| [1] |
Rutten M.G.T.A., et al., Encoding information into polymers. Nature Reviews Chemistry, 2018. 2(11): p. 365-381.
|
| [2] |
Zhirnov V., et al., Nucleic acid memory. Nature Materials, 2016. 15(4): p. 366-370.
|
| [3] |
Organick L., et al., Random access in large-scale DNA data storage. Nature Biotechnology, 2018. 36(3): p. 242-248.
|
| [4] |
Davis J., Microvenus. Art Journal, 1996. 55(1): p. 70-74.
|
| [5] |
Church G.M., Gao Y., and Kosuri S., Next-Generation Digital Information Storage in DNA. Science, 2012. 337(6102): p. 1628-1628.
|
| [6] |
Goldman N., et al., Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature, 2013. 494(7435): p. 77-80.
|
| [7] |
Oligonucleotides and Analogues: A Practical Approach, ed. F. Eckstein. 1991: Oxford University Press.
|
| [8] |
Rayner S., et al., MerMade: an oligodeoxyribonucleotide synthesizer for high throughput oligonucleotide production in dual 96-well plates. Genome Res, 1998. 8(7): p. 741-7.
|
| [9] |
Cheng J.Y., et al., High throughput parallel synthesis of oligonucleotides with 1536 channel synthesizer. Nucleic Acids Research, 2002. 30(18): p. e93-e93.
|
| [10] |
Richmond K.E., et al., Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Research, 2004. 32(17): p. 5011-5018.
|
| [11] |
Lee H.H., et al., Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nature Communications, 2019. 10(1): p. 2383.
|
| [12] |
Chang L.M.S., B.F. J., and R.C. and Gallo, Molecular Biology of Terminal Transferas. Critical Reviews in Biochemistry, 1986. 21(1): p. 27-52.
|
| [13] |
Motea E.A. and Berdis A.J., Terminal deoxynucleotidyl transferase: The story of a misguided DNA polymerase. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2010. 1804(5): p. 1151-1166.
|
| [14] |
Rasool A., et al., BO-DNA: Biologically optimized encoding model for a highly-reliable DNA data storage. Computers in Biology and Medicine, 2023. 165.
|
| [15] |
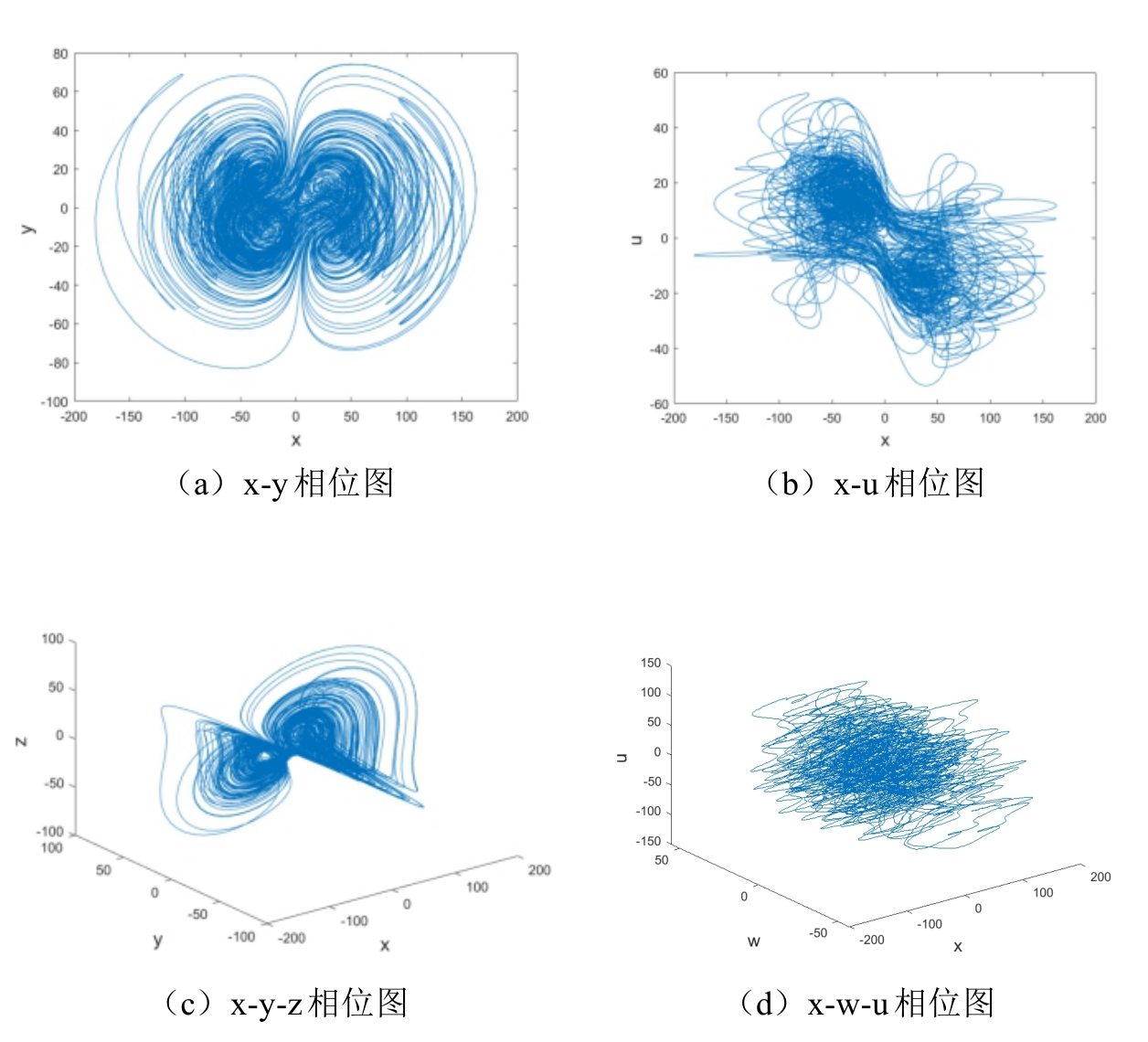
袁, 涛., 强. 曲, and 姜青山, 基于混沌系统和喷泉码的 DNA 加密编码方法. 集成技术, 2024. 13(3): p. 4-24.
|
| [16] |
Abbasi F., et al., SNCA: Semi-Supervised Node Classification for Evolving Large Attributed Graphs. Big Data Mining and Analytics, 2024. 7(3): p. 794-808.
|
| [17] |
Li M., et al., A self-contained and self-explanatory DNA storage system. Scientific Reports, 2021. 11(1).
|
| [18] |
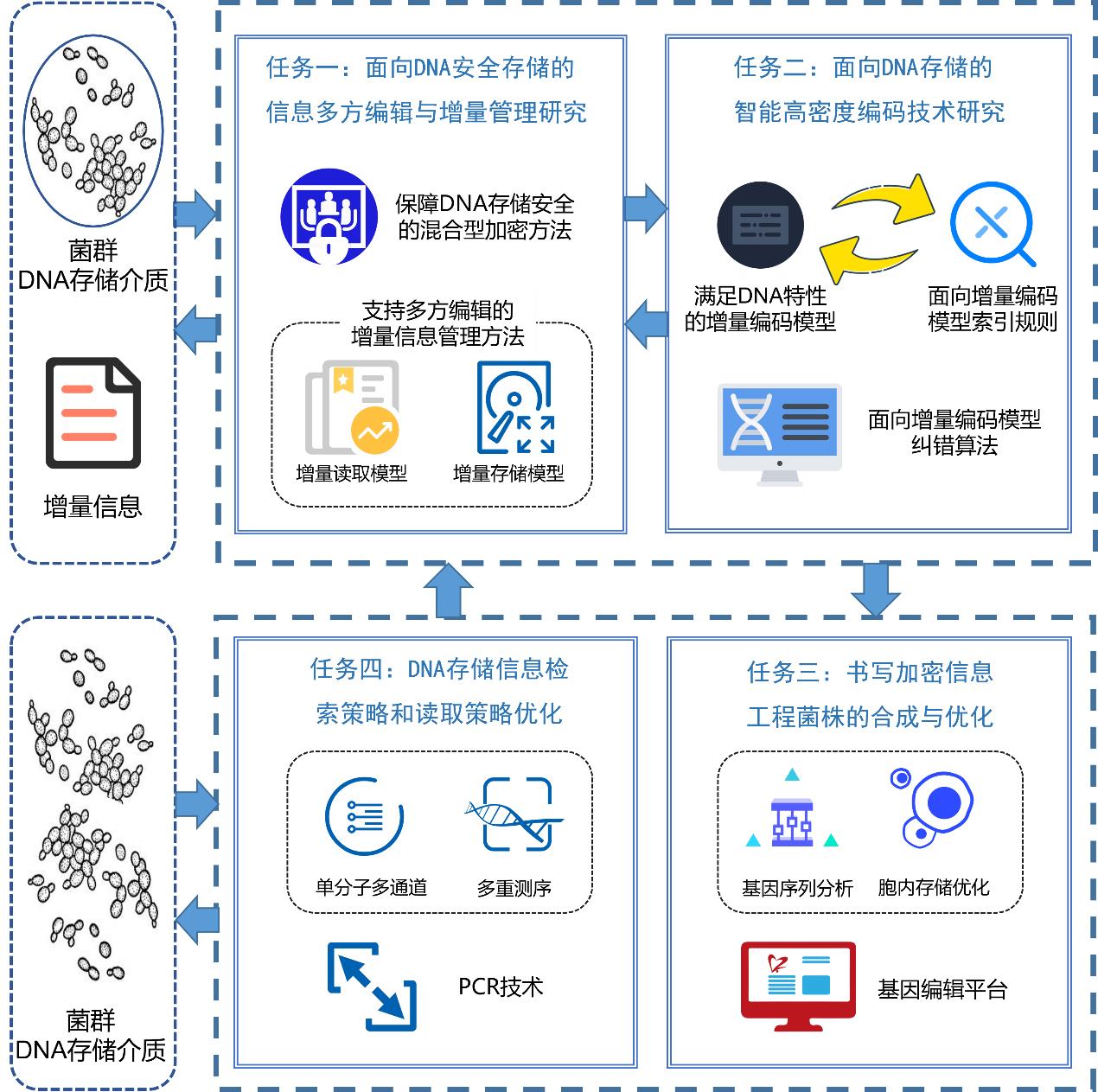
袁涛 and 曲强, 面向DNA安全存储的信息增量管理方法. 集成技术, 2025. 14(3): p. 38-50.
|
| [19] |
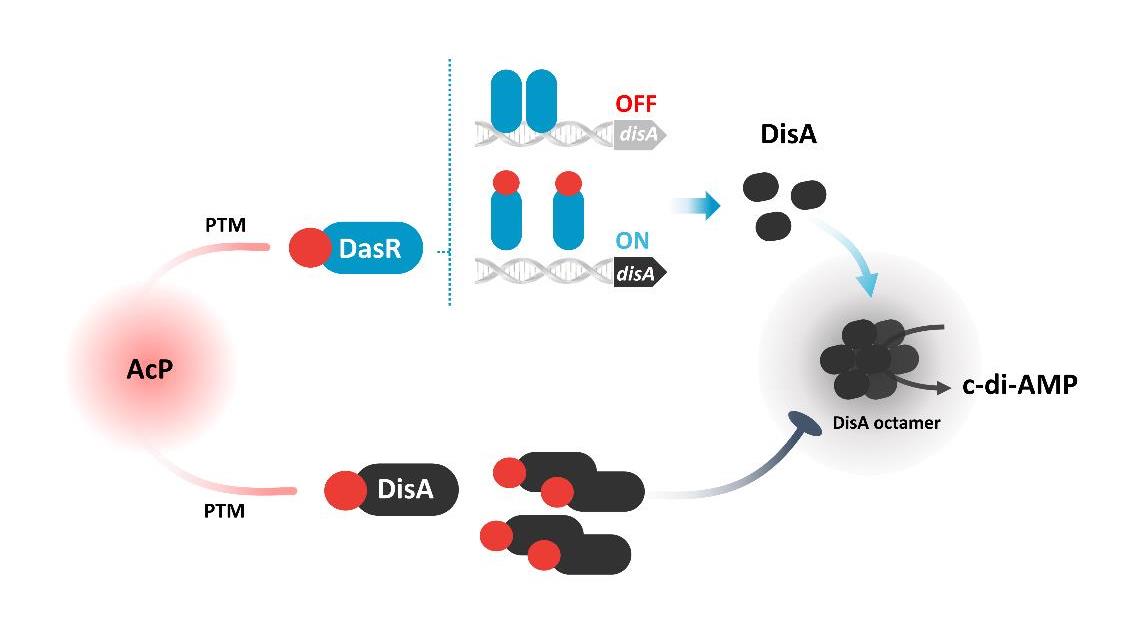
Fu Y., et al., A network of acetyl phosphate-dependent modification modulates c-di-AMP homeostasis in <i>Actinobacteria</i>. mBio, 2024. 15(8): p. e01411-24.
|
| [20] |
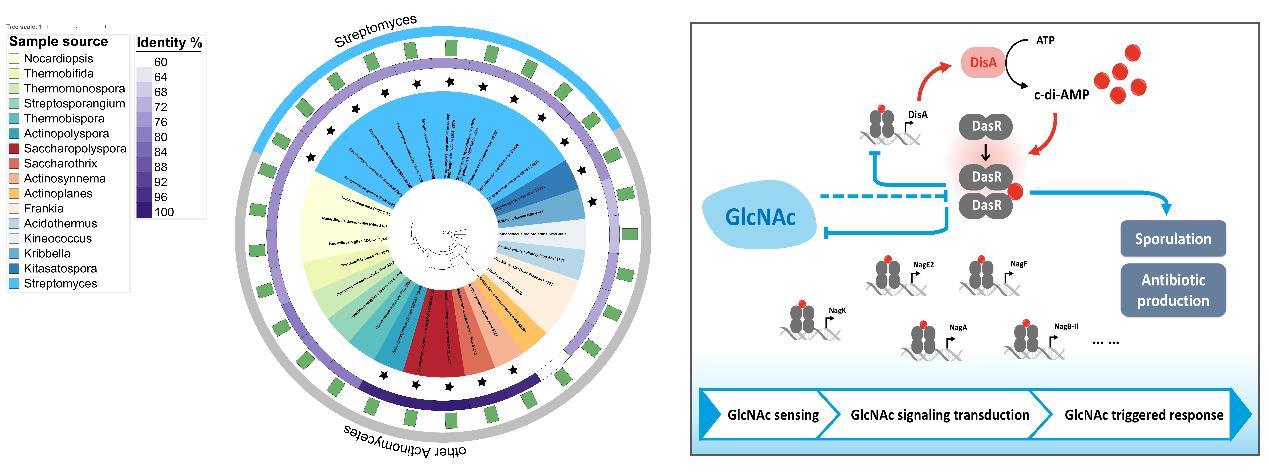
You D., et al., Allosteric regulation by c-di-AMP modulates a complete N-acetylglucosamine signaling cascade in Saccharopolyspora erythraea. Nature Communications, 2024. 15(1): p. 3825.
|
| [21] |
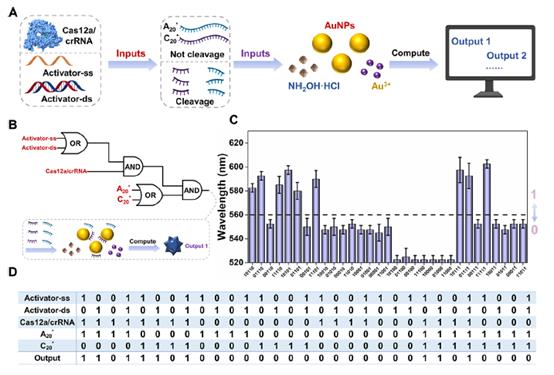
Fu R., et al., DNA Molecular Computation Using the CRISPR-Mediated Reaction and Surface Growth of Gold Nanoparticles. ACS Nano, 2024. 18(22): p. 14754-14763.
|
| [22] |
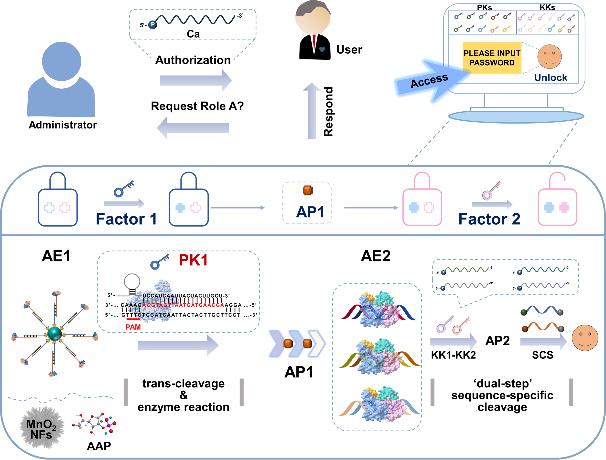
Fu R., et al., A CRISPR-Cas and Argonaute-Driven Two-Factor Authentication Strategy for Information Security. ACS Nano, 2025. 19(4): p. 4983-4992.
|
| [23] |
Rasool A., et al., An Effective DNA‐Based File Storage System for Practical Archiving and Retrieval of Medical MRI Data. Small Methods, 2024. 8(10):2301585.
|
| [24] |
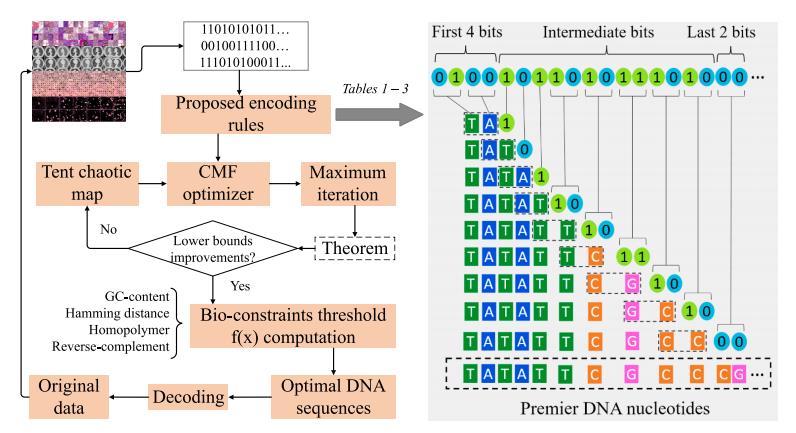
Rasool A., Qu Q., Wang Y., and Jiang Q., Bio-constrained codes with neural network for density-based DNA data storage. Mathematics, 2022. 10 (5): 845.
|