Nucleic acid biosensing
YAO Linxin, SONG Lu, LI Min, ZUO Xiaolei
- Institute of Molecular Medicine,Renji Hospital,School of Medicine,Shanghai Jiao Tong University,Shanghai 200127,China
-
Received:2025-03-13Revised:2025-04-15Published:2025-04-16 -
Contact:ZUO Xiaolei
核酸生物传感
姚林欣, 宋璐, 李敏, 左小磊
- 上海交通大学医学院附属仁济医院,分子医学研究院,上海 200127
-
通讯作者:左小磊 -
作者简介:姚林欣 (2001—),女,硕士研究生。研究方向为框架核酸、临床检验诊断。 E-mail:ylx888@sjtu.edu.cn左小磊 (1980—),男,研究员,博士生导师。研究方向为框架核酸、临床检验诊断、DNA信息存储等。E-mail:zuoxiaolei@sjtu.edu.cn -
基金资助:国家重点研发计划(2021YFF1200300);国家自然科学基金项目(22025404);上海市自然科学基金(23ZR1438700);上海市高水平大学创新研究团队(SHSMU-ZLCX20212602);上海市卫生健康委员会(2022JC027);国家资助博士后研究人员计划(GZB20230436)
CLC Number:
Cite this article
YAO Linxin, SONG Lu, LI Min, ZUO Xiaolei. Nucleic acid biosensing[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-015.
姚林欣, 宋璐, 李敏, 左小磊. 核酸生物传感[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-015.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-015
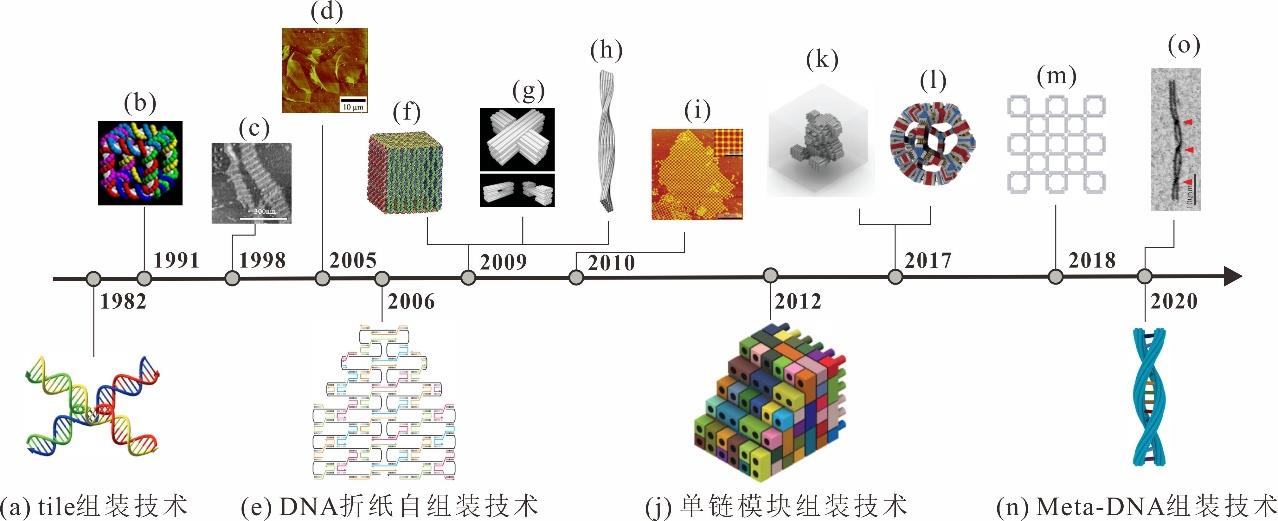
Fig. 1 History of DNA self-assembly technology(a) Four-arm junction, origins of DNA nanotechnology[1,6]; (b) The first 3D DNA nanostructure[7]; (c) The first 2D periodic DNA lattices[8]; (d) The 2D lattices breaking through the millimeter scale[9]; (e) DNA origami self-assembly technique[3]; (f) 3D hollow origami box[10]; (g) Three-dimensional structures built by multi-layer origami stack[11]; (h) 3D twisted origami[12]; (i) Two-dimensional arrays constructed using origami as a tile[13]; (j)Single strand tiles assembly technique[4]; (k) 3D teddy bear[14]; (l) Gigadalton-scale dodecahedron built by V-brick[15]; (m) The proposal of the concept of framework nucleic acids (FNAs)[16-17]; (n) Meta-DNA assembly technique[5]; (o) Right-handed dsM-DNA[5]

Fig. 2 2D DNA nanostructures(a) DNA tile: DX (DNA Double Crossover) (left), 4 × 4 tile (center), 6 × 4 tile (right) and the images of 2D arrays constructed from these structures (bottom)[34]; (b) 2D DNA origami smiley face[3]; (c) Asymmetric Chinese map origami[29]and concentric circle origami with curvature[28]; (d) Grid origami constructed by four-arm junctions[31] and bird and flower patterns constructed by multi-arm junctions[32]; (e) Mona Lisa image constructed with origami as a tile[33]; (f) Synthesized Chinese characters constructed by Single Strand Tile (SST) self-assembly strategy[4]
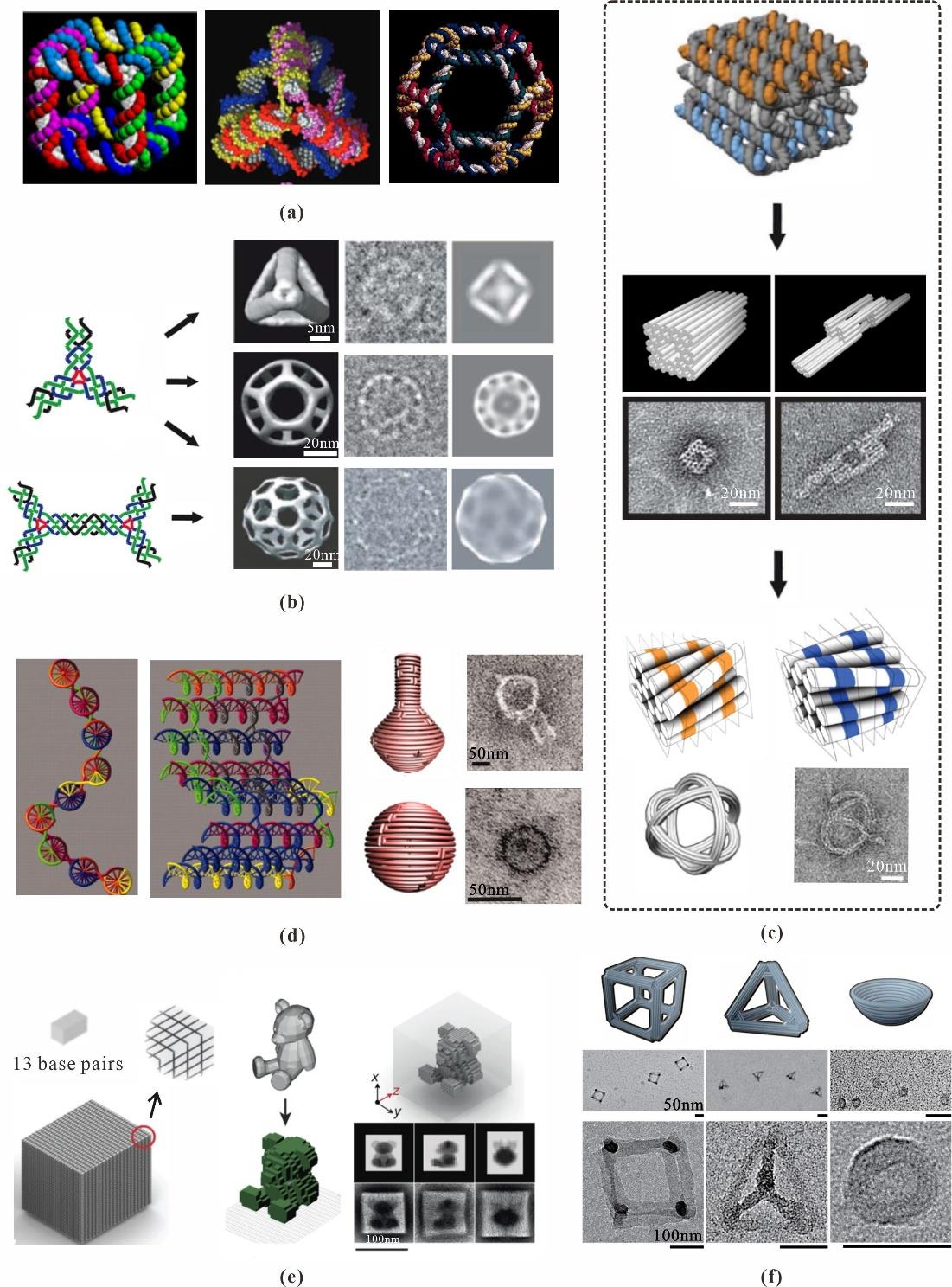
Fig. 3 3D DNA nanostructures(a) DNA cube[7] (left), DNA tetrahedron[35] (center), DNA-truncated octahedron[36] (right); (b) DNA polyhedra constructed by N-arms junctions[37]; (c) Square nut and railed bridge built based on honeycomb-pleated DNA origami strategy[11] (top), beachballs constructed by base pair insertions and deletions in the honeycomb lattice[12] (bottom); (d) Introduction of dihedral angles between adjacent planes (left), 3D nanoflask and sphere (right)[28]; (e) Single strand tile self-assembly strategy for synthesizing 3D teddy bear[14]; (f) Various siliconized framework nucleic acids[42]
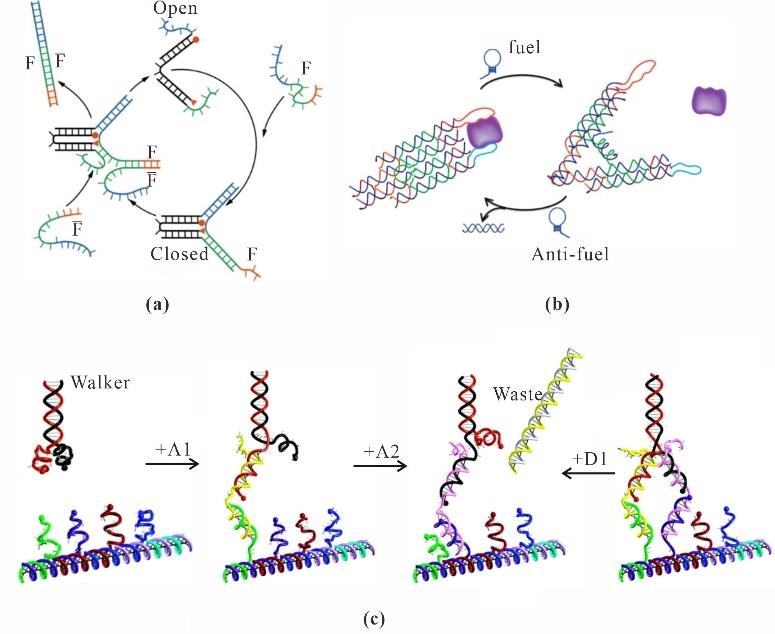
Fig. 4 Strand displacement-driven dynamic variational configurations(a) DNA nanotweezer[43]; (b) Nanotweezer using two DX as dual arms[44]; (c) DNA walker[47]
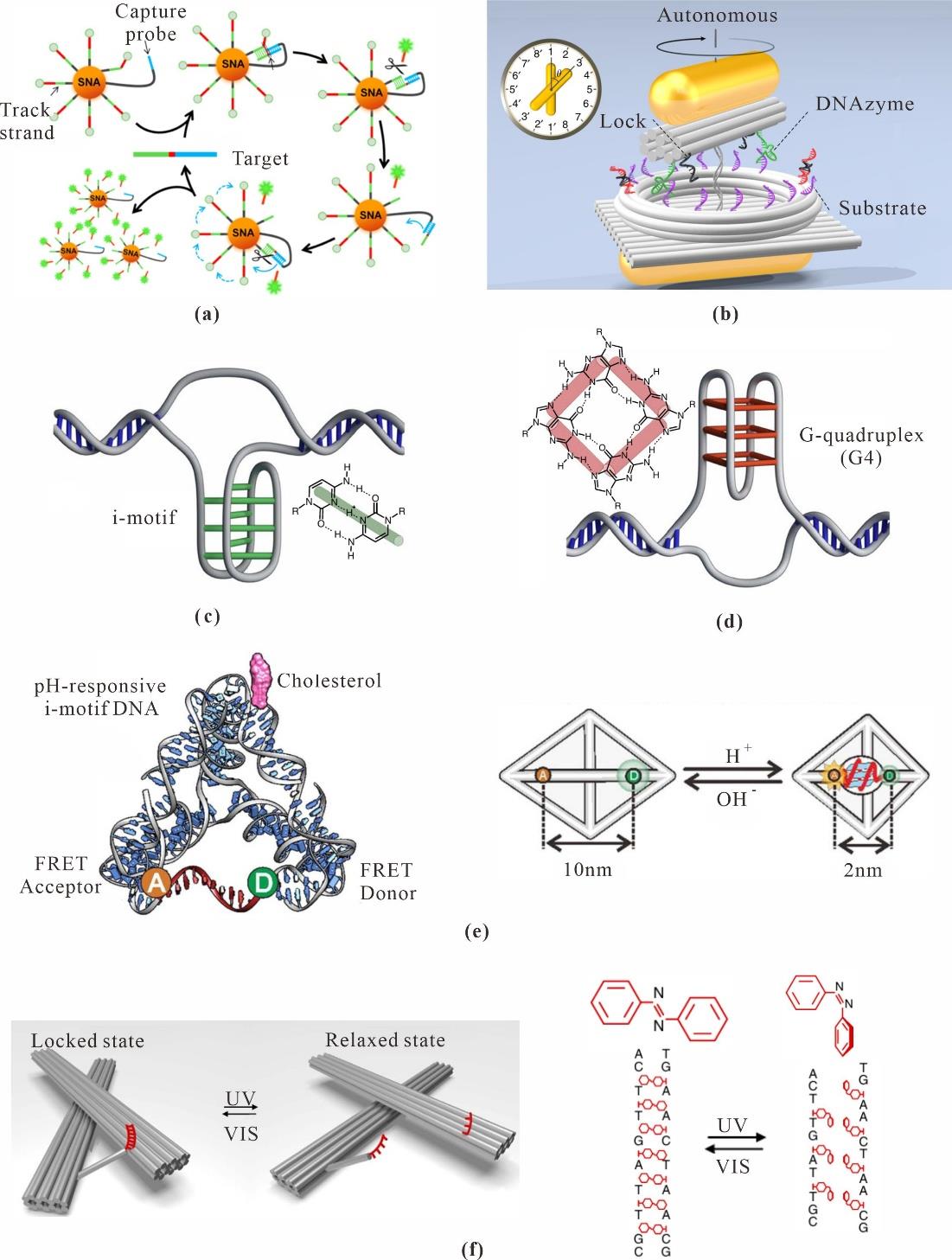
Fig. 5 Environmental stimulus-driven dynamic deformation(a) Enzyme-driven DNA walker [50]; (b) DNA plasmonic nanoclock[51]; (c) I-motif structure[57]; (d) G-quadruplex structure[57]; (e) pH-responsive TDF structure[54]; (f) Light-induced conformational changes of origami nanostructures[56]
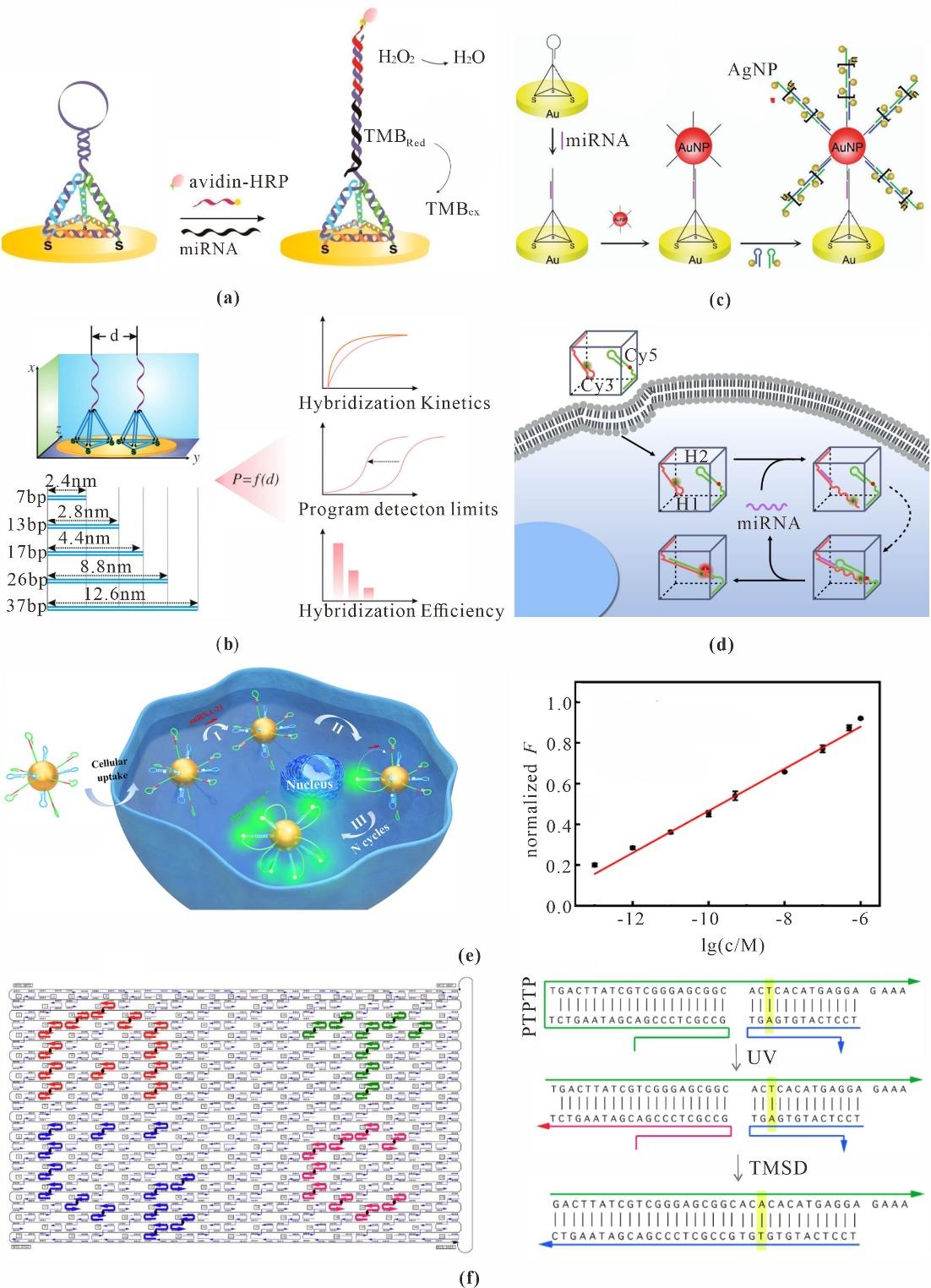
Fig. 6 Nucleic acids sensing based on DNA nanotechnology(a) TDF probes for detection of miRNA-141[62]; (b) A "soft lithography" approach to engineer the interface of electrochemical sensors[64]; (c) Detection of miRNAs using the HCR (Hybridization Chain Reaction) amplification strategy[65]; (d) DNA nanocubes for detection of miRNA[67]; (e) Ultrafast dual-layer 3D DNA nanosensor[69]; (f) The DNA origami for visual detection of SNP (Single Nucleotide Polymorphism)[79]
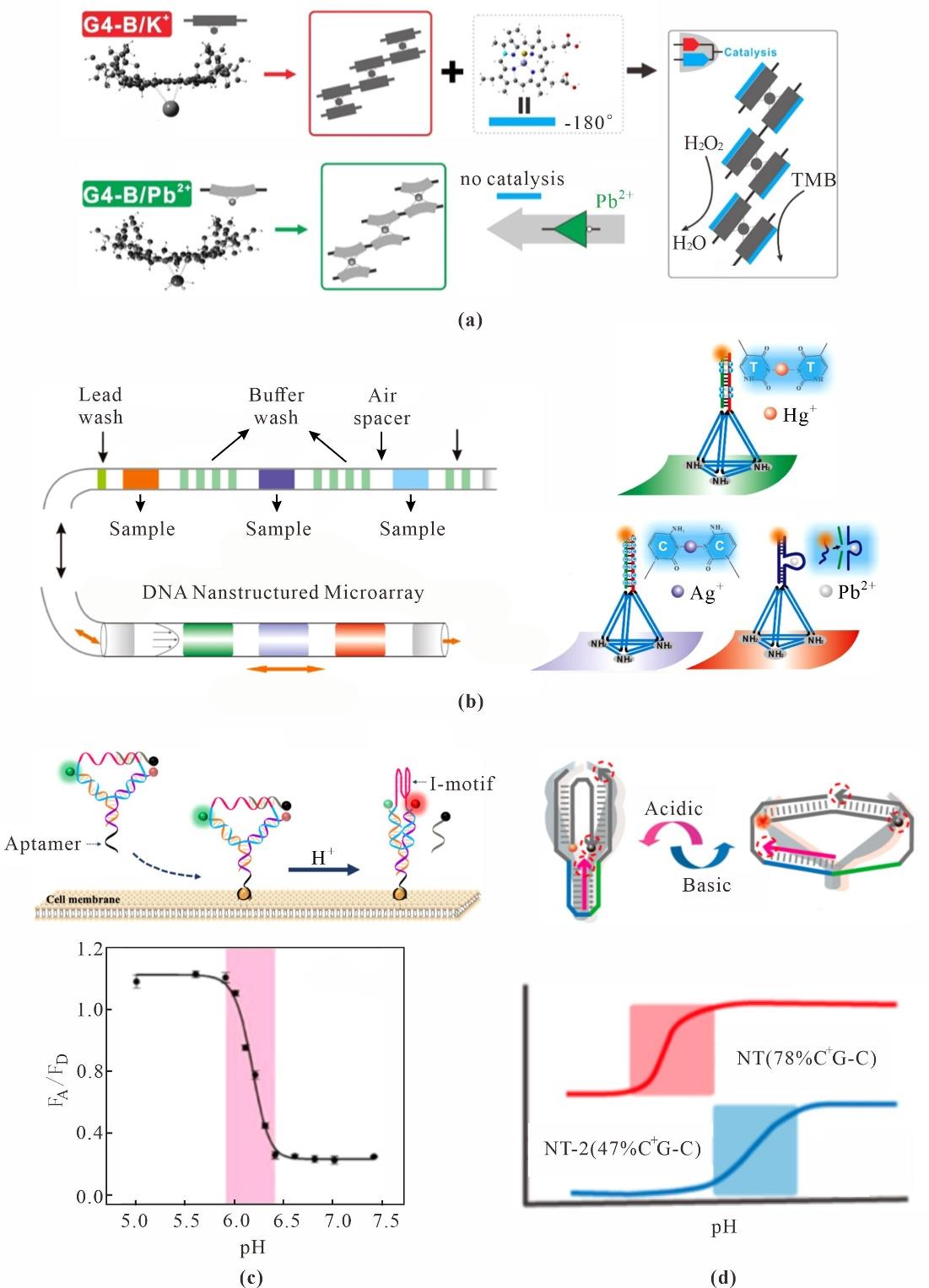
Fig. 7 Ion sensing based on DNA nanotechnology(a) Artificial enzyme hydrogel for detection of Pb2+[88]; (b) Multiplex detection of Metal Ions in TDF-encoded microchannels[89]; (c) Hybridization of i-motif with complementary chains narrows pH response range for more sensitive detection of extracellular pH changes[99]; (d) DNA triplexes with different levels of C-G-C enable programmable pH detection range[100]
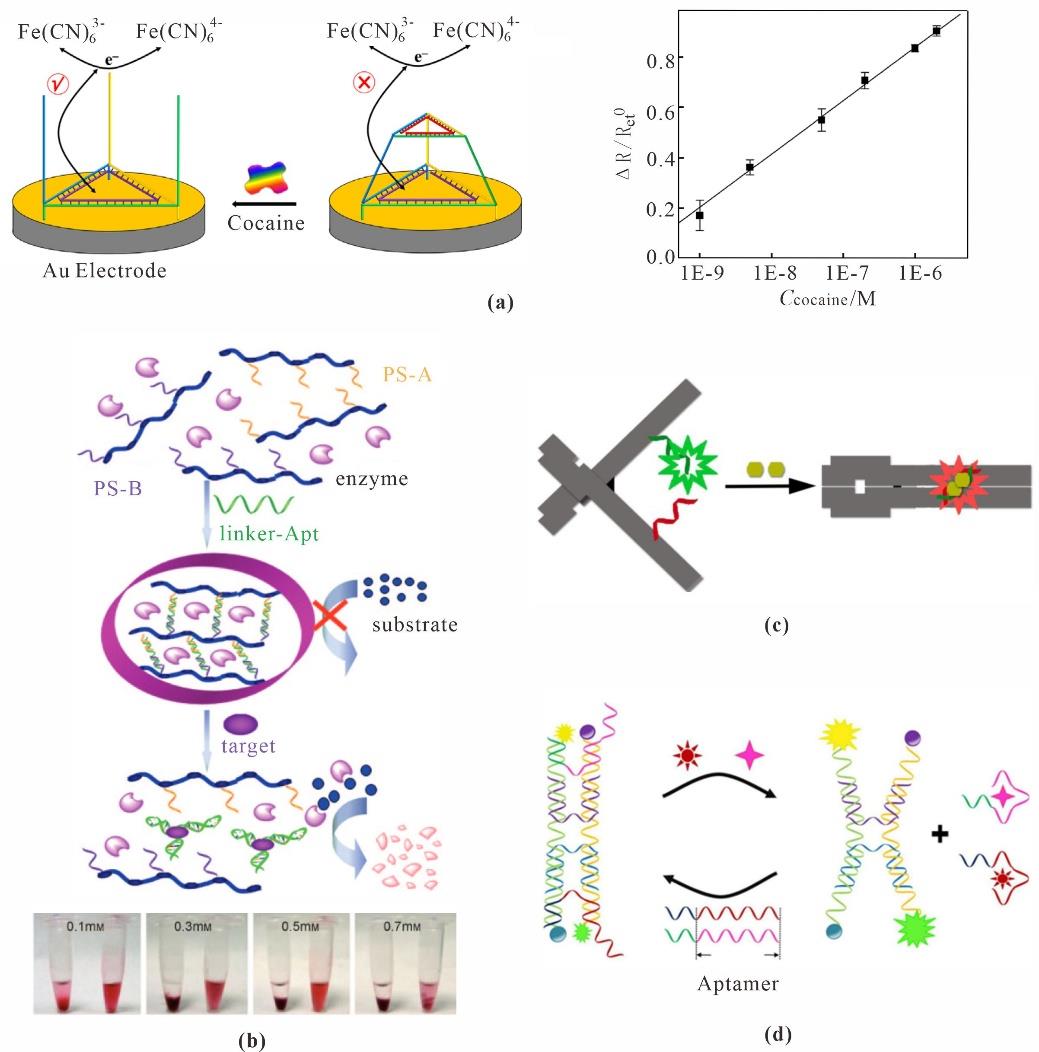
Fig. 8 Small molecules sensing based on DNA nanotechnology(a) TPF for detection of Cocaine[107]; (b) Artificial enzyme hydrogel for detection of Cocaine[108]; (c) DNA nanotweezer for detection of ATP[112]; (d) Simultaneous detection of AFB1 and OTA by dual DNA nanotweezers[113]
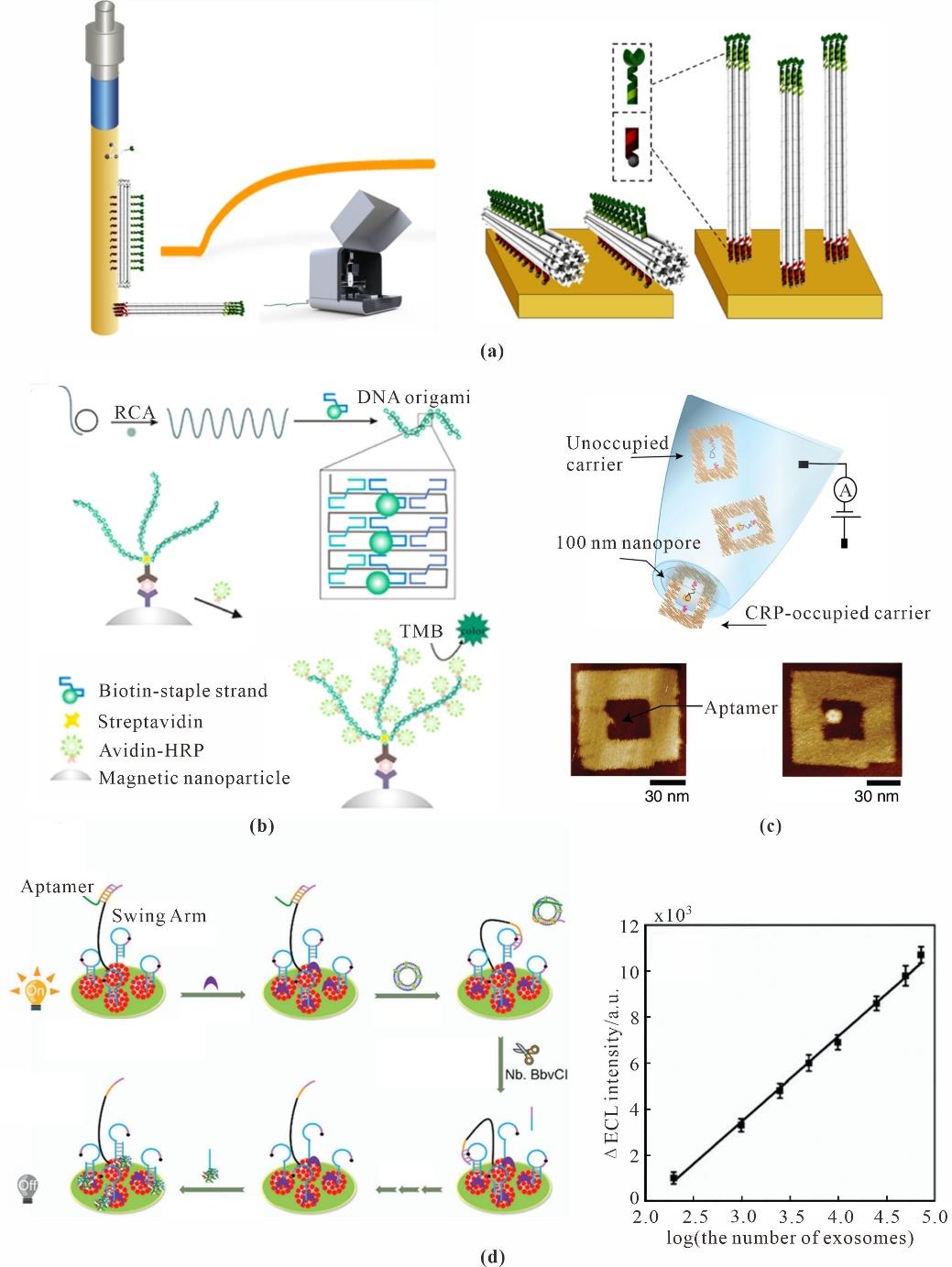
Fig. 9 Proteins sensing based on DNA nanotechnology(a) Fiber optic surface plasmon resonance biosensor for detection of thrombin[122]; (b) DNA nanobelt based on the ELASA method for PSA detection[126]; (c) Concentric square DNA origami for single molecule C-reactive protein biosensing[133]; (d) DNA walker for exosomes detection[139]
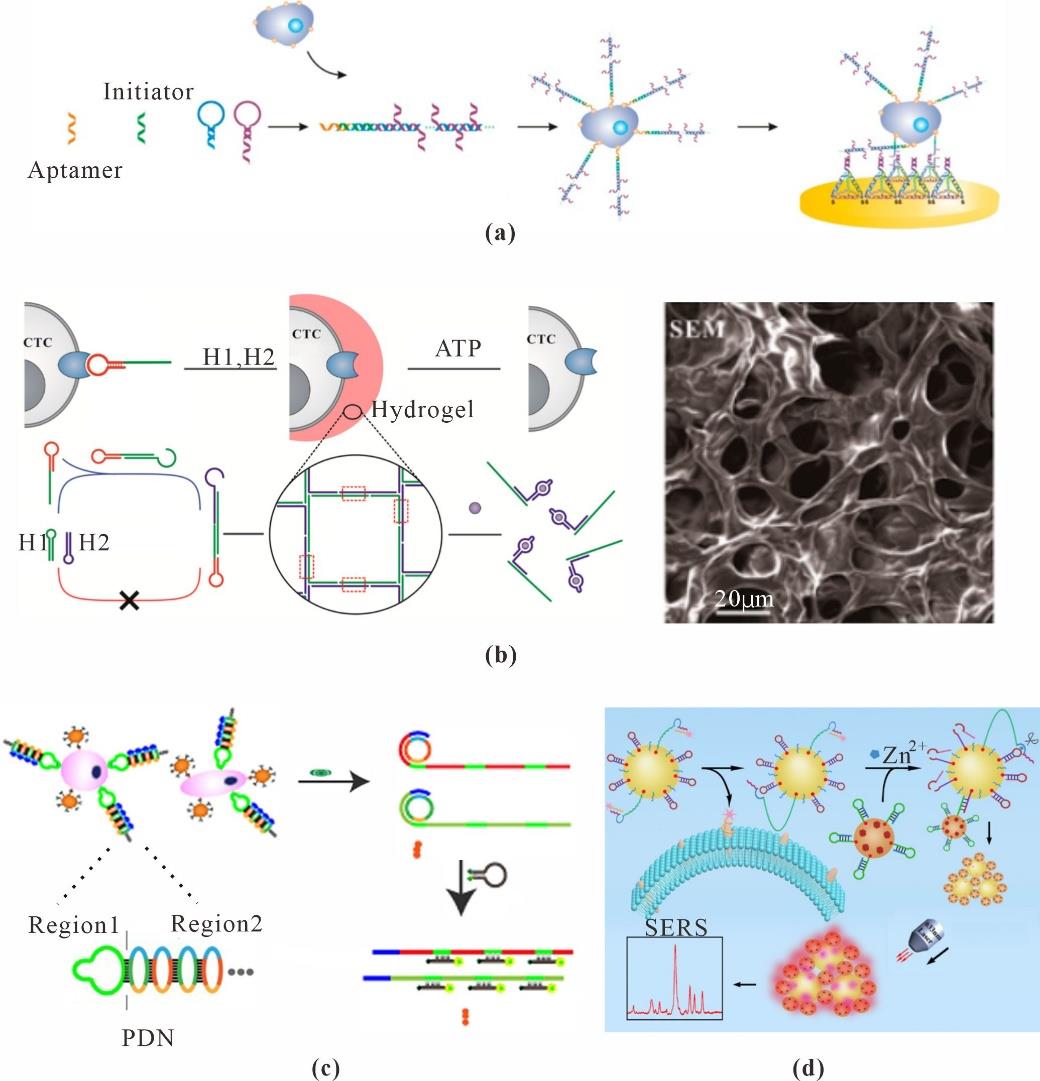
Fig. 10 Cells sensing based on DNA nanotechnology(a) Detection of cancer cells with TDF probes and multibranched hybridization chain reaction amplification[144]; (b) Hydrogel used for cloaking and decloaking circulating tumor cells[145]; (c) PDN-based CTC fluorescence analysis[146]; (d) DNA walker-powered ratiometric SERS cytosensor of circulating tumor cells[147]
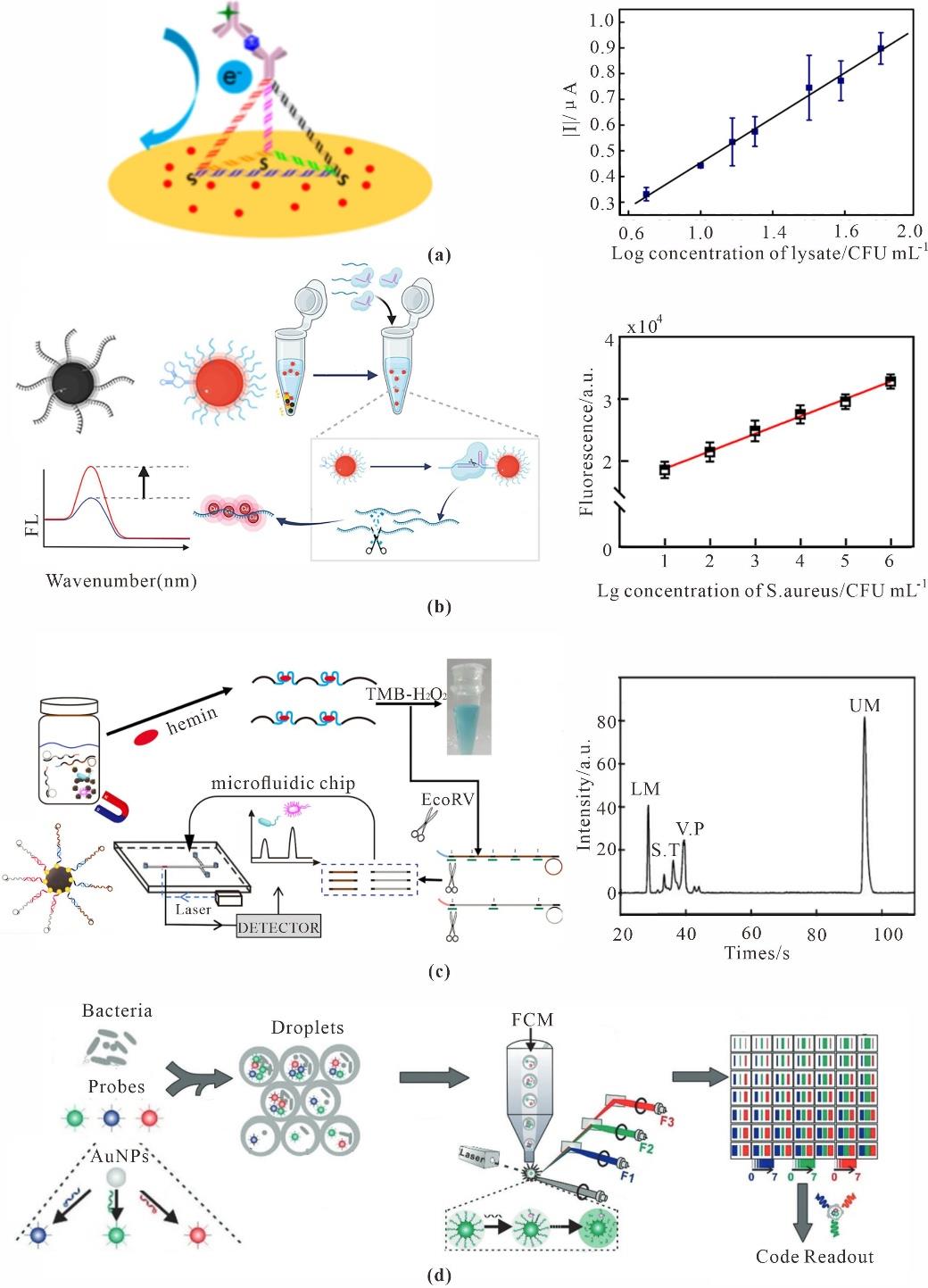
Fig. 11 Bacteria sensing based on DNA nanotechnology(a) Streptococcus pneumoniae biosensor based on TDF[152]; (b) Sensitive detection of Staphylococcus aureus by spherical nucleic acid triggered CRISPR/Cas12a and Poly T-CuNP[154]; (c) Simultaneous determination of different bacteria based on colorimetric enzyme assay[156]; (d) Super-multiplexed, high-throughput bacterial sensor[158]
| 59 | BARTEL D P. MicroRNAs: genomics, biogenesis, mechanism, and function[J]. Cell, 2004, 116(2): 281-297. |
| 60 | LI M, YIN F, SONG L, et al. Nucleic acid tests for clinical translation[J]. Chemical Reviews, 2021, 121(17): 10469-10558. |
| 61 | CHEN Y J, GROVES B, MUSCAT R A, et al. DNA nanotechnology from the test tube to the cell[J]. Nature Nanotechnology, 2015, 10(9): 748-760. |
| 62 | LIN M, WEN Y, LI L, et al. Target-responsive, DNA nanostructure-based E-DNA sensor for microRNA analysis[J]. Analytical Chemistry, 2014, 86(5): 2285-2288. |
| 63 | WANG X, CHEN F, ZHANG D, et al. Single copy-sensitive electrochemical assay for circulating methylated DNA in clinical samples with ultrahigh specificity based on a sequential discrimination–amplification strategy[J]. Chemical Science, 2017, 8(7): 4764-4770. |
| 64 | LIN M, WANG J, ZHOU G, et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection[J]. Angewandte Chemie International Edition, 2015, 54(7): 2151-2155. |
| 65 | MIAO P, TANG Y, YIN J. MicroRNA detection based on analyte triggered nanoparticle localization on a tetrahedral DNA modified electrode followed by hybridization chain reaction dual amplification[J]. Chemical Communications, 2015, 51(86): 15629-15632. |
| 66 | LIU H, LUO J, FANG L, et al. An electrochemical strategy with tetrahedron rolling circle amplification for ultrasensitive detection of DNA methylation[J]. Biosensors and Bioelectronics, 2018, 121: 47-53. |
| 67 | LIU L, RONG Q, KE G, et al. Efficient and reliable microRNA imaging in living cells via a FRET-based localized hairpin-DNA cascade amplifier[J]. Analytical Chemistry, 2019, 91(5): 3675-3680. |
| 68 | WANG S, XIA M, LIU J, et al. Simultaneous imaging of three tumor-related mRNAs in living cells with a DNA tetrahedron-based multicolor nanoprobe[J]. ACS Sensors, 2017, 2(6): 735-739. |
| 69 | MENG R, ZHANG X, LIU J, et al. Dual-layer 3D DNA nanostructure: The next generation of ultrafast DNA nanomachine for microRNA sensing and intracellular imaging[J]. Biosensors and Bioelectronics, 2023, 237: 115517. |
| 70 | ZHANG P, LI Z, WANG H, et al. DNA nanomachine-based regenerated sensing platform: a novel electrochemiluminescence resonance energy transfer strategy for ultra-high sensitive detection of microRNA from cancer cells[J]. Nanoscale, 2017, 9(6): 2310-2316. |
| 71 | QIU X, HILDEBRANDT N. Rapid and multiplexed microRNA diagnostic assay using quantum dot-based förster resonance energy transfer[J]. ACS Nano, 2015, 9(8): 8449-8457. |
| 72 | TSANG M K, YE W, WANG G, et al. Ultrasensitive detection of ebola virus oligonucleotide based on upconversion nanoprobe/nanoporous membrane system[J]. ACS Nano, 2016, 10(1): 598-605. |
| 73 | LIU F, CHOI J Y, SEO T S. Graphene oxide arrays for detecting specific DNA hybridization by fluorescence resonance energy transfer[J]. Biosensors and Bioelectronics, 2010, 25(10): 2361-2365. |
| 74 | RESCH-GENGER U, GRABOLLE M, CAVALIERE-JARICOT S, et al. Quantum dots versus organic dyes as fluorescent labels[J]. Nature Methods, 2008, 5(9): 763-775. |
| 75 | SAPSFORD K E, BERTI L, MEDINTZ I L. Materials for fluorescence resonance energy transfer analysis: beyond traditional donor–acceptor combinations[J]. Angewandte Chemie International Edition, 2006, 45(28): 4562-4589. |
| 76 | LI J M, ZHAO M X, SU H, et al. Multifunctional quantum-dot-based siRNA delivery for HPV18 E6 gene silence and intracellular imaging[J]. Biomaterials, 2011, 32(31): 7978-7987. |
| 77 | ANDO T. High-speed AFM imaging[J]. Current opinion in structural biology, 2014, 28: 63-68. |
| 78 | FU W, ZHANG W. Hybrid AFM for nanoscale physicochemical characterization: recent development and emerging applications[J]. Small, 2017, 13(11): 1603525. |
| 79 | SUBRAMANIAN H K K, CHAKRABORTY B, SHA R, et al. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami[J]. Nano Letters, 2011, 11(2): 910-913. |
| 80 | ZHANG H, CHAO J, PAN D, et al. DNA origami-based shape IDs for single-molecule nanomechanical genotyping[J]. Nature Communications, 2017, 8(1): 14738. |
| 81 | LIU K, PAN D, WEN Y, et al. Identifying the genotypes of hepatitis B virus (HBV) with DNA origami Label[J]. Small, 2018, 14(6): 1701718. |
| 82 | MUKHERJEE S, GHOSH R N, MAXFIELD F R. Endocytosis[J]. Physiological Reviews, 1997, 77(3): 759-803. |
| 1 | SEEMAN N C, KALLENBACH N R. Design of immobile nucleic acid junctions[J]. Biophysical Journal, 1983, 44(2): 201-209. |
| 2 | FU T J, SEEMAN N C. DNA double-crossover molecules[J]. Biochemistry, 1993, 32(13): 3211-3220. |
| 3 | ROTHEMUND P W K. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440(7082): 297-302. |
| 4 | WEI B, DAI M, YIN P. Complex shapes self-assembled from single-stranded DNA tiles[J]. Nature, 2012, 485(7400): 623-626. |
| 5 | YAO G, ZHANG F, WANG F, et al. Meta-DNA structures[J]. Nature Chemistry, 2020, 12(11): 1067-1075. |
| 6 | PINHEIRO A V, HAN D, SHIH W M, et al. Challenges and opportunities for structural DNA nanotechnology[J]. Nature Nanotechnology, 2011, 6(12): 763-772. |
| 7 | CHEN J, SEEMAN N C. Synthesis from DNA of a molecule with the connectivity of a cube[J]. Nature, 1991, 350(6319): 631-633. |
| 8 | WINFREE E, LIU F, WENZLER L A, et al. Design and self-assembly of two-dimensional DNA crystals[J]. Nature, 1998, 394(6693): 539-544. |
| 9 | HE Y, TIAN Y, CHEN Y, et al. Sequence symmetry as a tool for designing DNA nanostructures[J]. Angewandte Chemie International Edition, 2005, 44(41): 6694-6696. |
| 10 | ANDERSEN E S, DONG M, NIELSEN M M, et al. Self-assembly of a nanoscale DNA box with a controllable lid[J]. Nature, 2009, 459(7243): 73-76. |
| 11 | DOUGLAS S M, DIETZ H, LIEDL T, et al. Self-assembly of DNA into nanoscale three-dimensional shapes[J]. Nature, 2009, 459(7245): 414-418. |
| 12 | DIETZ H, DOUGLAS S M, SHIH W M. Folding DNA into twisted and curved nanoscale shapes[J]. Science, 2009, 325(5941): 725-730. |
| 83 | MONTESANO R, ROTH J, ROBERT A, et al. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins[J]. Nature, 1982, 296(5858): 651-653. |
| 84 | SHI W, CHANCE M R. Metallomics and metalloproteomics[J]. Cellular and Molecular Life Sciences, 2008, 65(19): 3040-3048. |
| 85 | HUANG Y, YAN J, FANG Z, et al. Highly sensitive and selective optical detection of lead( ii ) using a label-free fluorescent aptasensor[J]. RSC Advances, 2016, 6(93): 90300-90304. |
| 86 | KOTCH F W, FETTINGER J C, DAVIS J T. A lead-filled G-Quadruplex: insight into the G-Quartet's selectivity for Pb2+ over K+ [J]. Organic Letters, 2000, 2(21): 3277-3280. |
| 87 | LI T, WANG E, DONG S. Potassium-lead-switched G-Quadruplexes: A new class of DNA logic gates[J]. Journal of the American Chemical Society, 2009, 131(42): 15082-15083. |
| 88 | ZHONG R, XIAO M, ZHU C, et al. Logic catalytic interconversion of G-molecular hydrogel[J]. ACS Applied Materials & Interfaces, 2018, 10(5): 4512-4518. |
| 89 | QU X, YANG F, CHEN H, et al. Bubble-mediated ultrasensitive multiplex detection of metal ions in three-dimensional DNA nanostructure-encoded microchannels[J]. ACS Applied Materials & Interfaces, 2017, 9(19): 16026-16034. |
| 90 | CHEN J, ZHOU S, WEN J. Disposable strip biosensor for visual detection of Hg2+ based on Hg2+ -triggered toehold binding and exonuclease III-assisted signal amplification[J]. Analytical Chemistry, 2014, 86(6): 3108-3114. |
| 91 | LANZ E, GREGOR M, SLAVÍK J, et al. Use of FITC as a fluorescent probe for intracellular pH measurement[J]. Journal of Fluorescence, 1997, 7(4): 317-319. |
| 92 | MORDON S, DEVOISSELLE J M, MAUNOURY V. In vivo pH measurement and imaging of tumor tissue using a pH-sensitive fluorescent probe (5,6–carboxyfluorescein): instrumental and experimental studies [J]. Photochemistry and Photobiology, 1994, 60(3): 274-279. |
| 93 | BAE W, YOON T Y, JEONG C. Direct evaluation of self-quenching behavior of fluorophores at high concentrations using an evanescent field[J]. PLOS ONE, 2021, 16(2): e0247326. |
| 94 | CHEN G, LIU D, HE C, et al. Enzymatic synthesis of periodic DNA nanoribbons for intracellular pH sensing and gene silencing[J]. Journal of the American Chemical Society, 2015, 137(11): 3844-3851. |
| 95 | LI N, WANG M, GAO X, et al. A DNA tetrahedron nanoprobe with controlled distance of dyes for multiple detection in living cells and in vivo[J]. Analytical Chemistry, 2017, 89(12): 6670-6677. |
| 13 | LIU W, ZHONG H, WANG R, et al. Crystalline two-dimensional DNA-origami arrays[J]. Angewandte Chemie International Edition, 2011, 50(1): 264-267. |
| 14 | ONG L L, HANIKEL N, YAGHI O K, et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components[J]. Nature, 2017, 552(7683): 72-77. |
| 15 | WAGENBAUER K F, SIGL C, DIETZ H. Gigadalton-scale shape-programmable DNA assemblies[J]. Nature, 2017, 552(7683): 78-83. |
| 16 | GE Z, GU H, LI Q, et al. Concept and development of framework nucleic acids[J]. Journal of the American Chemical Society, 2018, 140(51): 17808-17819. |
| 17 | LIU Y, DAI Z, XIE X, et al. Spacer-programmed two-dimensional DNA origami assembly[J]. Journal of the American Chemical Society, 2024. |
| 18 | GEHRING K, LEROY J L, GUÉRON M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs[J]. Nature, 1993, 363(6429): 561-565. |
| 19 | DHAMODHARAN V, PRADEEPKUMAR P I. Specific recognition of promoter G-Quadruplex DNAs by small molecule ligands and light-up probes[J]. ACS Chemical Biology, 2019: acschembio.9b00475. |
| 20 | MA R I, KALLENBACH N R, SHEARDY R D, et al. Three-arm nucleic acid junctions are flexible[J]. Nucleic Acids Research, 1986, 14(24): 9745-9753. |
| 21 | WANG Y, MUELLER J E, KEMPER B, et al. Assembly and characterization of five-arm and six-arm DNA branched junctions[J]. Biochemistry, 1991, 30(23): 5667-5674. |
| 22 | WANG X, SEEMAN N C. Assembly and characterization of 8-arm and 12-arm DNA branched junctions[J]. Journal of the American Chemical Society, 2007, 129(26): 8169-8176. |
| 23 | LABEAN T H, YAN H, KOPATSCH J, et al. Construction, analysis, ligation, and self-asembly of DNA triple crossover complexes[J]. Journal of the American Chemical Society, 2000, 122(9): 1848-1860. |
| 24 | MATHIEU F, LIAO S, KOPATSCH J, et al. Six-helix bundles designed from DNA[J]. Nano Letters, 2005, 5(4): 661-665. |
| 96 | ZHAO J, LI Y, YU M, et al. Time-resolved activation of pH sensing and imaging in vivo by a remotely controllable DNA nanomachine[J]. Nano Letters, 2020, 20(2): 874-880. |
| 97 | CARDONE R A, CASAVOLA V, RESHKIN S J. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis[J]. Nature Reviews Cancer, 2005, 5(10): 786-795. |
| 98 | ANDERSON M, MOSHNIKOVA A, ENGELMAN D M, et al. Probe for the measurement of cell surface pH in vivo and ex vivo[J]. Proceedings of the National Academy of Sciences, 2016, 113(29): 8177-8181. |
| 99 | CHEN B, WANG Y, MA W, et al. A mimosa-inspired cell-surface-anchored ratiometric DNA nanosensor for high-resolution and sensitive response of target tumor extracellular pH[J]. Analytical Chemistry, 2020, 92(22): 15104-15111. |
| 100 | LIU L, DOU C X, LIU J W, et al. Cell Surface-anchored DNA nanomachine for dynamically tunable sensing and imaging of extracellular pH[J]. Analytical Chemistry, 2018, 90(19): 11198-11202. |
| 101 | SAHA S, CHAKRABORTY K, KRISHNAN Y. Tunable, colorimetric DNA-based pH sensors mediated by A-motif formation[J]. Chemical Communications, 2012, 48(19): 2513. |
| 102 | RICH A, DAVIES D R, CRICK F H C, et al. The molecular structure of polyadenylic acid[J]. Journal of Molecular Biology, 1961, 3(1): 71-IN19. |
| 103 | FRESCO J R. Polynucleotides II : The X-ray diffraction patterns of solutions of the randomly coiled and helical forms of polyriboadenylic acid[J]. Journal of Molecular Biology, 1959, 1(2): 106-IN3. |
| 104 | TS'O P O P, HELMKAMP G K, SANDER C. Interaction of nucleosides and related compounds with nucleic acids as indicated by the change of helix-coil transition temperature[J]. Proceedings of the National Academy of Sciences, 1962, 48(4): 686-698. |
| 105 | LI X, LIU Y, LI Y, et al. High glucose concentration induces endothelial cell proliferation by regulating cyclin‐D2‐related miR‐98[J]. Journal of Cellular and Molecular Medicine, 2016, 20(6): 1159-1169. |
| 106 | SANG F, ZHANG X, LIU J, et al. A label-free hairpin aptamer probe for colorimetric detection of adenosine triphosphate based on the anti-aggregation of gold nanoparticles[J]. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2019, 217: 122-127. |
| 107 | SHENG Q, LIU R, ZHANG S, et al. Ultrasensitive electrochemical cocaine biosensor based on reversible DNA nanostructure[J]. Biosensors and Bioelectronics, 2014, 51: 191-194. |
| 25 | YAN H, PARK S H, FINKELSTEIN G, et al. DNA-templated self-assembly of protein arrays and highly conductive nanowires[J]. Science, 2003, 301(5641): 1882-1884. |
| 26 | HE Y, CHEN Y, LIU H, et al. Self-assembly of hexagonal DNA two-dimensional (2D) arrays[J]. Journal of the American Chemical Society, 2005, 127(35): 12202-12203. |
| 27 | HE Y, TIAN Y, RIBBE A E, et al. Highly connected two-dimensional crystals of DNA Six-Point-Stars[J]. Journal of the American Chemical Society, 2006, 128(50): 15978-15979. |
| 28 | HAN D, PAL S, NANGREAVE J, et al. DNA origami with complex curvatures in three-dimensional space[J]. Science, 2011, 332(6027): 342-346. |
| 29 | QIAN L, WANG Y, ZHANG Z, et al. Analogic China map constructed by DNA[J]. Chinese Science Bulletin, 2006, 51(24): 2973-2976. |
| 30 | ANDERSEN E S, DONG M, NIELSEN M M, et al. DNA origami design of dolphin-shaped structures with flexible tails[J]. ACS Nano, 2008, 2(6): 1213-1218. |
| 31 | HAN D, PAL S, YANG Y, et al. DNA gridiron nanostructures based on four-arm junctions[J]. Science, 2013, 339(6126): 1412-1415. |
| 32 | ZHANG F, JIANG S, WU S, et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices[J]. Nature Nanotechnology, 2015, 10(9): 779-784. |
| 33 | TIKHOMIROV G, PETERSEN P, QIAN L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns[J]. Nature, 2017, 552(7683): 67-71. |
| 34 | ZHANG F, NANGREAVE J, LIU Y, et al. Structural DNA nanotechnology: state of the art and future perspective[J]. Journal of the American Chemical Society, 2014, 136(32): 11198-11211. |
| 35 | GOODMAN R P, SCHAAP I A T, TARDIN C F, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication[J]. Science, 2005, 310(5754): 1661-1665. |
| 36 | ZHANG Y, SEEMAN N C. Construction of a DNA-truncated octahedron[J]. Journal of the American Chemical Society, 1994, 116(5): 1661-1669. |
| 108 | ZHU Z, WU C, LIU H, et al. An aptamer cross‐linked hydrogel as a colorimetric platform for visual detection[J]. Angewandte Chemie International Edition, 2010, 49(6): 1052-1056. |
| 109 | YANG H, LIU H, KANG H, et al. Engineering target-responsive hydrogels based on aptamer-target interactions[J]. Journal of the American Chemical Society, 2008, 130(20): 6320-6321. |
| 110 | MA Y, MAO Y, AN Y, et al. Target-responsive DNA hydrogel for non-enzymatic and visual detection of glucose[J]. The Analyst, 2018, 143(7): 1679-1684. |
| 111 | MENG H M, ZHANG X, LV Y, et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing[J]. ACS Nano, 2014, 8(6): 6171-6181. |
| 112 | WALTER H K, BAUER J, STEINMEYER J, et al. "DNA origami traffic lights" with a split aptamer sensor for a bicolor fluorescence readout[J]. Nano Letters, 2017, 17(4): 2467-2472. |
| 113 | XIONG Z, WANG Q, XIE Y, et al. Simultaneous detection of aflatoxin B1 and ochratoxin A in food samples by dual DNA tweezers nanomachine[J]. Food Chemistry, 2021, 338: 128122. |
| 114 | WANG Y, SONG W, ZHAO H, et al. DNA walker-assisted aptasensor for highly sensitive determination of Ochratoxin A[J]. Biosensors and Bioelectronics, 2021, 182: 113171. |
| 115 | VISCO G. [Immunoglobulins][J]. Il Policlinico. Sezione Pratica, 1971, 78(6): 245-250. |
| 116 | JENKINS W J, PETERS T J. Mitochondrial enzyme activities in liver biopsies from patients with alcoholic liver disease.[J]. Gut, 1978, 19(5): 341-344. |
| 117 | YAN H, ZHONG G, XU G, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus[J]. ELife, 2012, 1: e00049. |
| 118 | LI J, WANG J, GREWAL Y S, et al. Multiplexed SERS detection of soluble cancer protein biomarkers with gold-silver alloy nanoboxes and nanoyeast single-chain variable fragments[J]. Analytical Chemistry, 2018, 90(17): 10377-10384. |
| 119 | HORI S S, GAMBHIR S S. Mathematical model identifies blood biomarker–based early cancer detection strategies and limitations[J]. Science Translational Medicine, 2011, 3(109). |
| 120 | CARDOZO K H M, LEBKUCHEN A, OKAI G G, et al. Establishing a mass spectrometry-based system for rapid detection of SARS-CoV-2 in large clinical sample cohorts[J]. Nature Communications, 2020, 11(1): 6201. |
| 37 | HE Y, YE T, SU M, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra[J]. Nature, 2008, 452(7184): 198-201. |
| 38 | ZHANG C, SU M, HE Y, et al. Conformational flexibility facilitates self-assembly of complex DNA nanostructures[J]. Proceedings of the National Academy of Sciences, 2008, 105(31): 10665-10669. |
| 39 | LI Y, TIAN C, LIU Z, et al. Structural transformation: assembly of an otherwise inaccessible DNA nanocage[J]. Angewandte Chemie International Edition, 2015, 54(20): 5990-5993. |
| 40 | BENSON E, MOHAMMED A, GARDELL J, et al. DNA rendering of polyhedral meshes at the nanoscale[J]. Nature, 2015, 523(7561): 441-444. |
| 41 | IINUMA R, KE Y, JUNGMANN R, et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT[J]. Science, 2014, 344(6179): 65-69. |
| 42 | LIU X, ZHANG F, JING X, et al. Complex silica composite nanomaterials templated with DNA origami[J]. Nature, 2018, 559(7715): 593-598. |
| 43 | YURKE B, TURBERFIELD A J, MILLS A P, et al. A DNA-fuelled molecular machine made of DNA[J]. Nature, 2000, 406(6796): 605-608. |
| 44 | ZHOU C, YANG Z, LIU D. Reversible regulation of protein binding affinity by a DNA machine[J]. Journal of the American Chemical Society, 2012, 134(3): 1416-1418. |
| 45 | LIU M, FU J, HEJESEN C, et al. A DNA tweezer-actuated enzyme nanoreactor[J]. Nature Communications, 2013, 4(1): 2127. |
| 46 | KOU B, CHAI Y, YUAN Y, et al. Dynamical regulation of enzyme cascade amplification by a regenerated DNA nanotweezer for ultrasensitive electrochemical DNA detection[J]. Analytical Chemistry, 2018, 90(18): 10701-10706. |
| 47 | SHIN J S, PIERCE N A. A synthetic DNA walker for molecular transport[J]. Journal of the American Chemical Society, 2004, 126(35): 10834-10835. |
| 48 | CHA T G, PAN J, CHEN H, et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes[J]. Nature Nanotechnology, 2014, 9(1): 39-43. |
| 121 | ZHOU X, ZHAO M, DUAN X, et al. Collapse of DNA tetrahedron nanostructure for "off–on" fluorescence detection of DNA methyltransferase activity[J]. ACS Applied Materials & Interfaces, 2017, 9(46): 40087-40093. |
| 122 | DAEMS D, PFEIFER W, RUTTEN I, et al. Three-dimensional DNA origami as programmable anchoring points for bioreceptors in fiber optic surface plasmon resonance biosensing[J]. ACS Applied Materials & Interfaces, 2018, 10(28): 23539-23547. |
| 123 | HOU J, LI X, XIE K P. Coupled liquid biopsy and bioinformatics for pancreatic cancer early detection and precision prognostication[J]. Molecular Cancer, 2021, 20(1): 34. |
| 124 | KARAMI P, KHOSHSAFAR H, JOHARI-AHAR M, et al. Colorimetric immunosensor for determination of prostate specific antigen using surface plasmon resonance band of colloidal triangular shape gold nanoparticles[J]. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2019, 222: 117218. |
| 125 | SELL S. AFP as a marker for liver cell injury: differentiation of tumor growth, hepatotoxicity, and carcinogenesis[J]. UCLA forum in medical sciences, 1978, 20: 51-58. |
| 126 | YAN J, HU C, WANG P, et al. Novel rolling circle amplification and DNA origami-based dna belt-involved signal amplification assay for highly sensitive detection of Prostate-Specific Antigen (PSA)[J]. ACS Applied Materials & Interfaces, 2014, 6(22): 20372-20377. |
| 127 | GHORBANI F, ABBASZADEH H, DOLATABADI J E N, et al. Application of various optical and electrochemical aptasensors for detection of human prostate specific antigen: A review[J]. Biosensors and Bioelectronics, 2019, 142: 111484. |
| 128 | SUN Y, FAN J, CUI L, et al. Fluorometric nanoprobes for simultaneous aptamer-based detection of carcinoembryonic antigen and prostate specific antigen[J]. Microchimica Acta, 2019, 186(3): 152. |
| 129 | CHEN C, FENG S, ZHOU M, et al. Development of a structure-switching aptamer-based nanosensor for salicylic acid detection[J]. Biosensors and Bioelectronics, 2019, 140: 111342. |
| 130 | TAGHDISI S M, DANESH N M, NAMEGHI M A, et al. A DNA triangular prism-based fluorescent aptasensor for ultrasensitive detection of prostate-specific antigen[J]. Analytica Chimica Acta, 2020, 1120: 36-42. |
| 131 | AHMADI-SANGACHIN E, MOHAMMADNEJAD J, HOSSEINI M. Fluorescence self-assembled DNA hydrogel for the determination of prostate specific antigen by aggregation induced emission[J]. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2023, 303: 123234. |
| 132 | QIN Y, LI D, YUAN R, et al. Netlike hybridization chain reaction assembly of DNA nanostructures enables exceptional signal amplification for sensing trace cytokines[J]. Nanoscale, 2019, 11(35): 16362-16367. |
| 133 | RAVEENDRAN M, LEE A J, SHARMA R, et al. Rational design of DNA nanostructures for single molecule biosensing[J]. Nature Communications, 2020, 11(1): 4384. |
| 49 | BAZRAFSHAN A, MEYER T A, SU H, et al. Tunable DNA origami motors translocate ballistically over μm distances at nm/s speeds[J]. Angewandte Chemie International Edition, 2020, 59(24): 9514-9521. |
| 50 | CHENG X, BAO Y, LIANG S, et al. Flap endonuclease 1-assisted DNA walkers for sensitively and specifically sensing ctDNAs[J]. Analytical Chemistry, 2021, 93(27): 9593-9601. |
| 51 | XIN L, ZHOU C, DUAN X, et al. A rotary plasmonic nanoclock[J/OL]. Nature Communications, 2019, 10(1): 5394. |
| 52 | DONG Y, YANG Z, LIU D. DNA nanotechnology based on i-Motif structures[J]. Accounts of Chemical Research, 2014, 47(6): 1853-1860. |
| 53 | CHEN C, LI M, XING Y, et al. Study of pH-Induced folding and unfolding kinetics of the DNA i-Motif by stopped-flow circular dichroism[J]. Langmuir, 2012, 28(51): 17743-17748. |
| 54 | LIU J, JING X, LIU M, et al. Mechano-fluorescence actuation in single synaptic vesicles with a DNA framework nanomachine[J]. Science Robotics, 2022, 7(73): eabq5151. |
| 55 | UEYAMA H, TAKAGI M, TAKENAKA S. A novel potassium sensing in aqueous media with a synthetic oligonucleotide derivative. fluorescence resonance energy transfer associated with guanine quartet-potassium ion complex formation[J]. Journal of the American Chemical Society, 2002, 124(48): 14286-14287. |
| 56 | KUZYK A, YANG Y, DUAN X, et al. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function[J]. Nature Communications, 2016, 7(1): 10591. |
| 57 | ZERAATI M, LANGLEY D B, SCHOFIELD P, et al. I-motif DNA structures are formed in the nuclei of human cells[J]. Nature Chemistry, 2018, 10(6): 631-637. |
| 58 | HELLER M J. DNA microarray technology: devices, systems, and applications[J]. Annual Review of Biomedical Engineering, 2002, 4(1): 129-153. |
| 134 | URABE F, KOSAKA N, ITO K, et al. Extracellular vesicles as biomarkers and therapeutic targets for cancer[J]. American Journal of Physiology-Cell Physiology, 2020, 318(1): C29-C39. |
| 135 | THÉRY C, ZITVOGEL L, AMIGORENA S. Exosomes: composition, biogenesis and function[J]. Nature Reviews Immunology, 2002, 2(8): 569-579. |
| 136 | WANG C, JIN D, YU Y, et al. A dual antibody-modified nanochannel biosensor for capture and identification of exosomes[J]. Sensors and Actuators B: Chemical, 2020, 314: 128056. |
| 137 | ZHOU Q, RAHIMIAN A, SON K, et al. Development of an aptasensor for electrochemical detection of exosomes[J]. Methods, 2016, 97: 88-93. |
| 138 | ZHANG H, WANG Z, ZHANG Q, et al. Ti3C2 MXenes nanosheets catalyzed highly efficient electrogenerated chemiluminescence biosensor for the detection of exosomes[J]. Biosensors and Bioelectronics, 2019, 124-125: 184-190. |
| 139 | FENG Q M, MA P, CAO Q H, et al. An aptamer-binding DNA walking machine for sensitive electrochemiluminescence detection of tumor exosomes[J]. Chemical Communications, 2020, 56(2): 269-272. |
| 140 | ZHAO L, SUN R, HE P, et al. Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and Exonuclease III -assisted electrochemical ratiometric biosensing[J]. Analytical Chemistry, 2019, 91(22): 14773-14779. |
| 141 | NAGRATH S, SEQUIST L V, MAHESWARAN S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology[J]. Nature, 2007, 450(7173): 1235-1239. |
| 142 | YOON H J, KIM T H, ZHANG Z, et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets[J]. Nature Nanotechnology, 2013, 8(10): 735-741. |
| 143 | ZHAO W, CUI C H, BOSE S, et al. Bioinspired multivalent DNA network for capture and release of cells[J]. Proceedings of the National Academy of Sciences, 2012, 109(48): 19626-19631. |
| 144 | ZHOU G, LIN M, SONG P, et al. Multivalent capture and detection of cancer cells with DNA nanostructured biosensors and multibranched hybridization chain reaction amplification[J]. Analytical Chemistry, 2014, 86(15): 7843-7848. |
| 145 | SONG P, YE D, ZUO X, et al. DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis[J]. Nano Letters, 2017, 17(9): 5193-5198. |
| 146 | WANG J, DONG H yan, ZHOU Y, et al. Immunomagnetic antibody plus aptamer pseudo-DNA nanocatenane followed by rolling circle amplication for highly-sensitive CTC detection[J]. Biosensors and Bioelectronics, 2018, 122: 239-246. |
| 147 | XIONG J, DONG C, ZHANG J, et al. DNA walker-powered ratiometric SERS cytosensor of circulating tumor cells with single-cell sensitivity[J]. Biosensors and Bioelectronics, 2022, 213: 114442. |
| 148 | MORRIS J G, ACHESON D. Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell[J]. Clinical Infectious Diseases, 2003, 37(2): 272-280. |
| 149 | BISHARA J, LEIBOVICI L, GARTMAN‐ISRAEL D, et al. Long-term outcome of infective endocarditis: the impact of early surgical intervention[J]. Clinical Infectious Diseases, 2001, 33(10): 1636-1643. |
| 150 | MUSHER D M, THORNER A R. Community-acquired Pneumonia[J]. New England Journal of Medicine, 2014, 371(17): 1619-1628. |
| 151 | GIOVANNI M, SETYAWATI M I, TAY C Y, et al. Electrochemical quantification of Escherichia coli with DNA nanostructure[J]. Advanced Functional Materials, 2015, 25(25): 3840-3846. |
| 152 | WANG J, LEONG M C, LEONG E Z W, et al. Clinically relevant detection of streptococcus pneumoniae with DNA-antibody nanostructures[J]. Analytical Chemistry, 2017, 89(12): 6900-6906. |
| 153 | WEI Y, TAO Z, WAN L, et al. Aptamer-based Cas14a1 biosensor for amplification-free live pathogenic detection[J]. Biosensors and Bioelectronics, 2022, 211: 114282. |
| 154 | ZHANG X, SUN R, ZHENG H, et al. Amplification-free sensitive detection of Staphylococcus aureus by spherical nucleic acid triggered CRISPR/Cas12a and Poly T-Cu reporter[J]. Microchimica Acta, 2025, 192(2): 76. |
| 155 | SUN Y, DUAN N, MA P, et al. Colorimetric Aptasensor based on truncated aptamer and trivalent DNAzyme for Vibrio parahemolyticus determination[J]. Journal of Agricultural and Food Chemistry, 2019, 67(8): 2313-2320. |
| 156 | YU J, WU H, HE L, et al. The universal dual-mode aptasensor for simultaneous determination of different bacteria based on naked eyes and microfluidic-chip together with magnetic DNA encoded probes[J]. Talanta, 2021, 225: 122062. |
| 157 | YANG H, XIAO M, LAI W, et al. Stochastic DNA dual-walkers for ultrafast colorimetric bacteria detection[J]. Analytical Chemistry, 2020, 92(7): 4990-4995. |
| 158 | XIAO M, ZOU K, LI L, et al. Stochastic DNA walkers in droplets for super‐multiplexed bacterial phenotype detection[J]. Angewandte Chemie International Edition, 2019, 58(43): 15448-15454. |
| 159 | BROWN T M, FAKIH H H, SALIBA D, et al. Stabilization of functional DNA structures with mild photochemical methods[J]. Journal of the American Chemical Society, 2023, 145(4): 2142-2151. |
| 160 | BAI T, ZHANG J, HUANG K, et al. Reconfiguration of DNA nanostructures induced by enzymatic ligation treatment[J]. Nucleic Acids Research, 2022, 50(14): 8392-8398. |
| 161 | HANG X M, LIU P F, TIAN S, et al. Rapid and sensitive detection of Ebola RNA in an unamplified sample based on CRISPR-Cas13a and DNA roller machine[J/OL]. Biosensors and Bioelectronics, 2022, 211: 114393. |
| 162 | YAO S, LIU Y, DING Y, et al. Three-dimensional DNA nanomachine biosensor coupled with CRISPR Cas12a cascade amplification for ultrasensitive detection of carcinoembryonic antigen[J/OL]. Journal of Nanobiotechnology, 2024, 22(1): 266. |
| 163 | GUO J, ZHU Y, MIAO P. Nano-impact electrochemical biosensing based on a CRISPR-responsive DNA hydrogel[J]. Nano Letters, 2023, 23(23): 11099-11104. |
| [1] | SONG Yidong, YUAN Qianmu, YANG Yuedong. Application of deep learning in protein function prediction [J]. Synthetic Biology Journal, 2023, 4(3): 488-506. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||