合成生物学 ›› 2022, Vol. 3 ›› Issue (6): 1235-1249.DOI: 10.12211/2096-8280.2022-028
非特异性过加氧酶(UPO)的研究综述
赖铭元, 韦健, 许建和, 郁惠蕾
- 华东理工大学,生物反应器工程国家重点实验室,上海 200237
-
收稿日期:2022-05-16修回日期:2022-07-07出版日期:2022-12-31发布日期:2023-01-17 -
通讯作者:郁惠蕾 -
作者简介:赖铭元 (1997—),男,博士研究生。研究方向为新酶挖掘与分子改造E-mail:y30200594@mail.ecust.edu.cn郁惠蕾 (1980—),女,博士,教授。研究方向为:(1)新酶基因挖掘和理性设计方法开;(2)新型萜类羟化酶的发现和分子改造;(3)非天然硫醚单加氧酶的进化和应用;(4)系统生物催化合成重要精细化学品E-mail:huileiyu@ecust.edu.cn -
基金资助:国家重点研发计划(2019YFA0905000)
Review of research on unspecific peroxygenases (UPOs)
LAI Mingyuan, WEI Jian, XU Jianhe, YU Huilei
- State Key Laboratory of Bioreactor Engineering,East China University of Science and Technology,Shanghai 200237,China
-
Received:2022-05-16Revised:2022-07-07Online:2022-12-31Published:2023-01-17 -
Contact:YU Huilei
摘要:
未活化的C—H键选择性插入活性氧是目前有机合成面临的最具挑战性之一。真菌非特异性过加氧酶(UPO)是一类高度糖基化的硫代血红素酶,催化的反应包括正构烷烃中未活化的C—H键的羟基化、烯烃和芳烃的环氧化、含杂原子(N、S)化合物的氧化、乙醚裂解、N-脱烷基化、脱酰化和酚类的单电子氧化。UPO以H2O2为氧供体与电子受体,不需要任何辅因子,是目前最具发展潜力的氧化酶之一。然而,UPO的异源表达困难与选择性差的问题仍限制着UPO的发展。近两年,通过信号肽改造或更换的方法在UPO的异源表达方面取得了重要突破,对UPO结构功能关系的深入研究以及蛋白结构预测算法的发展也将助力UPO的分子改造,为解决UPO选择性差的问题奠定基础。本文聚焦于UPO的异源表达、选择性问题与H2O2原位再生,综述了UPO的最新发展以及存在的技术瓶颈,并对解决这些瓶颈问题的方案做出展望。
中图分类号:
引用本文
赖铭元, 韦健, 许建和, 郁惠蕾. 非特异性过加氧酶(UPO)的研究综述[J]. 合成生物学, 2022, 3(6): 1235-1249.
LAI Mingyuan, WEI Jian, XU Jianhe, YU Huilei. Review of research on unspecific peroxygenases (UPOs)[J]. Synthetic Biology Journal, 2022, 3(6): 1235-1249.
| 名称 | 编号 | 辅因子 | 电子受体 | 催化作用 |
|---|---|---|---|---|
| 氧化酶 | EC 1.x.3.x | 金属离子(Cu等) | O2 | 将电子从底物或辅因子转移到分子氧上 |
| 黄素 | O2、Fe3+、醌-等 | |||
| 过氧化物酶 | EC 1.11.1.x | 血红素 | H2O2 | 催化两个电子从底物转移到H2O2 |
| 过加氧酶 | EC 1.11.2.1 | 血红素 | H2O2 | 催化氧原子从H2O2转移到底物上 |
| 单加氧酶 | EC 1.13.12.x | 血红素、黄素等 | O2 | 催化1个氧原子从O2转移到底物中 |
| 双加氧酶 | EC 1.13.11.x EC 1.14.11.x EC 1.14.12.x | 黄素、金属离子等 | O2 | 催化2个氧原子从O2转移到底物中 |
表1 氧化还原酶分类
Tab. 1 Classification of oxidoreductases (according to the webpage Enzyme Nomenclature of the NC-IUBMB https://iubmb.qmul.ac.uk/enzyme/EC1/)
| 名称 | 编号 | 辅因子 | 电子受体 | 催化作用 |
|---|---|---|---|---|
| 氧化酶 | EC 1.x.3.x | 金属离子(Cu等) | O2 | 将电子从底物或辅因子转移到分子氧上 |
| 黄素 | O2、Fe3+、醌-等 | |||
| 过氧化物酶 | EC 1.11.1.x | 血红素 | H2O2 | 催化两个电子从底物转移到H2O2 |
| 过加氧酶 | EC 1.11.2.1 | 血红素 | H2O2 | 催化氧原子从H2O2转移到底物上 |
| 单加氧酶 | EC 1.13.12.x | 血红素、黄素等 | O2 | 催化1个氧原子从O2转移到底物中 |
| 双加氧酶 | EC 1.13.11.x EC 1.14.11.x EC 1.14.12.x | 黄素、金属离子等 | O2 | 催化2个氧原子从O2转移到底物中 |
| UPO | 来源 | 异源表达宿主 | 方法 |
|---|---|---|---|
| MroUPO | M. rotula | E. coli | 原序列,自诱导培养基[ |
| DcaUPO | D. caldariorum | E. coli | 原序列[ |
| CviUPO | C. virescens | E. coli | 原序列[ |
| HinUPO | H. insolens | Aspergillus oryzae | 原序列[ |
| CciUPO | C. cinerea | Aspergillus oryzae | 原序列[ |
| PviUPO | P.virgatula | Aspergillus oryzae | 原序列[ |
| ThyUPO | T.hyrcaniae | Aspergillus oryzae | 原序列[ |
| LfuUPO | C. fumago | Aspergillus niger | 原序列[ |
| AaeUPO (wild-type) | A. aegerita | 无细胞 | 无细胞蛋白表达系统[ |
| AaeUPO (PADAⅠ) | A. aegerita | S. cerevisiae & P. pastoris | 信号肽改造[ |
| MroUPO | M. rotula | S. cerevisiae & P. pastoris | 更换信号肽[ |
| CglUPO | C. globosum | S. cerevisiae & P. pastoris | 更换信号肽[ |
| MthUPO | M. thermophila | S. cerevisiae & P. pastoris | 更换信号肽[ |
| TteUPO | T. terrestris | S. cerevisiae & P. pastoris | 更换信号肽[ |
| PabUPO | P. aberdarensis | S. Cerevisiae & P. pastoris | 更换信号肽[ |
| MfeUPO | M. fergusii | P. pastoris | 更换信号肽[ |
| MhiUPO | M. hinnulea | P. pastoris | 更换信号肽[ |
| DcaUPO | D. caldariorum | P. pastoris | 更换信号肽[ |
| HspUPO | Hypoxylon sp | P. pastoris | 原序列[ |
| AniUPO | Aspergillus niger | P. pastoris | 信号肽预测[ |
| CabUPO 1 | C. aberdarensis | P. pastoris | 信号肽预测[ |
| CabUPO 2 | C. aberdarensis | P. pastoris | 信号肽预测[ |
表2 已成功异源表达的UPO
Tab. 2 UPO that have been successfully heterologously expressed
| UPO | 来源 | 异源表达宿主 | 方法 |
|---|---|---|---|
| MroUPO | M. rotula | E. coli | 原序列,自诱导培养基[ |
| DcaUPO | D. caldariorum | E. coli | 原序列[ |
| CviUPO | C. virescens | E. coli | 原序列[ |
| HinUPO | H. insolens | Aspergillus oryzae | 原序列[ |
| CciUPO | C. cinerea | Aspergillus oryzae | 原序列[ |
| PviUPO | P.virgatula | Aspergillus oryzae | 原序列[ |
| ThyUPO | T.hyrcaniae | Aspergillus oryzae | 原序列[ |
| LfuUPO | C. fumago | Aspergillus niger | 原序列[ |
| AaeUPO (wild-type) | A. aegerita | 无细胞 | 无细胞蛋白表达系统[ |
| AaeUPO (PADAⅠ) | A. aegerita | S. cerevisiae & P. pastoris | 信号肽改造[ |
| MroUPO | M. rotula | S. cerevisiae & P. pastoris | 更换信号肽[ |
| CglUPO | C. globosum | S. cerevisiae & P. pastoris | 更换信号肽[ |
| MthUPO | M. thermophila | S. cerevisiae & P. pastoris | 更换信号肽[ |
| TteUPO | T. terrestris | S. cerevisiae & P. pastoris | 更换信号肽[ |
| PabUPO | P. aberdarensis | S. Cerevisiae & P. pastoris | 更换信号肽[ |
| MfeUPO | M. fergusii | P. pastoris | 更换信号肽[ |
| MhiUPO | M. hinnulea | P. pastoris | 更换信号肽[ |
| DcaUPO | D. caldariorum | P. pastoris | 更换信号肽[ |
| HspUPO | Hypoxylon sp | P. pastoris | 原序列[ |
| AniUPO | Aspergillus niger | P. pastoris | 信号肽预测[ |
| CabUPO 1 | C. aberdarensis | P. pastoris | 信号肽预测[ |
| CabUPO 2 | C. aberdarensis | P. pastoris | 信号肽预测[ |

图1 硫代血红素酶的氧化机理[细胞色素P450单加氧酶氧化过程包含6个步骤(Step 1-6),且需要NAD(P)H与O2的参与。UPO氧化机理以红色线条表示(Step a-d),UPO不催化分子氧的还原活化,而是直接利用过氧化氢形成具有催化活性的氧-铁基阳离子自由基配合物,因此不依赖于昂贵的NAD(P)H以及拥有更为简洁的电子传递链]
Fig. 1 Mechanism of thioheme-catalyzed oxidation[The process of cytochrome P450 monooxygenase catalyzation consists of six steps and requires the participation of NAD(P)H and O2(Steps 1-6). The catalytic oxidation of UPO is indicated by red lines(Steps a-d). UPO do not catalyze the reduction and activation of molecular oxygen, but directly use hydrogen peroxide to form catalytically active ferric oxide groups, so it does not rely on the expensive NAD(P)H and has a more concise electron transport chain.]

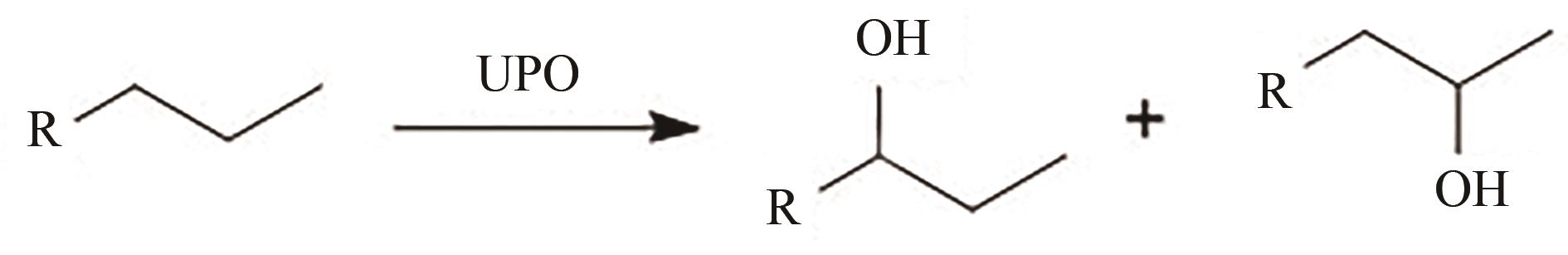
图3 UPO氧化不同烷烃与脂肪酸(a)~(d) AaeUPO氧化直链(支链)烷烃、环烷烃;(e) AaeUPO羟化脂肪酸;(f) MroUPO氧化脂肪酸发生脱羧反应;(g) AaeUPO、CciUPO催化脂肪酸碳链缩短(脱去2个碳原子)
Fig. 3 Oxidation of alkanes and fatty acids by UPO(a)~(d) AaeUPO oxidizes linear (branched) alkanes and cycloalkanes; (e) AaeUPO hydroxylates fatty acids; (f) MroUPO oxidizes fatty acids for decarboxylation; (g) AaeUPO/CciUPO catalyze fatty acid carbon chain shortening (removal of two carbon atom)

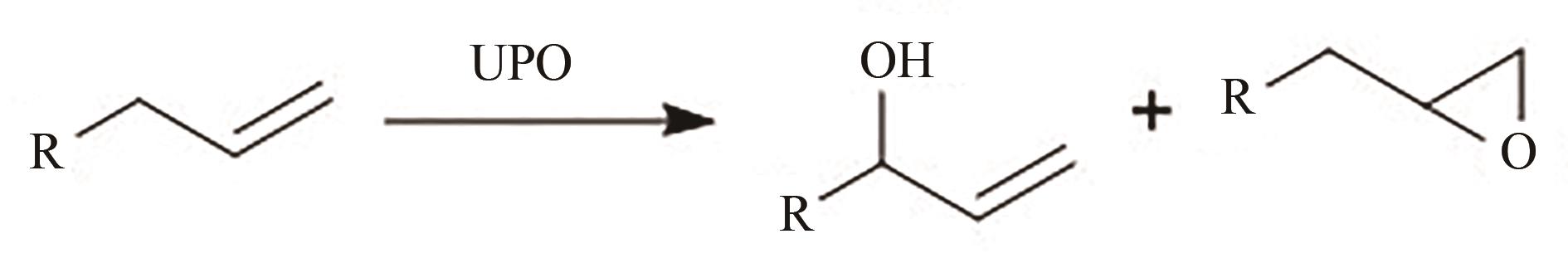
图5 UPO氧化不同烯烃(a)~(d) AaeUPO氧化不同烯烃,烯烃结构的不同会影响羟化产物与环氧化产物的比例
Fig. 5 Oxidation of Olefins by UPO(a)~(d) AaeUPO oxidizes different olefins, and the difference in olefin structure will affect the ratio of hydroxylation products to epoxidation products

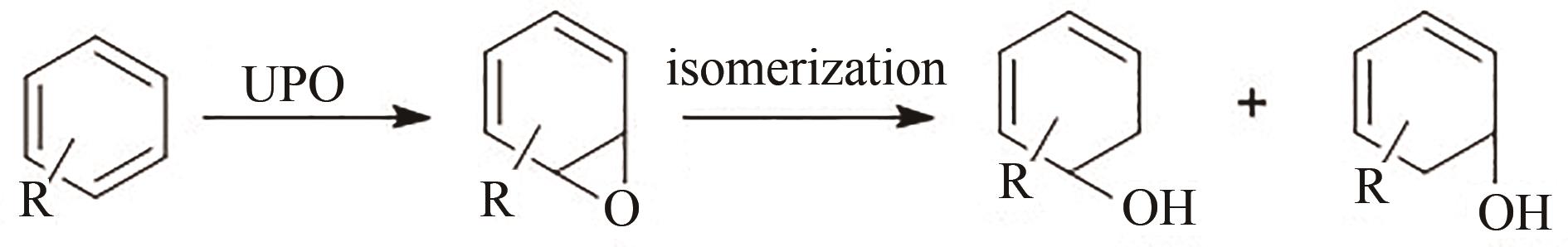
图7 UPO氧化不同芳烃化合物(a) AaeUPO氧化苯[56];(b) AaeUPO氧化萘[55];(c) AaeUPO氧化黄酮[57]
Fig. 7 Oxidation of aromatic hydrocarbons by UPO(a) AaeUPO oxidized benzene[56]; (b) AaeUPO oxidized naphthalene[55]; (c) AaeUPO oxidized flavonoids[57]
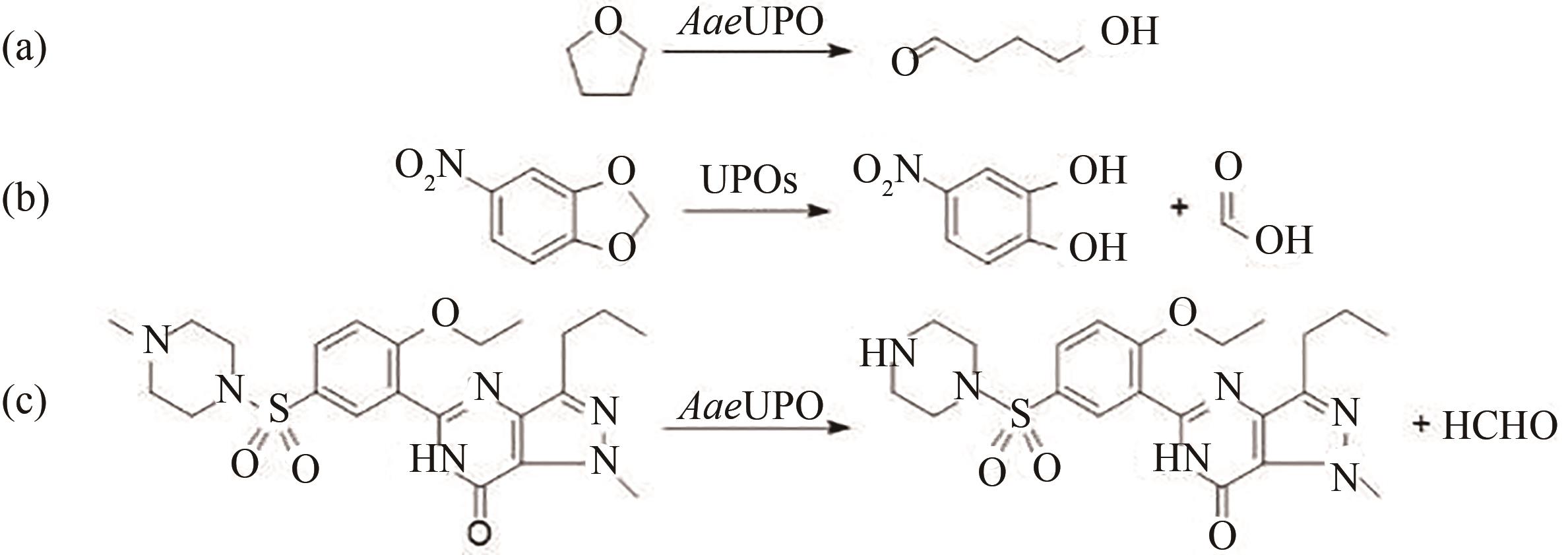
图10 UPO催化裂解反应(a) AaeUPO催化四氢呋喃发生开环反应;(b) 5-硝基-1,3-苯并二氧戊环(NBD)在UPO作用下脱烷基化生成甲酸和4-硝基邻苯二酚;(c) AaeUPO选择性地催化药物西地那非的N-甲基哌嗪环N-脱烷基化
Fig. 10 UPO catalyzes the cleavage reaction(a) AaeUPO catalyzed the ring-opening reaction of tetrahydrofuran; (b) 5-nitro-1,3-benzodioxole (NBD) was dealkylated under the action of UPO to form formic acid and 4-nitrocatechol; (c) AaeUPO selectively catalyzes the N-dealkylation of the N-methylpiperazine ring of the drug sildenafil

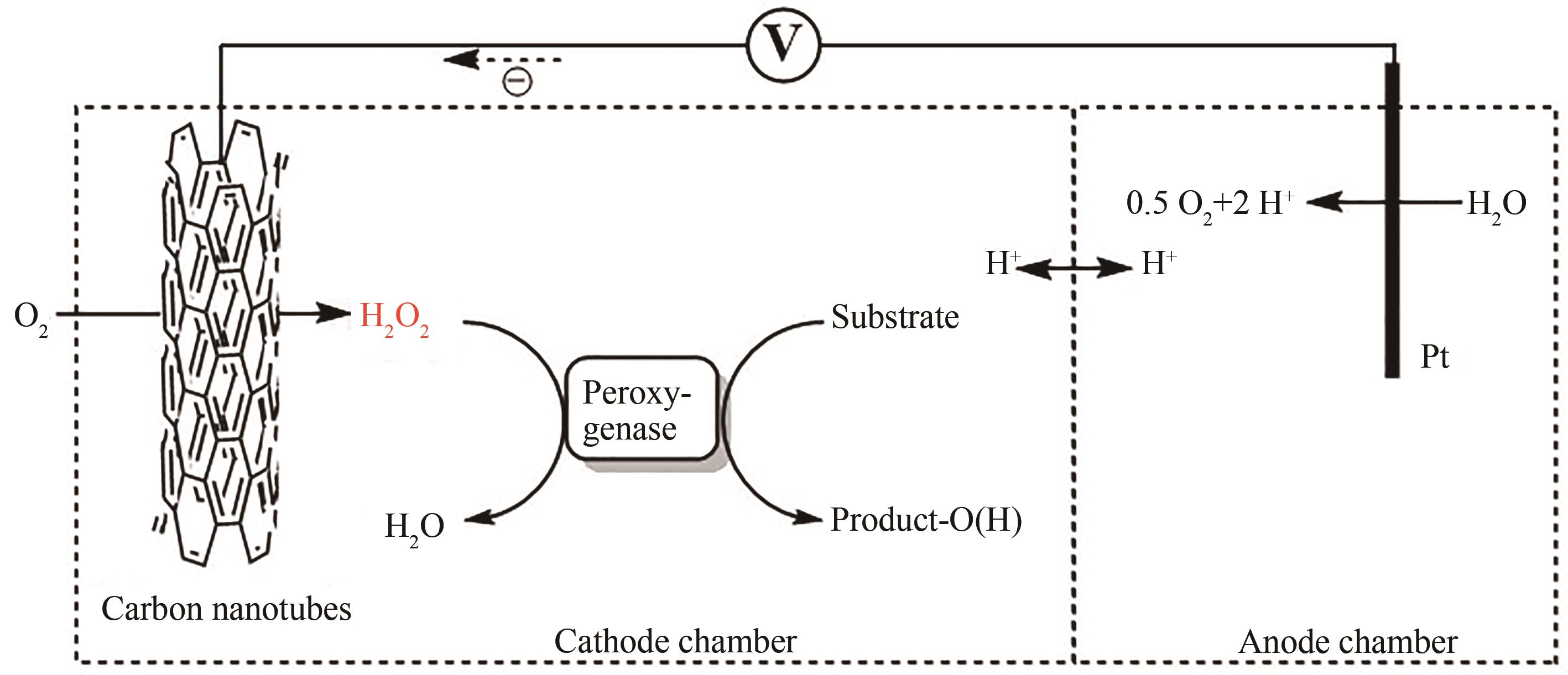
图12 Au-TiO2介导的光催化水氧化反应原位生成H2O2驱动AaeUPO催化氧化反应
Fig. 12 Au-TiO2-mediated photocatalytic water oxidation to generate H2O2in situ for driving AaeUPO-catalyzed oxidation reaction
| 1 | GYGLI G, VAN BERKEL W J H. Oxizymes for biotechnology[J]. Current Biotechnology, 2015, 4(2): 100-110. |
| 2 | MARTÍNEZ A T, RUIZ-DUEÑAS F J, CAMARERO S, et al. Oxidoreductases on their way to industrial biotransformations[J]. Biotechnology Advances, 2017, 35(6): 815-831. |
| 3 | YAMAGUCHI J, YAMAGUCHI A D, ITAMI K. C—H bond functionalization: emerging synthetic tools for natural products and pharmaceuticals[J]. Angewandte Chemie (International Ed in English), 2012, 51(36): 8960-9009. |
| 4 | ZHANG R K, HUANG X Y, ARNOLD F H. Selective C-H bond functionalization with engineered heme proteins: new tools to generate complexity [J]. Current Opinion in Chemical Biology, 2019, 49: 67-75. |
| 5 | BATHE U, TISSIER A. Cytochrome P450 enzymes: a driving force of plant diterpene diversity[J]. Phytochemistry, 2019, 161: 149-162. |
| 6 | HOLTMANN D, HOLLMANN F. The oxygen dilemma: a severe challenge for the application of monooxygenases? [J]. ChemBioChem, 2016, 17(15): 1391-1398. |
| 7 | WANG Y H, LAN D M, DURRANI R, et al. Peroxygenases en route to becoming dream catalysts. What are the opportunities and challenges? [J]. Current Opinion in Chemical Biology, 2017, 37: 1-9. |
| 8 | HOFRICHTER M, ULLRICH R. Heme-thiolate haloperoxidases: versatile biocatalysts with biotechnological and environmental significance[J]. Applied Microbiology and Biotechnology, 2006, 71(3): 276-288. |
| 9 | KLEBANOFF S J. Myeloperoxidase: friend and foe[J]. Journal of Leukocyte Biology, 2005, 77(5): 598-625. |
| 10 | LEE D S, YAMADA A, SUGIMOTO H, et al. Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis: crystallographic, spectroscopic, and mutational studies[J]. The Journal of Biological Chemistry, 2003, 278(11): 9761-9767. |
| 11 | HANANO A, BURCKLEN M, FLENET M, et al. Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif[J]. The Journal of Biological Chemistry, 2006, 281(44): 33140-33151. |
| 12 | TANG M C, FU C Y, TANG G L. Characterization of SfmD as a heme peroxidase that catalyzes the regioselective hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine in saframycin A biosynthesis[J]. The Journal of Biological Chemistry, 2012, 287(7): 5112-5121. |
| 13 | CONNOR K L, COLABROY K L, GERRATANA B. A heme peroxidase with a functional role as an L-tyrosine hydroxylase in the biosynthesis of anthramycin[J]. Biochemistry, 2011, 50(41): 8926-8936. |
| 14 | ULLRICH R, NÜSKE J, SCHEIBNER K, et al. Novel haloperoxidase from the Agrocybe aegerita oxidizes aryl alcohols and aldehydes[J]. Applied & Environmental Microbiology, 2004, 70(8): 4575-4581. |
| 15 | MORRIS D R, HAGER L P. Chloroperoxidase. I. Isolation and properties of the crystalline glycoprotein[J]. The Journal of Biological Chemistry, 1966, 241(8): 1763-1768. |
| 16 | MANOJ K M, HAGER L P. Chloroperoxidase, a janus enzyme[J]. Biochemistry, 2008, 47(9): 2997-3003. |
| 17 | SANFILIPPO C, NICOLOSI G. Catalytic behaviour of chloroperoxidase from Caldariomyces fumago in the oxidation of cyclic conjugated dienes[J]. Cheminform, 2003, 13(17): 1889-1892. |
| 18 | KLUGE M G, ULLRICH R, SCHEIBNER K, et al. Spectrophotometric assay for detection of aromatic hydroxylation catalyzed by fungal haloperoxidase-peroxygenase[J]. Applied Microbiology and Biotechnology, 2007, 75(6): 1473-1478. |
| 19 | ULLRICH R, HOFRICHTER M. The haloperoxidase of the agaric fungus Agrocybe aegerita hydroxylates toluene and naphthalene[J]. FEBS Letters, 2005, 579(27): 6247-6250. |
| 20 | HOFRICHTER M, ULLRICH R. Oxidations catalyzed by fungal peroxygenases[J]. Current Opinion in Chemical Biology, 2014, 19: 116-125. |
| 21 | FAIZA M, LAN D M, HUANG S F, et al. UPObase: an online database of unspecific peroxygenases[J]. Database-the Journal of Biological Databases and Curation, 2019, 2019(8): baz122. |
| 22 | KELLNER H, LUIS P, PECYNA M J, et al. Widespread occurrence of expressed fungal secretory peroxidases in forest soils[J]. PLoS One, 2014, 9(4): e95557. |
| 23 | HOFRICHTER M, KELLNER H, PECYNA M J, et al. Fungal unspecific peroxygenases: heme-thiolate proteins that combine peroxidase and cytochrome p450 properties[J]. Advances in Experimental Medicine and Biology, 2015, 851: 341-368. |
| 24 | KENIGSBERG P, FANG G H, HAGER L P. Post-translational modifications of chloroperoxidase from Caldariomyces fumago [J]. Archives of Biochemistry and Biophysics, 1987, 254(2): 409-415. |
| 25 | ZONG Q, OSMULSKI P A, HAGER L P. High-pressure-assisted reconstitution of recombinant chloroperoxidase[J]. Biochemistry, 1995, 34(38): 12420-12425. |
| 26 | CARRO J, GONZÁLEZ-BENJUMEA A, FERNÁNDEZ-FUEYO E, et al. Modulating fatty acid epoxidation vs hydroxylation in a fungal peroxygenase[J]. ACS Catalysis, 2019, 9(7): 6234-6242. |
| 27 | LINDE D, OLMEDO A, GONZÁLEZ-BENJUMEA A, et al. Two new unspecific peroxygenases from heterologous expression of fungal genes in Escherichia coli [J]. Applied and Environmental Microbiology, 2020, 86(7): e02899-e02819. |
| 28 | GONZÁLEZ-BENJUMEA A, CARRO J, RENAU-MÍNGUEZ C, et al. Fatty acid epoxidation by Collariella virescensperoxygenase and heme-channel variants[J]. Catalysis Science & Technology, 2020, 10(3): 717-725. |
| 29 | PÜLLMANN P, KNORRSCHEIDT A, MÜNCH J, et al. A modular two yeast species secretion system for the production and preparative application of unspecific peroxygenases[J]. Communications Biology, 2021, 4: 562. |
| 30 | MOLINA-ESPEJA P, GARCIA-RUIZ E, GONZALEZ-PEREZ D, et al. Directed evolution of unspecific peroxygenase from Agrocybe aegerita [J]. Applied & Environmental Microbiology, 2014, 80(11): 3496-3507. |
| 31 | MOLINA-ESPEJA P, MA S, MATE D M, et al. Tandem-yeast expression system for engineering and producing unspecific peroxygenase[J]. Enzyme and Microbial Technology, 2015, 73/74: 29-33. |
| 32 | PECYNA M, SCHNORR K M, ULLRICH R, et al. Fungal peroxygenases and methords of application. WO/2008/119780[P]. 2008-09-10. |
| 33 | BABOT E D, DEL RÍO J C, KALUM L, et al. Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea [J]. Biotechnology & Bioengineering, 2013, 110(9): 2323-2332. |
| 34 | LUND H, KALUM L, HOFRICHTER M. US Pat.,9908860B2 [P], 2018. |
| 35 | ARANDA C, MUNICOY M, GUALLAR V, et al. Selective synthesis of 4-hydroxyisophorone and 4-ketoisophorone by fungal peroxygenases[J]. Catalysis Science & Technology, 2019, 9(6): 1398-1405. |
| 36 | PÜLLMANN P, WEISSENBORN M J. Improving the heterologous production of fungal peroxygenases through an episomal pichia pastoris promoter and signal peptide shuffling system[J]. ACS Synthetic Biology, 2021, 10(6): 1360-1372. |
| 37 | GOMEZ DE SANTOS P, HOANG M D, KIEBIST J, et al. Functional expression of two unusual acidic peroxygenases from Candolleomyces aberdarensis in yeasts by adopting evolved secretion mutations[J]. Applied and Environmental Microbiology, 2021, 87(19): e0087821. |
| 38 | SCHRAMM M, FRIEDRICH S, SCHMIDTKE K U, et al. Cell-free protein synthesis with fungal lysates for the rapid production of unspecific peroxygenases[J]. Antioxidants, 2022, 11(2): 284. |
| 39 | CONESA A, VAN DE VELDE F, VAN RANTWIJK F, et al. Expression of the Caldariomyces fumagochloroperoxidase in Aspergillus niger and characterization of the recombinant enzyme[J]. Journal of Biological Chemistry, 2001, 276(21): 17635-17640. |
| 40 | ROTILIO L, SWOBODA A, EBNER K, et al. Structural and biochemical studies enlighten the unspecific peroxygenase from Hypoxylon sp. EC38 as an efficient oxidative biocatalyst[J]. ACS Catalysis, 2021, 11(18): 11511-11525. |
| 41 | BORMANN S, KELLNER H, HERMES J, et al. Broadening the biocatalytic toolbox-screening and expression of new unspecific peroxygenases[J]. Antioxidants, 2022, 11(2): 223. |
| 42 | PIONTEK K, STRITTMATTER E, ULLRICH R, et al. Structural basis of substrate conversion in a new aromatic peroxygenase: Cytochrome P450 functionality with benefits[J]. The Journal of Biological Chemistry, 2013, 288(48): 34767-34776. |
| 43 | PECYNA M J, ULLRICH R, BITTNER B, et al. Molecular characterization of aromatic peroxygenase from Agrocybe aegerita [J]. Applied Microbiology and Biotechnology, 2009, 84(5): 885-897. |
| 44 | LEWIS D F V, ITO Y. Cytochrome P450 structure and function: an evolutionary perspective[M]//Issues in Toxicology. Cambridge: Royal Society of Chemistry, 2008: 3-45. |
| 45 | PIONTEK K, ULLRICH R, LIERS C, et al. Crystallization of a 45 kDa peroxygenase/peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron[J]. Acta Crystallographica Section F, 2010, 66(6): 693-698. |
| 46 | HOFRICHTER M, KELLNER H, HERZOG R, et al. Fungal peroxygenases: a phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions[M]. Grand Challenges in Fungal Biotechnology, 2020, 369-403. |
| 47 | RAMIREZ-ESCUDERO M, MOLINA-ESPEJA P, GOMEZ DE SANTOS P, et al. Structural insights into the substrate promiscuity of a laboratory-evolved peroxygenase[J]. ACS Chemical Biology, 2018, 13(12): 3259-3268. |
| 48 | LECINA D, GILABERT J F, GUALLAR V. Adaptive simulations, towards interactive protein-ligand modeling[J]. Scientific Reports, 2017, 7: 8466. |
| 49 | PETER S, KINNE M, WANG X S, et al. Selective hydroxylation of alkanes by an extracellular fungal peroxygenase[J]. The FEBS Journal, 2011, 278(19): 3667-3675. |
| 50 | PETER S, KARICH A, ULLRICH R, et al. Enzymatic one-pot conversion of cyclohexane into cyclohexanone: comparison of four fungal peroxygenases[J]. Journal of Molecular Catalysis B: Enzymatic, 2014, 103: 47-51. |
| 51 | GUTIÉRREZ A, BABOT E D, ULLRICH R, et al. Regioselective oxygenation of fatty acids, fatty alcohols and other aliphatic compounds by a basidiomycete heme-thiolate peroxidase[J]. Archives of Biochemistry and Biophysics, 2011, 514(1/2): 33-43. |
| 52 | OLMEDO A, RÍO J C D, KIEBIST J, et al. Fatty acid chain shortening by a fungal peroxygenase[J]. Chemistry, 2017, 23(67): 16985-16989. |
| 53 | OLMEDO A, ULLRICH R, HOFRICHTER M, et al. Novel fatty acid chain-shortening by fungal peroxygenases yielding 2C-shorter dicarboxylic acids[J]. Antioxidants, 2022, 11(4): 744. |
| 54 | PETER S, KINNE M, ULLRICH R, et al. Epoxidation of linear, branched and cyclic alkenes catalyzed by unspecific peroxygenase[J]. Enzyme & Microbial Technology, 2013, 52(6/7): 370-376. |
| 55 | KLUGE M, ULLRICH R, DOLGE C, et al. Hydroxylation of naphthalene by aromatic peroxygenase from Agrocybe aegerita proceeds via oxygen transfer from H2O2 and intermediary epoxidation[J]. Applied Microbiology and Biotechnology, 2009, 81(6): 1071-1076. |
| 56 | KARICH A, KLUGE M, ULLRICH R, et al. Benzene oxygenation and oxidation by the peroxygenase of Agrocybe aegerita [J]. AMB Express, 2013, 3(1): 5. |
| 57 | BARKOVÁ K, KINNE M, ULLRICH R, et al. Regioselective hydroxylation of diverse flavonoids by an aromatic peroxygenase[J]. Tetrahedron, 2011, 67(26): 4874-4878. |
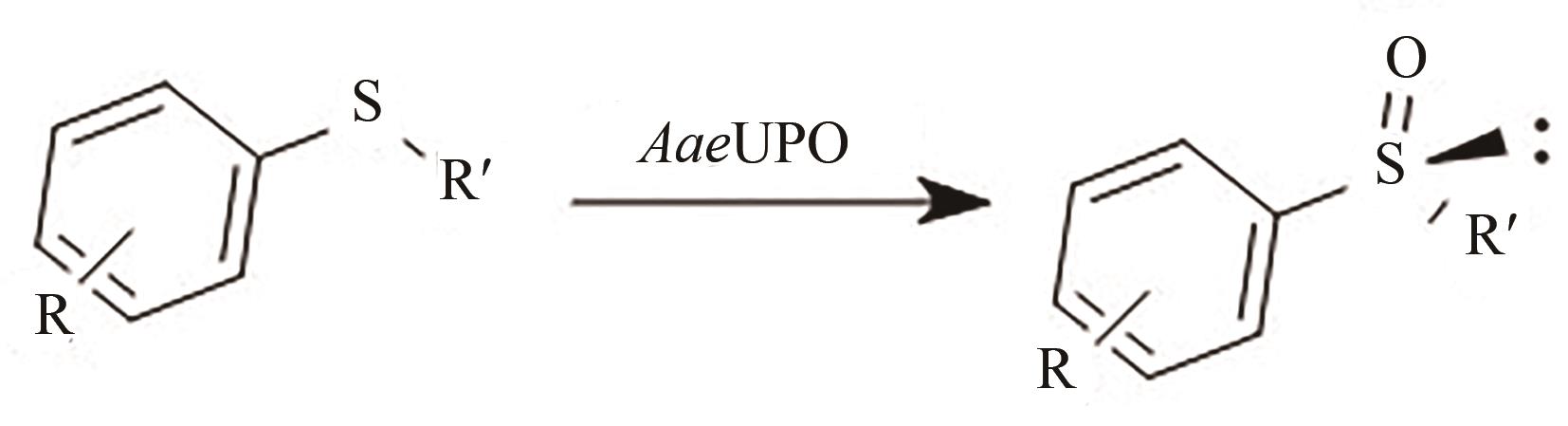
| 58 | BASSANINI I, FERRANDI E E, VANONI M, et al. Peroxygenase-catalyzed enantioselective sulfoxidations[J]. European Journal of Organic Chemistry, 2017, 2017(47): 7186-7189. |
| 59 | KINNE M, PORAJ-KOBIELSKA M, RALPH S A, et al. Oxidative cleavage of diverse ethers by an extracellular fungal peroxygenase[J]. Journal of Biological Chemistry, 2009, 284(43): 29343-29349. |
| 60 | PORAJ-KOBIELSKA M, KINNE M, ULLRICH R, et al. A spectrophotometric assay for the detection of fungal peroxygenases[J]. Analytical Biochemistry, 2012, 421(1): 327-329. |
| 61 | PORAJ-KOBIELSKA M, KINNE M, ULLRICH R, et al. Preparation of human drug metabolites using fungal peroxygenases[J]. Biochemical Pharmacology, 2011, 82(7): 789-796. |
| 62 | HYLAND R, ROE E G H, JONES B C, et al. Identification of the cytochrome P450 enzymes involved in the N-demethylation of sildenafil[J]. British Journal of Clinical Pharmacology, 2001, 51(3): 239-248. |
| 63 | ARANDA E, KINNE M, KLUGE M, et al. Conversion of dibenzothiophene by the mushrooms Agrocybe aegerita and Coprinellus radians and their extracellular peroxygenases[J]. Applied Microbiology and Biotechnology, 2009, 82(6): 1057-1066. |
| 64 | CARRO J, FERREIRA P, RODRÍGUEZ L, et al. 5-hydroxymethylfurfural conversion by fungal aryl-alcohol oxidase and unspecific peroxygenase[J]. The FEBS Journal, 2015, 282(16): 3218-3229. |
| 65 | KLUGE M, ULLRICH R, SCHEIBNER K, et al. Stereoselective benzylic hydroxylation of alkylbenzenes and epoxidation of styrene derivatives catalyzed by the peroxygenase of Agrocybe aegerita [J]. Green Chemistry, 2012, 14(2): 440-446. |
| 66 | ULLRICH R, HOFRICHTER M. Enzymatic hydroxylation of aromatic compounds[J]. Cellular and Molecular Life Sciences, 2007, 64(3): 271-293. |
| 67 | LUCAS F, BABOT E D, CAÑELLAS M, et al. Molecular determinants for selective C25-hydroxylation of vitamins D2 and D3 by fungal peroxygenases[J]. Catalysis Science & Technology, 2016, 6(1): 288-295. |
| 68 | BORMANN S, GOMEZ BARAIBAR A, NI Y, et al. Specific oxyfunctionalisations catalysed by peroxygenases: Opportunities, challenges and solutions[J]. Catalysis Science & Technology, 2015, 5(4): 2038-2052. |
| 69 | ZHANG W Y, FERNÁNDEZ-FUEYO E, NI Y, et al. Selective aerobic oxidation reactions using a combination of photocatalytic water oxidation and enzymatic oxyfunctionalizations[J]. Nature Catalysis, 2018, 1(1): 55-62. |
| 70 | WILLOT S J P, FERNÁNDEZ-FUEYO E, TIEVES F, et al. Expanding the spectrum of light-driven peroxygenase reactions[J]. ACS Catalysis, 2019, 9(2): 890-894. |
| 71 | YUAN B, MAHOR D, FEI Q, et al. Water-soluble anthraquinone photocatalysts enable methanol-driven enzymatic halogenation and hydroxylation reactions[J]. ACS Catalysis, 2020, 10(15): 8277-8284. |
| 72 | WAPSHOTT-STEHLI H L, GRUNDEN A M. In situ H2O2 generation methods in the context of enzyme biocatalysis[J]. Enzyme and Microbial Technology, 2021, 145: 109744. |
| 73 | HOLTMANN D, KRIEG T, GETREY L, et al. Electroenzymatic process to overcome enzyme instabilities[J]. Catalysis Communications, 2014, 51: 82-85. |
| 74 | GUILLET N, ROUÉ L, MARCOTTE S, et al. Electrogeneration of hydrogen peroxide in acid medium using pyrolyzed cobalt-based catalysts: Influence of the cobalt content on the electrode performance[J]. Journal of Applied Electrochemistry, 2006, 36(8): 863-870. |
| 75 | SAHA M S, NISHIKI Y, FURUTA T, et al. Electrolytic synthesis of peroxyacetic acid using in situ generated hydrogen peroxide on gas diffusion electrodes[J]. Journal of the Electrochemical Society, 2004, 151(9): D93. |
| 76 | CHOI D S, NI Y, FERNÁNDEZ-FUEYO E, et al. Photoelectroenzymatic oxyfunctionalization on flavin-hybridized carbon nanotube electrode platform[J]. ACS Catalysis, 2017, 7(3): 1563-1567. |
| 77 | BORMANN S, VAN SCHIE M M C H, DE ALMEIDA T P, et al. H2O2 production at low overpotentials for electroenzymatic halogenation reactions[J]. ChemSusChem, 2019, 12(21): 4759-4763. |
| 78 | NGUYEN L T, YANG K L. Combined cross-linked enzyme aggregates of horseradish peroxidase and glucose oxidase for catalyzing cascade chemical reactions[J]. Enzyme & Microbial Technology, 2017, 100: 52-59. |
| 79 | NI Y, FERNÁNDEZ-FUEYO E, BARAIBAR A G, et al. Peroxygenase-catalyzed oxyfunctionalization reactions promoted by the complete oxidation of methanol[J]. Angewandte Chemie International Edition, 2016, 55(2): 798-801. |
| 80 | TIEVES F, WILLOT S J P, VAN SCHIE M M C H, et al. Formate oxidase (FOx) from Aspergillus oryzae: one catalyst enables diverse H2O2-dependent biocatalytic oxidation reactions[J]. Angewandte Chemie International Edition, 2019, 58(23): 7873-7877. |
| 81 | VAITHYANATHAN V K, RAVI S, LEDUC R, et al. Utilization of biosolids for glucose oxidase production: a potential bio-fenton reagent for advanced oxidation process for removal of pharmaceutically active compounds[J]. Journal of Environmental Management, 2020, 271: 110995. |
| 82 | KARICH A, ULLRICH R, SCHEIBNER K, et al. Fungal unspecific peroxygenases oxidize the majority of organic EPA priority pollutants[J]. Frontiers in Microbiology, 2017, 8: 1463. |
| 83 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 84 | 卞佳豪, 杨广宇. 人工智能辅助的蛋白质工程[J]. 合成生物学, 2022, 3(3): 429-444. |
| BIAN J H, YANG G Y. Artificial intelligence-assisted protein engineering[J]. Synthetic Biology Journal, 2022, 3(3): 429-444. |
| [1] | 程中玉, 李付琸. 基于P450选择性氧化的天然产物化学-酶法合成进展[J]. 合成生物学, 2024, 5(5): 960-980. |
| [2] | 郑梦梦, 刘犇犇, 林芝, 瞿旭东. 重要甾体化合物的化学酶法合成研究进展[J]. 合成生物学, 2024, 5(5): 941-959. |
| [3] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [4] | 林继聪, 邹根, 刘宏民, 魏勇军. CRISPR/Cas基因组编辑技术在丝状真菌次级代谢产物合成中的应用[J]. 合成生物学, 2023, 4(4): 738-755. |
| [5] | 董佳钰, 李敏, 肖宗华, 胡明, 松田侑大, 汪伟光. 米曲霉异源表达天然产物研究进展[J]. 合成生物学, 2022, 3(6): 1126-1149. |
| [6] | 楼玉姣, 徐鉴, 吴起. 生物催化惰性碳氢键的氘代反应研究进展[J]. 合成生物学, 2022, 3(3): 530-544. |
| [7] | 孙文涛, 张昕哲, 万盛通, 王茹雯, 李春. Ⅱ型细胞色素P450酶氧化β-香树脂醇的选择性调控研究[J]. 合成生物学, 2021, 2(5): 804-814. |
| [8] | 王凤姣, 徐海洋, 闫建斌, 李伟. 植物天然农药除虫菊酯的生物合成和应用研究进展[J]. 合成生物学, 2021, 2(5): 751-763. |
| [9] | 周海波, 申琪瑶, 陈汉娜, 王宗杰, 李越中, 张友明, 卞小莹. 利用异源表达挖掘纤维堆囊菌So0157-2的新型天然产物[J]. 合成生物学, 2021, 2(5): 837-849. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||