Engineering an in vivo directed evolution system for developing genetic switches
ZHOU Yujie1,2, YI Xiao1,2
- 1.Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen Institute of Synthetic Biology,Key Laboratory of Quantitative Synthetic Biology,Shenzhen 518055,Guangdong,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2025-03-24Revised:2025-04-23Published:2025-04-26 -
Contact:YI Xiao
遗传开关的活细胞定向进化平台的建立及应用
周玉洁1,2, 易啸1,2
- 1.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,定量合成生物学全国重点实验室,广东 深圳 518055
2.中国科学院大学,北京 100049
-
通讯作者:易啸 -
作者简介:周玉洁 (2000—),女,硕士研究生。研究方向为合成生物学。E-mail:yj.zhou@siat.ac.cn易啸 (1986—),男,研究员,博士,博士生导师。研究方向为合成生物学,定向进化。E-mail:xiao.yi@siat.ac.cn
CLC Number:
Cite this article
ZHOU Yujie, YI Xiao. Engineering an in vivo directed evolution system for developing genetic switches[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-023.
周玉洁, 易啸. 遗传开关的活细胞定向进化平台的建立及应用[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-023.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-023
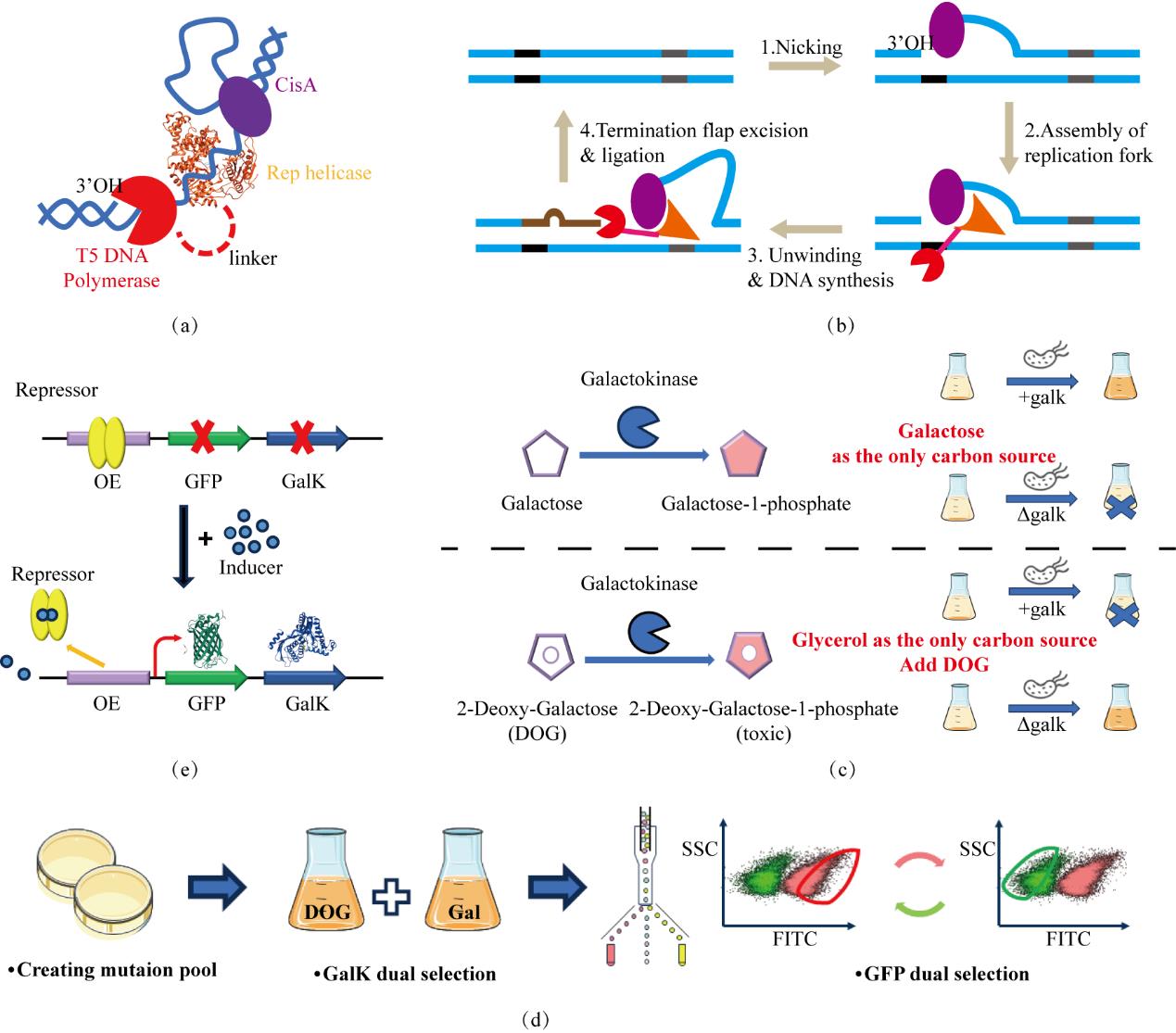
Fig. 1 Construction of the directed evolution experimental platform(a) Components of the TADR system; (b) The process of introducing mutations by the TADR system, the black and gray segments represent the initiation and termination; (c) Taking the repressor protein as an example, in the screening system, the expression of GFP-GalK is regulated by the repressor protein, and positive and negative screening are achieved by changing the induction conditions; (d) The working principle of the positive and negative screening markers of GalK; (e) The working process of the TADR-GalK-GFP experimental platform
RBS名称 RBS name | RBS序列 RBS sequence |
|---|---|
| RBS1 | TCGAGGT |
| RBS2 | TCCTGGT |
| RBS4 | AGGAGGT |
| RBS5 | GAGGAGG |
| RBS28 | CTCGTGA |
| RBS166 | TCCTGGA |
| RBS1036 | TCCAGGA |
Table 1 The RBS sequence used in the experiment
RBS名称 RBS name | RBS序列 RBS sequence |
|---|---|
| RBS1 | TCGAGGT |
| RBS2 | TCCTGGT |
| RBS4 | AGGAGGT |
| RBS5 | GAGGAGG |
| RBS28 | CTCGTGA |
| RBS166 | TCCTGGA |
| RBS1036 | TCCAGGA |
启动子-RBS组合 Promoter-RBS groups | 启动子预测强度 Promoter strength | RBS预测强度 RBS strength |
|---|---|---|
| Ptac-RBS2 | 16000 | 500 |
| Pbla-RBS1 | 2000 | 3000 |
| Pbla-RBS2 | 2000 | 500 |
| Ptac-RBS1 | 16000 | 3000 |
Table 2 Prediction strength of multiple promoter and RBS sequences
启动子-RBS组合 Promoter-RBS groups | 启动子预测强度 Promoter strength | RBS预测强度 RBS strength |
|---|---|---|
| Ptac-RBS2 | 16000 | 500 |
| Pbla-RBS1 | 2000 | 3000 |
| Pbla-RBS2 | 2000 | 500 |
| Ptac-RBS1 | 16000 | 3000 |
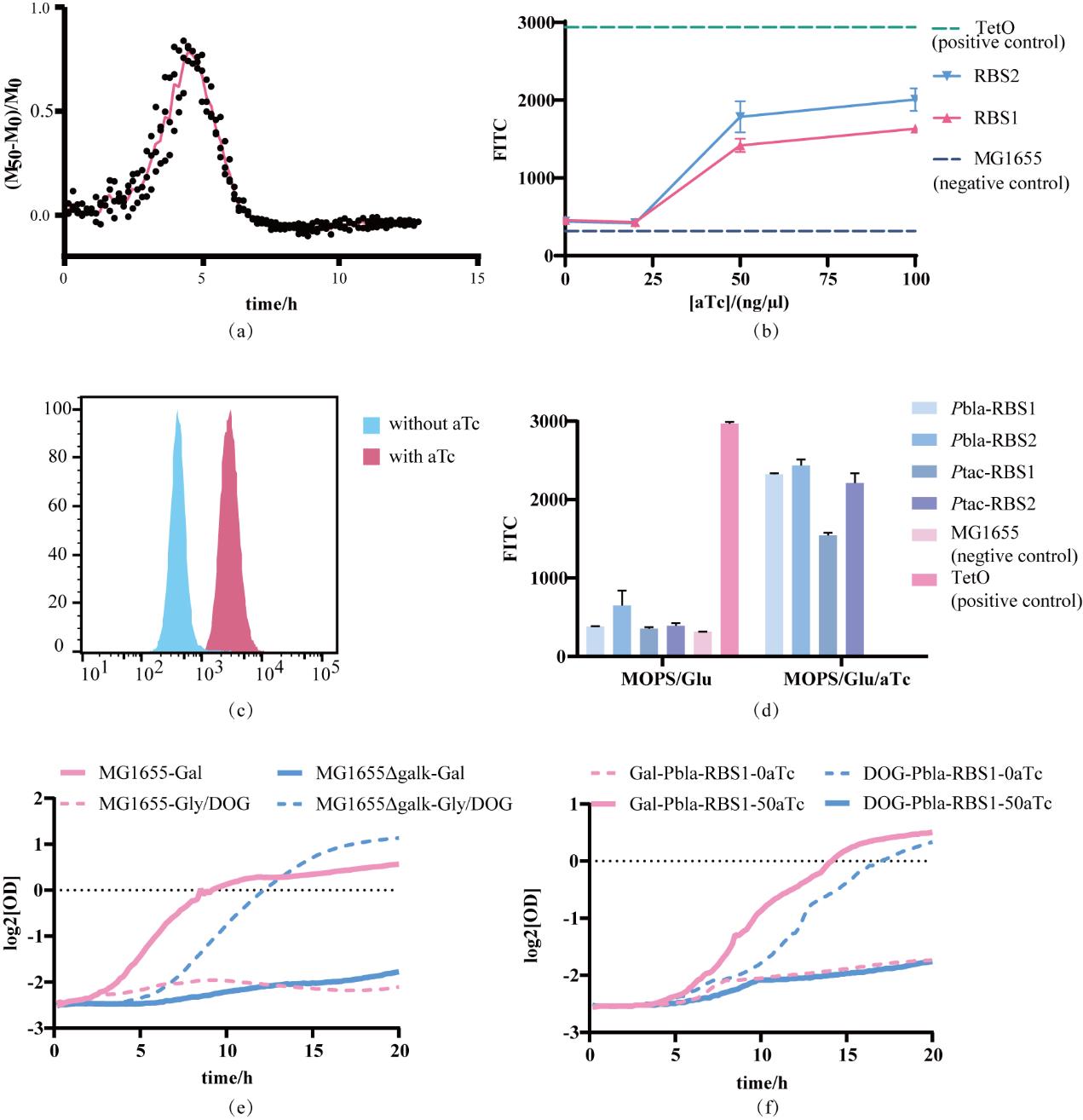
Fig. 2 Construction of the TetR evolution experimental platform(a) The average fluorescence intensity of the population, denoted as M, can be calculated from the fluorescence intensity and absorbance value measured by the microplate reader. Compare the difference of M before and after induction to reflect the regulatory state of the repressor protein; (b) The response curve of TetR to the aTc inducer, with the inducer concentrations being 0, 20, 50, and 100 ng/µl respectively. The dashed lines represent the two control groups. The TetO group consists of PL-tetO-GFP chassis cells, in which GFP is expressed constitutively. The MG1655 group, by contrast, lacks GFP. These two groups represent the upper and lower limits of fluorescence expression intensity within this system, respectively; (c) When the induction concentration is 50 ng/µl, the population fluorescence distribution before and after induction; (d) The variations in fluorescence intensity before and after the induction of TetR proteins with different expression levels, as well as the comparison of these fluorescence intensities with those of the control groups; (e) Use two strains, MG1655 (containing galk) and MG1655Δgalk, to verify the positive and negative screening performance of GalK; (f) The response of Pbla-RBS1-TetR to the positive and negative screening of GalK
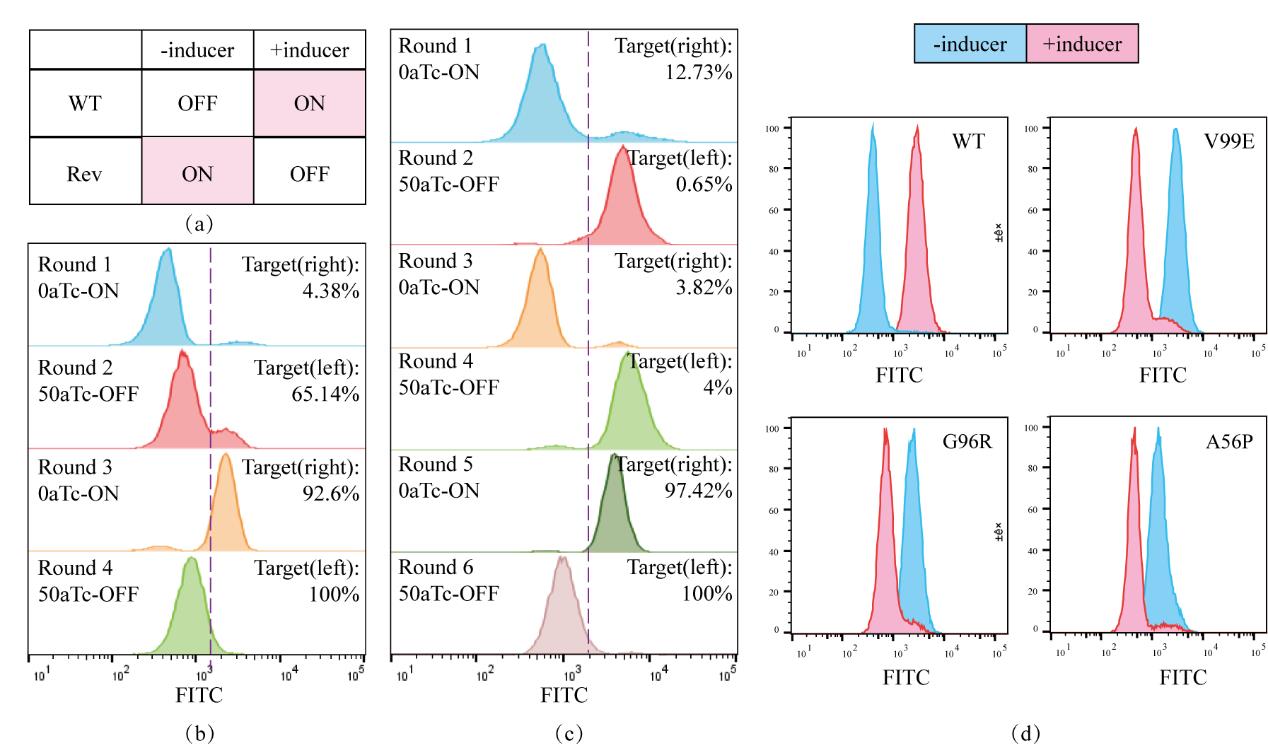
Fig. 3 Screening of the TetR-OFF system(a) The response of the wild-type TetR and the reversed phenotype to the inducer. (b) The screening process using the GalK-GFP dual positive and negative screening. (c) The screening process using only GFP as the screening marker. (d) Characterization of the regulatory performance of the wild-type and TetR-OFF mutants. The blue represents the population fluorescence distribution without the inducer, and the red represents the population fluorescence distribution after induction with 50 ng/µl aTc.
突变工具 Mutation Tool | 组别 Groups | 位点突变 Mutation sites | ||||
|---|---|---|---|---|---|---|
| WT | N18Y | V20D | I22T | L60S | Others | |
| 易错PCR | 1 | 18N | 20V | 22I | 60L | R28H, T202A |
| 2 | 18N | 20V | 22I | |||
| TADR | 3 | 18N | 20V | 22I | Y93C, D95E | |
| 4 | 22I | |||||
Table 3 The mutation sites where revTetR evolved into TetR-ON.
突变工具 Mutation Tool | 组别 Groups | 位点突变 Mutation sites | ||||
|---|---|---|---|---|---|---|
| WT | N18Y | V20D | I22T | L60S | Others | |
| 易错PCR | 1 | 18N | 20V | 22I | 60L | R28H, T202A |
| 2 | 18N | 20V | 22I | |||
| TADR | 3 | 18N | 20V | 22I | Y93C, D95E | |
| 4 | 22I | |||||
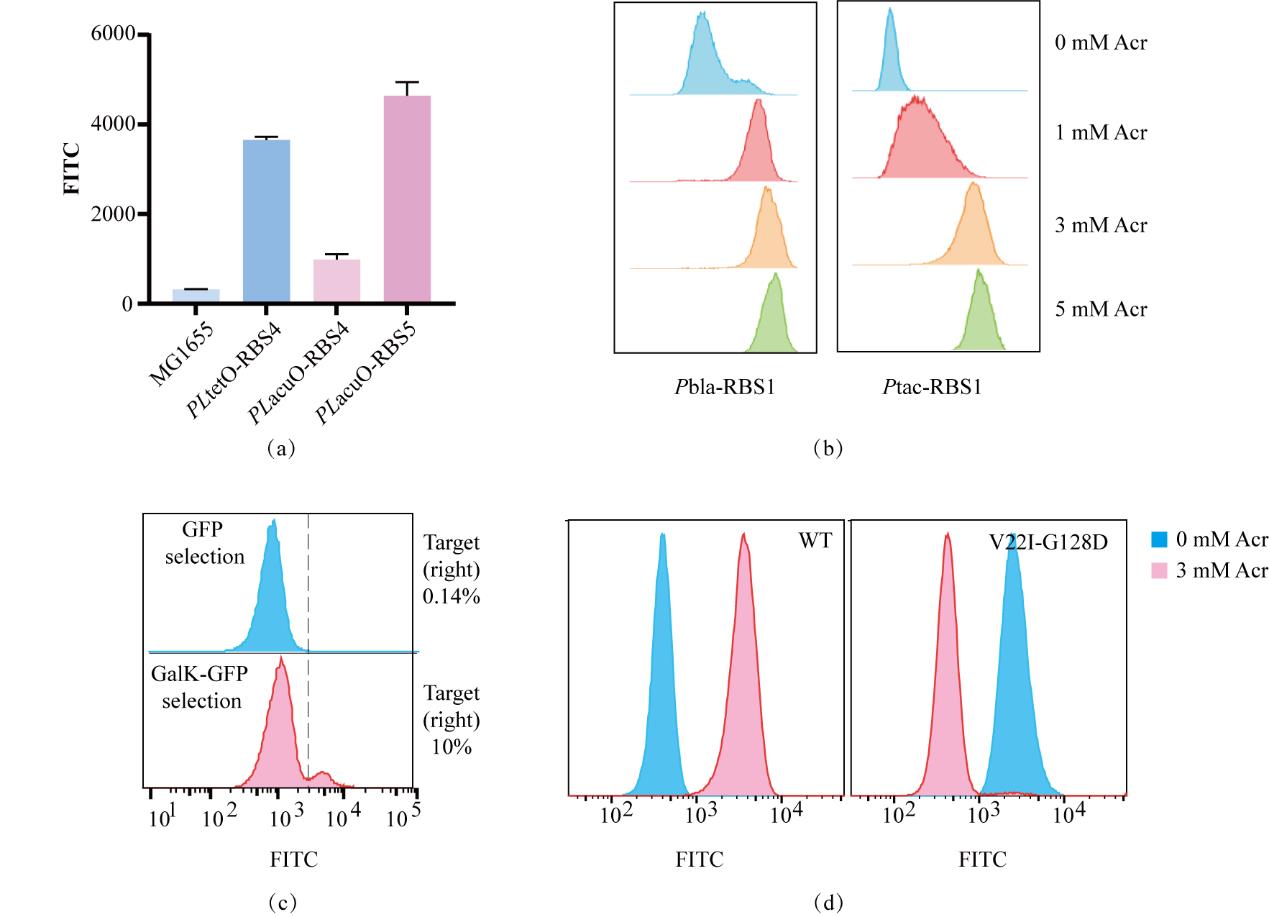
Fig. 4 Construction of the experimental platform for the development of AcuR-OFF.(a) Using MG1655 as the fluorescence negative control and the TetO group as the fluorescence positive control, the AcuO regulatory system was optimized. (b) The fluorescence population distributions of two groups of TetR with different expression intensities were measured under the induction concentration gradients respectively to select the optimal induction conditions. (c) The changes in the proportion of the target population in the absence of the inducer before and after GalK screening. The blue color represents the population fluorescence distribution before GalK screening, and the red color represents the population fluorescence distribution after GalK screening. (d) Characterization of the regulatory performance of the wild type and the AcuR-OFF mutant. The blue color represents the population fluorescence distribution in the absence of the inducer, and the red color represents the population fluorescence distribution after induction with 3 mM Acr.
| 1 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 2 | CAMPS M, NAUKKARINEN J, JOHNSON B P, et al. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(17): 9727-9732. |
| 3 | CROOK N, ABATEMARCO J, SUN J, et al. In vivo continuous evolution of genes and pathways in yeast[J]. Nature Communications, 2016, 7: 13051. |
| 4 | ENGLISH J G, OLSEN R H J, LANSU K, et al. VEGAS as a platform for facile directed evolution in mammalian cells[J]. Cell, 2019, 178(3): 748-761.e17. |
| 5 | ESVELT K M, CARLSON J C, LIU D R. A system for the continuous directed evolution of biomolecules[J]. Nature, 2011, 472(7344): 499-503. |
| 6 | FAURE G, SAITO M, WILKINSON M E, et al. TIGR-Tas: a family of modular RNA-guided DNA-targeting systems in prokaryotes and their viruses[J]. Science, 2025: eadv9789. |
| 7 | HALPERIN S O, TOU C J, WONG E B, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560(7717): 248-252. |
| 8 | HESS G T, FRÉSARD L, HAN K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells[J]. Nature Methods, 2016, 13(12): 1036-1042. |
| 9 | MOORE C L, PAPA L J 3 RD, SHOULDERS M D. A processive protein Chimera introduces mutations across defined DNA regions in vivo [J]. Journal of the American Chemical Society, 2018, 140(37): 11560-11564. |
| 10 | NYERGES Á, CSÖRGŐ B, DRASKOVITS G, et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(25): E5726-E5735. |
| 11 | RAVIKUMAR A, ARZUMANYAN G A, OBADI M K A, et al. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds[J]. Cell, 2018, 175(7): 1946-1957.e13. |
| 12 | SIMON A J, MORROW B R, ELLINGTON A D. Retroelement-based genome editing and evolution[J]. ACS Synthetic Biology, 2018, 7(11): 2600-2611. |
| 13 | TIAN R Z, ZHAO R Z, GUO H Y, et al. Engineered bacterial orthogonal DNA replication system for continuous evolution[J]. Nature Chemical Biology, 2023, 19(12): 1504-1512. |
| 14 | YI X, KAZLAUSKAS R, TRAVISANO M. Evolutionary innovation using EDGE, a system for localized elevated mutagenesis[J]. PLoS One, 2020, 15(4): e0232330. |
| 15 | YI X, KHEY J, KAZLAUSKAS R J, et al. Plasmid hypermutation using a targeted artificial DNA replisome[J]. Science Advances, 2021, 7(29): eabg8712. |
| 16 | WILSON D S, KEEFE A D. Random mutagenesis by PCR[J]. Current Protocols in Molecular Biology, 2001, Chapter 8: Unit8.3. |
| 17 | MCCULLUM E O, WILLIAMS B A R, ZHANG J L, et al. Random mutagenesis by error-prone PCR[M/OL]//BRAMAN J. Methods in molecular biology: in vitro mutagenesis protocols. third edition. Totowa, NJ: Humana Press, 2010, 634: 103-109. (2010-03-19)[2024-02-01]. . |
| 18 | IKE K, ARASAWA Y, KOIZUMI S, et al. Evolutionary design of choline-inducible and-repressible T7-based induction systems[J]. ACS Synthetic Biology, 2015, 4(12): 1352-1360. |
| 19 | RKENES T P, LAMARK T, STRØM A R. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation[J]. Journal of Bacteriology, 1996, 178(6): 1663-1670. |
| 20 | LI X T, THOMASON L C, SAWITZKE J A, et al. Positive and negative selection using the TetA-sacB cassette: recombineering and P1 transduction in Escherichia coli [J]. Nucleic Acids Research, 2013, 41(22): e204. |
| 21 | RYU Y S, CHANDRAN S P, KIM K, et al. Oligo- and dsDNA-mediated genome editing using a tetA dual selection system in Escherichia coli [J]. PLoS One, 2017, 12(7): e0181501. |
| 22 | STAVROPOULOS T A, STRATHDEE C A. Synergy between tetA and rpsL provides high-stringency positive and negative selection in bacterial artificial chromosome vectors[J]. Genomics, 2001, 72(1): 99-104. |
| 23 | BISWAS K, STAUFFER S, SHARAN S K. Using recombineering to generate point mutations: galK-based positive-negative selection method[M/OL]// PECCOUD J. Methods in molecular biology: gene synthesis. Totowa, NJ: Humana Press, 2012, 852: 121-131. (2012-01-01)[2024-02-01]. . |
| 24 | POELWIJK F J, DE VOS M G J, TANS S J. Tradeoffs and optimality in the evolution of gene regulation[J]. Cell, 2011, 146(3): 462-470. |
| 25 | SAEKI K, TOMINAGA M, KAWAI-NOMA S, et al. Rapid diversification of BetI-based transcriptional switches for the control of biosynthetic pathways and genetic circuits[J]. ACS Synthetic Biology, 2016, 5(11): 1201-1210. |
| 26 | WARMING S, COSTANTINO N, COURT D L, et al. Simple and highly efficient BAC recombineering using galK selection[J]. Nucleic Acids Research, 2005, 33(4): e36. |
| 27 | DABIRIAN Y, GONÇALVES TEIXEIRA P, NIELSEN J, et al. FadR-based biosensor-assisted screening for genes enhancing fatty acyl-CoA pools in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2019, 8(8): 1788-1800. |
| 28 | XIAO Y, BOWEN C H, LIU D, et al. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis[J]. Nature Chemical Biology, 2016, 12(5): 339-344. |
| 29 | XU X H, LI X L, LIU Y F, et al. Pyruvate-responsive genetic circuits for dynamic control of central metabolism[J]. Nature Chemical Biology, 2020, 16(11): 1261-1268. |
| 30 | DAS A T, TENENBAUM L, BERKHOUT B. Tet-on systems for doxycycline-inducible gene expression[J]. Current Gene Therapy, 2016, 16(3): 156-167. |
| 31 | GOSSEN M, BUJARD H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters[J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89(12): 5547-5551. |
| 32 | RAMOS J L, MARTÍNEZ-BUENO M, MOLINA-HENARES A J, et al. The TetR family of transcriptional repressors[J]. Microbiology and Molecular Biology Reviews, 2005, 69(2): 326-356. |
| 33 | CUTHBERTSON L, NODWELL J R. The TetR family of regulators[J]. Microbiology and Molecular Biology Reviews, 2013, 77(3): 440-475. |
| 34 | BHUKYA H, ANAND R. TetR regulators: a structural and functional perspective[J]. Journal of the Indian Institute of Science, 2017, 97(2): 245-259. |
| 35 | BERENS C, HILLEN W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes[J]. European Journal of Biochemistry, 2003, 270(15): 3109-3121. |
| 36 | BERTRAM R, HILLEN W. The application of Tet repressor in prokaryotic gene regulation and expression[J]. Microbial Biotechnology, 2008, 1(1): 2-16. |
| 37 | MOL A A, GROHER F, SCHREIBER B, et al. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module[J]. Nucleic Acids Research, 2019, 47(20): e132. |
| 38 | HE S F, ZHANG Z W, LU W Y. Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae [J]. Journal of Industrial Microbiology & Biotechnology, 2023, 50(1): kuac029. |
| 39 | SCHOLZ O, HENßLER E M, BAIL J, et al. Activity reversal of Tet repressor caused by single amino acid exchanges[J]. Molecular Microbiology, 2004, 53(3): 777-789. |
| 40 | SULLIVAN M J, CURSON A R J, SHEARER N, et al. Unusual regulation of a leaderless operon involved in the catabolism of dimethylsulfoniopropionate in Rhodobacter sphaeroides [J]. PLoS One, 2011, 6(1): e15972. |
| 41 | HARMON D E, RUIZ C. The multidrug efflux regulator AcrR of Escherichia coli responds to exogenous and endogenous ligands to regulate efflux and detoxification[J]. mSphere, 2022, 7(6): e00474-22. |
| 42 | GRKOVIC S, BROWN M H, SKURRAY R A. Transcriptional regulation of multidrug efflux pumps in bacteria[J]. Seminars in Cell & Developmental Biology, 2001, 12(3): 225-237. |
| 43 | ROGERS J K, GUZMAN C D, TAYLOR N D, et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites[J]. Nucleic Acids Research, 2015, 43(15): 7648-7660. |
| 44 | JONES K A, SNODGRASS H M, BELSARE K, et al. Phage-assisted continuous evolution and selection of enzymes for chemical synthesis[J]. ACS Central Science, 2021, 7(9): 1581-1590. |
| 45 | ROGERS J K, CHURCH G M. Genetically encoded sensors enable real-time observation of metabolite production[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(9): 2388-2393. |
| [1] | GUO Liang, GAO Cong, LIU Yadi, CHEN Xiulai, LIU Liming. Advances in bioproduction of feed amino acid by Escherichia coli [J]. Synthetic Biology Journal, 2021, 2(6): 964-981. |
| [2] | WEI Xiaolian, QIAN Zhiling, CHEN Qiaoqiao, YU Hongwei. Regulatory requirements for food and feed produced with genetically modified microorganisms and case studies for EU authorization [J]. Synthetic Biology Journal, 2021, 2(1): 121-133. |
| [3] | CHU Pan, ZHU Jingwen, HUANG Wenqi, LIU Chenli, FU Xiongfei. Host-circuit coupling: toward a new framework for genetic circuit design [J]. Synthetic Biology Journal, 2021, 2(1): 91-105. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||