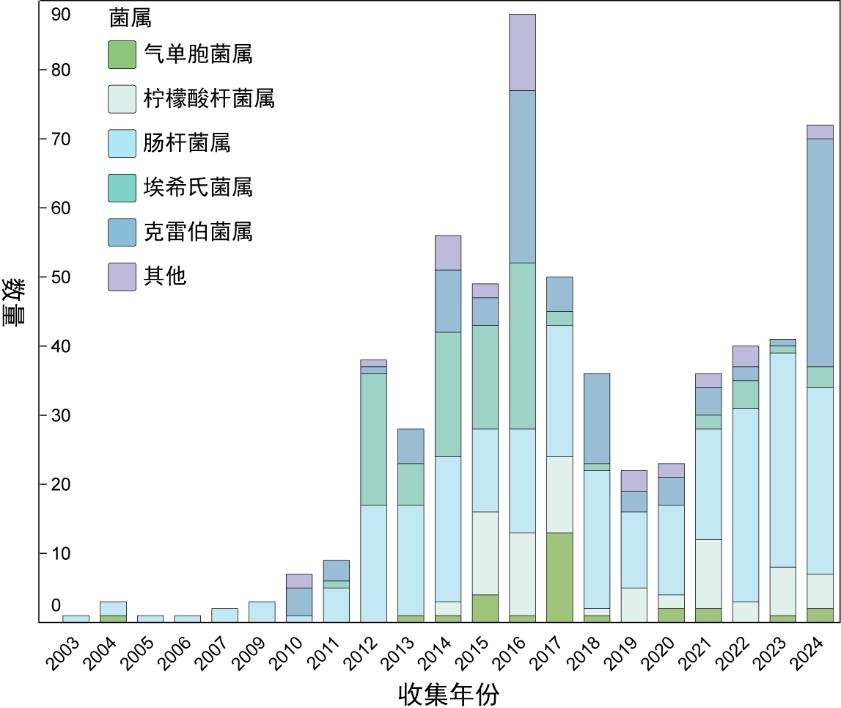
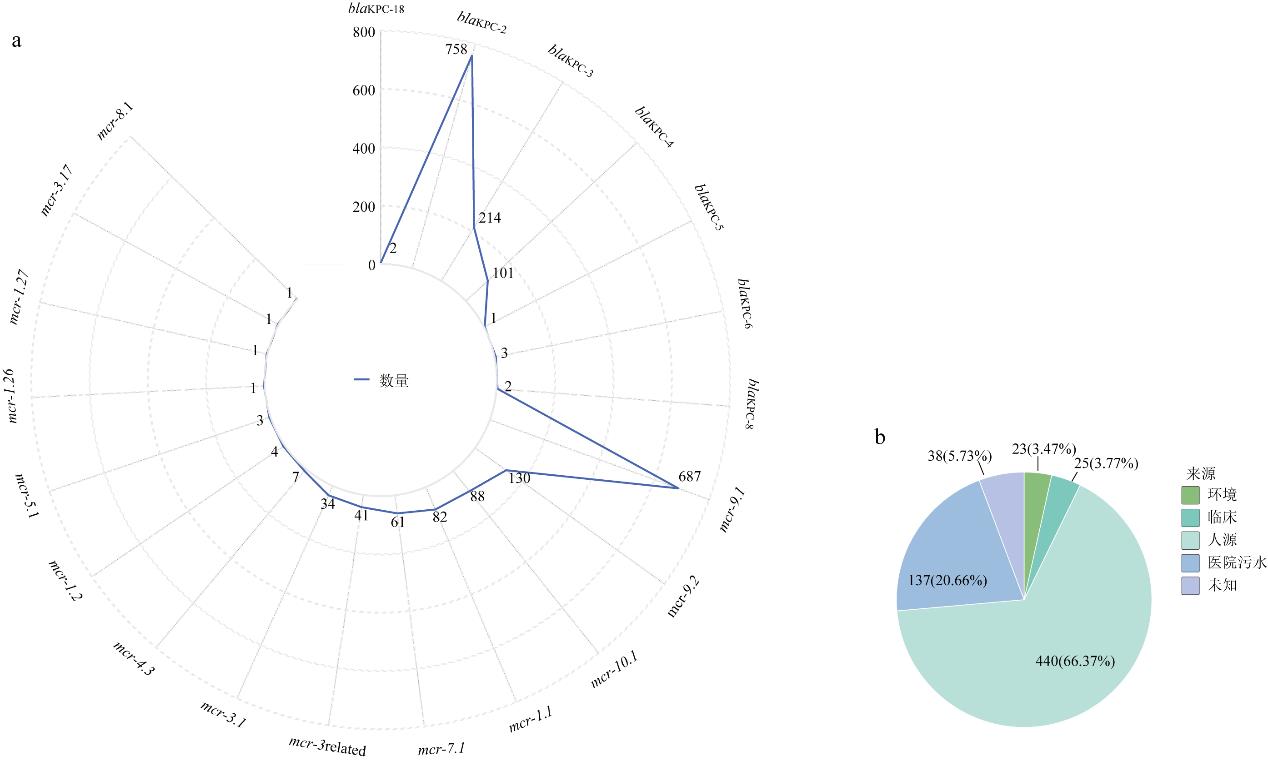
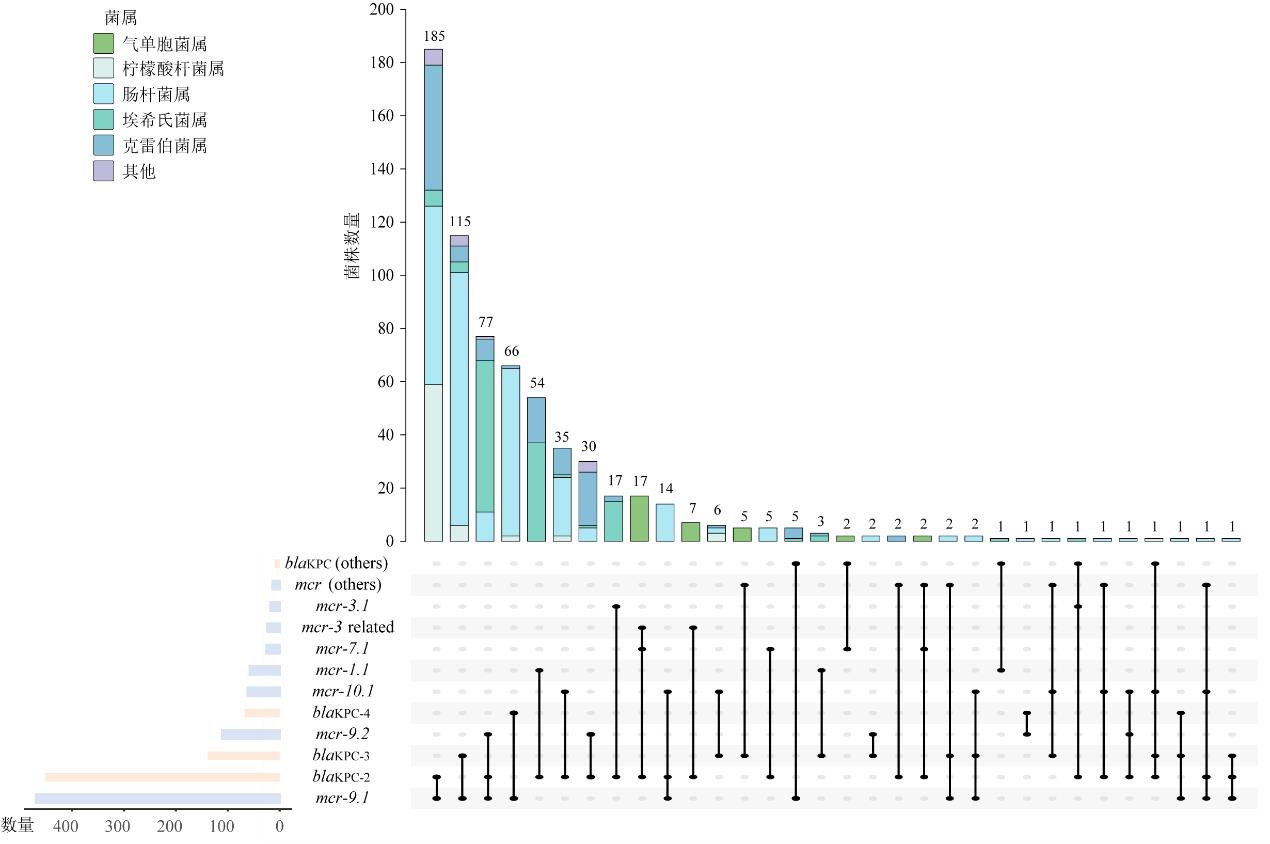
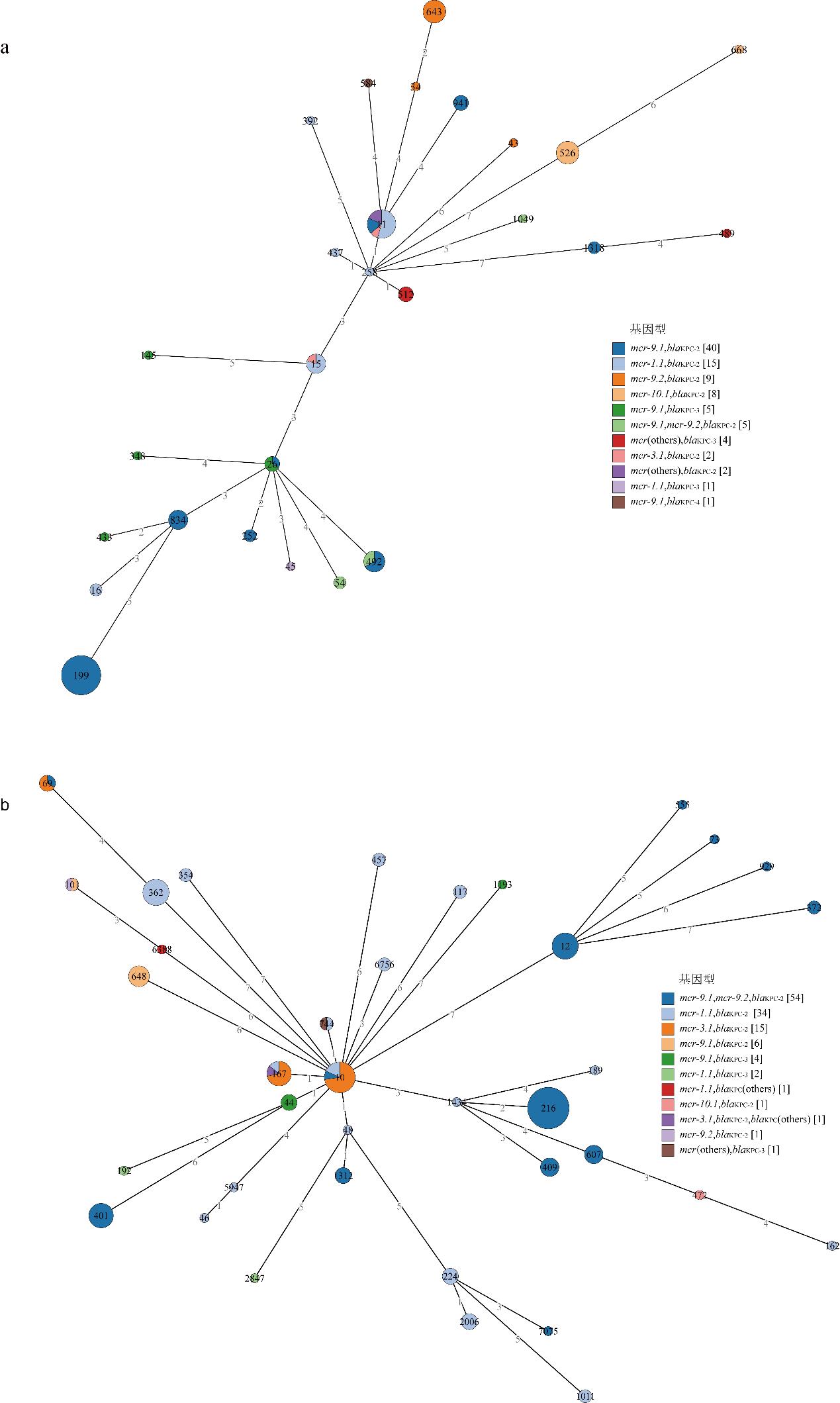
| [1] |
ZHU L, LI P, ZHANG G, et al. Role of the ISKpn element in mediating mgrB gene mutations in ST11 hypervirulent colistin-resistant Klebsiella pneumoniae [J]. Frontiers in Microbiology, 2023, 14: 1277320.
|
| [2] |
HUSSEIN NH, AL-KADMY IMS, TAHA BM, et al. Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review[J]. Molecular Biology Reports, 2021, 48(3): 2897-2907.
|
| [3] |
YU H, QU F, SHAN B, et al. Detection of the mcr-1 Colistin Resistance Gene in Carbapenem-Resistant Enterobacteriaceae from Different Hospitals in China[J]. Antimicrobial Agents and Chemotherapy, 2016, 60(8): 5033-5.
|
| [4] |
FALGENHAUER L, WAEZSADA SE, YAO Y, et al. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany[J]. The Lancet Infectious Diseases, 2016, 16(3): 282-3.
|
| [5] |
POIREL L, KIEFFER N, LIASSINE N, et al. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli [J]. The Lancet Infectious Diseases, 2016, 16(3): 281.
|
| [6] |
HASMAN H, HAMMERUM AM, HANSEN F, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015[J]. Eurosurveillance, 2015, 20(49).
|
| [7] |
BRATU S, LANDMAN D, ALAM M, et al. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York[J]. Antimicrobial Agents and Chemotherapy, 2005, 49(2): 776-8.
|
| [8] |
MUNOZ-PRICE LS, POIREL L, BONOMO RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases[J]. The Lancet Infectious Diseases, 2013, 13(9): 785-96.
|
| [9] |
DING L, SHEN S, CHEN J, et al. Klebsiella pneumoniae carbapenemase variants: the new threat to global public health[J]. Clinical Microbiology Reviews, 2023, 36(4): e0000823.
|
| [10] |
CHEN L, MATHEMA B, CHAVDA KD, et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding[J]. Trends in Microbiology, 2014, 22(12): 686-96.
|
| [11] |
CAI M, SONG K, WANG R, et al. Tracking intra-species and inter-genus transmission of KPC through global plasmids mining[J]. Cell Reports, 2024, 43(6): 114351.
|
| [12] |
WU R, YI LX, YU LF, et al. Fitness Advantage of mcr-1-Bearing IncI2 and IncX4 Plasmids in Vitro[J]. Frontiers in Microbiology, 2018, 9: 331.
|
| [13] |
HADJADJ L, RIZIKI T, ZHU Y, et al. Study of mcr-1 Gene-Mediated Colistin Resistance in Enterobacteriaceae Isolated from Humans and Animals in Different Countries[J]. Genes (Basel), 2017, 8(12).
|
| [14] |
MATAMOROS S, VAN HATTEM JM, ARCILLA MS, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction[J]. Scientific Reports, 2017, 7(1): 15364.
|
| [15] |
MACESIC N, BLAKEWAY LV, STEWART JD, et al. Silent spread of mobile colistin resistance gene mcr-9.1 on IncHI2 'superplasmids' in clinical carbapenem-resistant Enterobacterales [J]. Clinical Microbiology and Infection, 2021, 27(12): 1856.e7-1856.e13.
|
| [16] |
KUMAR S, STECHER G, LI M, et al. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547-1549.
|
| [17] |
FELDGARDEN M, BROVER V, HAFT DH, et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates[J]. Antimicrobial Agents and Chemotherapy, 2019, 63(11).
|
| [18] |
KRAWCZYK PS, LIPINSKI L, DZIEMBOWSKI A. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures[J]. Nucleic Acids Research, 2018, 46(6): e35.
|
| [19] |
CARATTOLI A, ZANKARI E, GARCÍA-FERNÁNDEZ A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing[J]. Antimicrobial Agents and Chemotherapy, 2014, 58(7): 3895-903.
|
| [20] |
SCHWENGERS O, JELONEK L, DIECKMANN MA, et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification[J]. Microbial Genomics, 2021, 7(11).
|
| [21] |
SULLIVAN MJ, PETTY NK, BEATSON SA. Easyfig: a genome comparison visualizer[J]. Bioinformatics, 2011, 27(7): 1009-10.
|
| [22] |
ZHOU Z, ALIKHAN NF, SERGEANT MJ, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens[J]. Genome Research, 2018, 28(9): 1395-1404.
|
| [23] |
DORMAN CJ. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria[J]. Plasmid, 2014, 75: 1-11.
|
| [24] |
NAVARRE WW, MCCLELLAND M, LIBBY SJ, et al. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA[J]. Genes & Development, 2007, 21(12): 1456-71.
|
| [25] |
HAN X, ZHOU J, YU L, et al. Genome sequencing unveils blaKPC-2-harboring plasmids as drivers of enhanced resistance and virulence in nosocomial Klebsiella pneumoniae [J]. Msystems, 2024, 9(2): e0092423.
|
| [26] |
ČUROVÁ K, SLEBODNÍKOVÁ R, KMEŤOVÁ M, et al. Virulence, phylogenetic background and antimicrobial resistance in Escherichia coli associated with extraintestinal infections[J]. Journal of Infection and Public Health, 2020, 13(10): 1537-1543.
|
| [27] |
TSENG C C, LIN W H, WU A B, et al. Escherichia coli FimH adhesins act synergistically with PapGII adhesins for enhancing establishment and maintenance of kidney infection[J]. Journal of Microbiology Immunology and Infection, 2022, 55(1): 44-50.
|
| [28] |
GARNETT J A, MARTÍNEZ-SANTOS V I, SALDAÑA Z, et al. Structural insights into the biogenesis and biofilm formation by the Escherichia coli common pilus[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(10): 3950-5.
|
| [29] |
GOOTZ TD, LESCOE MK, DIB-HAJJ F, et al. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital[J]. Antimicrobial Agents and Chemotherapy, 2009, 53(5): 1998-2004.
|
| [30] |
LING Z, YIN W, SHEN Z, et al. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9 [J]. Journal of Antimicrobial Chemotherapy, 2020, 75(11): 3087-3095.
|
| [31] |
LUO Q, WU Y, BAO D, et al. Genomic epidemiology of mcr carrying multidrug-resistant ST34 Salmonella enterica serovar Typhimurium in a one health context: The evolution of a global menace[J]. Science of The Total Environment, 2023, 896: 165203.
|
| [32] |
WANG C, FENG Y, LIU L, et al. Identification of novel mobile colistin resistance gene mcr-10 [J]. Emerging Microbes & Infections, 2020, 9(1): 508-516.
|
| [33] |
CASTILLO F, BENMOHAMED A, SZATMARI G. Xer Site Specific Recombination: Double and Single Recombinase Systems[J]. Frontiers in Microbiology, 2017, 8: 453.
|
| [34] |
ZHU Y, GALANI I, KARAISKOS I, et al. Multifaceted mechanisms of colistin resistance revealed by genomic analysis of multidrug-resistant Klebsiella pneumoniae isolates from individual patients before and after colistin treatment[J]. Journal of Infectious Diseases, 2019, 79(4): 312-321.
|
| [35] |
LI W, HE Z, DI W, et al. Transposition mechanism of ISApl1-the determinant of colistin resistance dissemination[J]. Antimicrobial Agents and Chemotherapy, 2024, 68(3): e0123123.
|
| [36] |
WANG Y, LIU H, WANG Q, et al. Fusion event mediated by IS903B between chromosome and plasmid in two MCR-9- and KPC-2-co-producing Klebsiella pneumoniae isolates[J]. Journal of Global Antimicrobial Resistance, 2024, 77: 101139.
|
| [37] |
YUAN Q, XIA P, XIONG L, et al. First report of coexistence of blaKPC-2-, blaNDM-1- and mcr-9-carrying plasmids in a clinical carbapenem-resistant Enterobacter hormaechei isolate[J]. Frontiers in Microbiology, 2023, 14: 1153366.
|
| [38] |
ZHU Z, WU S, ZHU J, et al. Emergence of Aeromonas veronii strain co-harboring blaKPC-2, mcr-3.17, and tmexC3.2-tmexD3.3-toprJ1b cluster from hospital sewage in China[J]. Frontiers in Microbiology, 2023, 14: 1115740.
|
| [39] |
WANG Y, LIU H, WANG Q, et al. Coexistence of blaKPC-2-IncN and mcr-1-IncX4 plasmids in a ST48 Escherichia coli strain in China[J]. Journal of Global Antimicrobial Resistance, 2020, 23: 149-153.
|
 ), HU Gui-fang1, HE Ya-qing1,2
), HU Gui-fang1, HE Ya-qing1,2
 ), 胡贵方1, 何雅青1,2
), 胡贵方1, 何雅青1,2