRISPR/Cas12a-Based Molecular Diagnosis Using a Deep Learning Enabled Microfluidic System [J]. ACS Nano, 2024, 18(35): 24236-24251. |
| 117 | LI X, LIU M, MEN D, et al. Rapid, portable, and sensitive detection of CaMV35S by RPA-CRISPR/Cas12a-G4 colorimetric assays with high accuracy deep learning object recognition and classification [J]. Talanta, 2024, 278: 126441. |
| 118 | ZHAO J, KONG D, ZHANG G, et al. An Efficient CRISPR/Cas Cooperative Shearing Platform for Clinical Diagnostics Applications [J]. Angewandte Chemie International Edition, 2024, 63(52): e202411705. |
| 119 | HUANG B, GUO L, YIN H, et al. Deep learning enhancing guide RNA design for CRISPR/Cas12a-based diagnostics [J]. Imeta, 2024, 3(4): e214. |
| 120 | H-H WESSELS, STIRN A, MéNDEZ-MANCILLA A, et al. Prediction of on-target and off-target activity of CRISPR-Cas13d guide RNAs using deep learning [J]. Nature Biotechnology, 2024, 42(4): 628-637. |
| 121 | XU T, ZHANG Y, LI S, et al. Deep Learning-Enhanced Hand-Driven Microfluidic Chip for Multiplexed Nucleic Acid Detection Based on RPA/CRISPR [J]. Advanced Science, 2025, 2414918. |
| 122 | LI Z, ZHAO W, MA S, et al. A chemical-enhanced system for CRISPR-Based nucleic acid detection [J]. Biosensors and Bioelectronics, 2021, 192: 113493. |
| 123 | MANTENA S, PILLAI P P, PETROS B A, et al. Model-directed generation of artificial CRISPR-Cas13a guide RNA sequences improves nucleic acid detection [J]. Nature Biotechnology, 2024, doi: 10.1038/s41587-024-02422-w . |
| 124 | HUANG Z, LYON C J, WANG J, et al. CRISPR Assays for Disease Diagnosis: Progress to and Barriers Remaining for Clinical Applications [J]. Advanced Science, 2023, 10(20): 2301697. |
| 125 | WENG Z, YOU Z, YANG J, et al. CRISPR-Cas Biochemistry and CRISPR-Based Molecular Diagnostics [J]. Angewandte Chemie International Edition, 2023, 62(17): e202214987. |
| 126 | ZHOU T, SHEN G, ZHONG L, et al. crRNA array-mediated CRISPR/Cas12a coupling with dual RPA for highly sensitive detection of Streptomyces aureofaciens Tü117 from hypertension with multi-signal output [J]. Biosensors and Bioelectronics, 2025, 282: 117493. |
| 127 | MAO Z, CHEN R, HUANG L, et al. CRISPR analysis based on Pt@MOF dual-modal signal for multichannel fluorescence and visual detection of norovirus [J]. Biosensors and Bioelectronics, 2025, 273: 117153. |
| 128 | HU M, YUAN C, TIAN T, et al. Single-Step, Salt-Aging-Free, and Thiol-Free Freezing Construction of AuNP-Based Bioprobes for Advancing CRISPR-Based Diagnostics [J]. Journal of the American Chemical Society, 2020, 142(16): 7506-7513. |
| 129 | XIONG E, JIANG L, TIAN T, et al. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay [J]. Angewandte Chemie International Edition, 2021, 60(10): 5307-5315. |
| 130 | ABUDAYYEH O O, GOOTENBERG J S. CRISPR diagnostics [J]. Science, 2021, 372(6545): 914-915. |
| 131 | ZUO X, FAN C, CHEN H-Y. Biosensing: CRISPR-powered diagnostics [J]. Nature Biomedical Engineering, 2017, 1(6): 0091. |
| 132 | 杜瑶, 高宏丹, 刘家坤 等. CRISPR-Cas系统在病原核酸检测中的研究进展[J]. 合成生物学, 2024, 5(1): 202-216. |
| DU Y, GAO H D, LIU J K, L, et al. Research progress of the CRISPR-Cas system in the detecting pathogen nucleic acids [J]. Synthetic Biology Journal, 2024, 5(1): 202-216. | |
| 133 | 李金成, 肖美娇, 陈渝萍. 基于CRISPR/Cas的体外核酸检测体系的研究进展 [J]. 中国生物工程杂志, 2024,44(9): 73-87. |
| LI J C, XIAO M J, CHEN Y P. Progress in in Vitro CRISPR/Cas-based Nucleic Acid Detection [J]. China Biotechnology, 2024, 44(9): 73-87. | |
| 134 | PACESA M, PELEA O, JINEK M. Past, present, and future of CRISPR genome editing technologies [J]. Cell, 2024, 187(5): 1076-1100. |
| 135 | TENG F, GUO L, CUI T, et al. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity [J]. Genome Biology, 2019, 20(1): 132. |
| 136 | CAO L, WANG Z, LEI C, et al. Engineered CRISPR/Cas Ribonucleoproteins for Enhanced Biosensing and Bioimaging [J]. Analytical Chemistry, 2025, 97(11): 5866-5879. |
| 137 | GUAN X, YANG R, ZHANG J, et al. Programmable Multiplexed Nucleic Acid Detection by Harnessing Specificity Defect of CRISPR-Cas12a [J]. Advanced Science, 2025, 12(4): 2411021. |
| 138 | MOLINA VARGAS ADRIAN M, SINHA S, OSBORN R, et al. New design strategies for ultra-specific CRISPR-Cas13a-based RNA detection with single-nucleotide mismatch sensitivity [J]. Nucleic Acids Research, 2023, 52(2): 921-939. |
| 139 | YE X, WU H, LIU J, et al. One-pot diagnostic methods based on CRISPR/Cas and Argonaute nucleases: strategies and perspectives [J]. Trends in Biotechnology, 2024, 42(11): 1410-1426. |
| 140 | DING X, YIN K, LI Z, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay [J]. Nature Communications, 2020, 11(1): 4711. |
| [1] | 林继聪, 邹根, 刘宏民, 魏勇军. CRISPR/Cas基因组编辑技术在丝状真菌次级代谢产物合成中的应用[J]. 合成生物学, 2023, 4(4): 738-755. |
| [2] | 龚仕涛, 王宇, 陈宇庭. CRISPR/Cas9及其衍生编辑器在衰老研究中的应用进展[J]. 合成生物学, 2022, 3(1): 66-77. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||