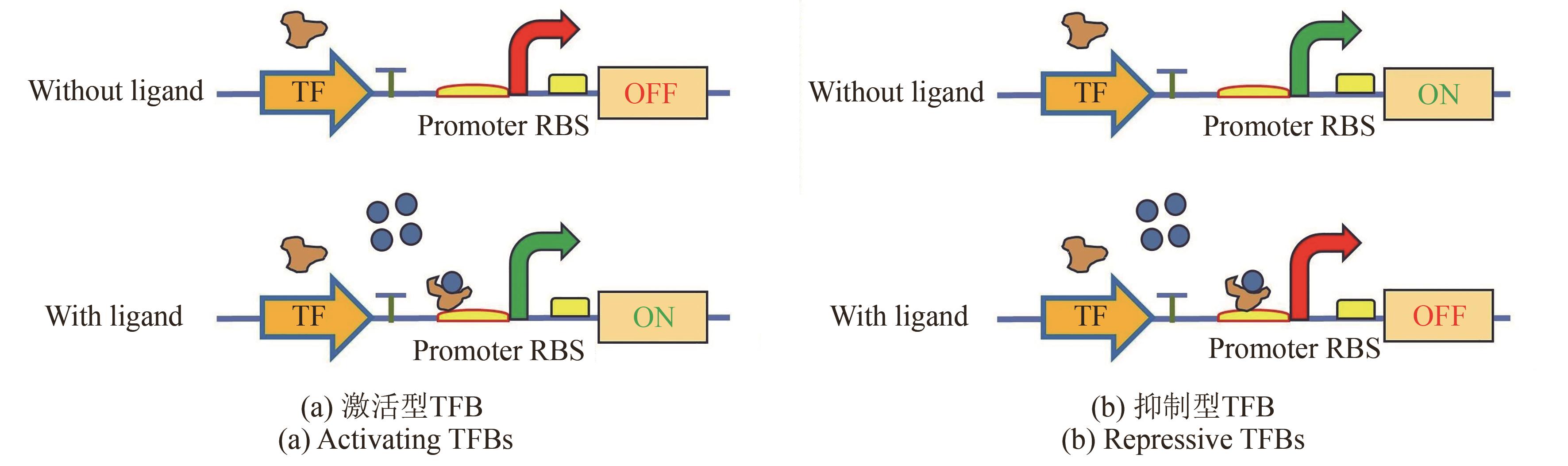
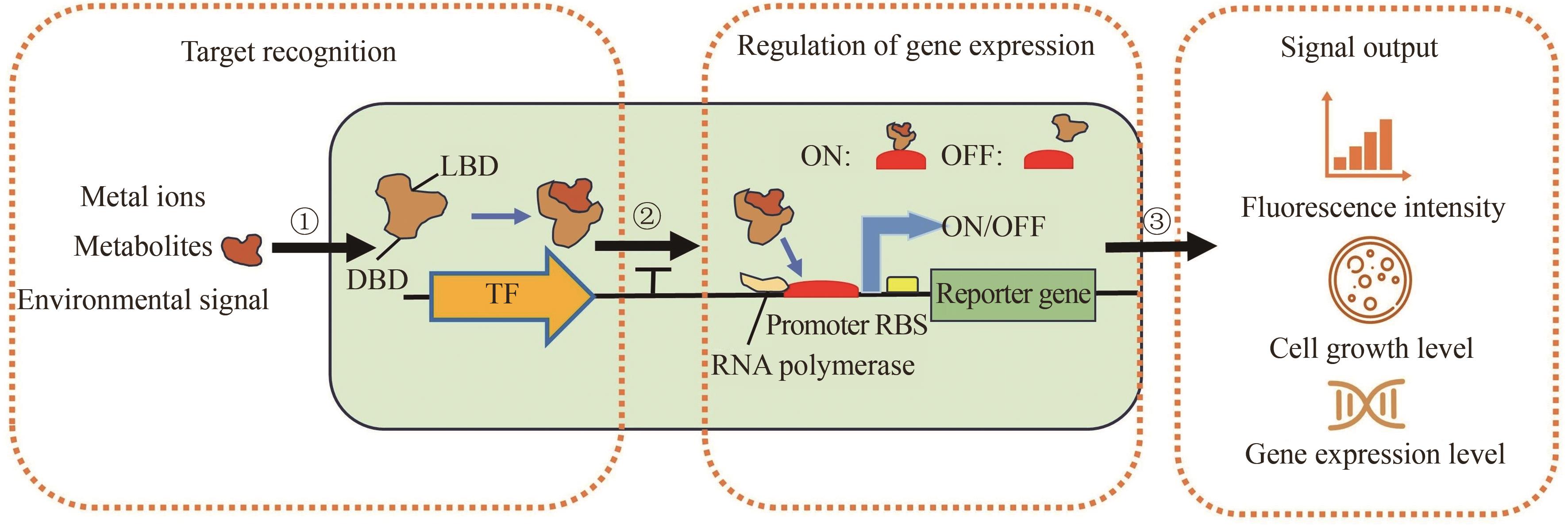
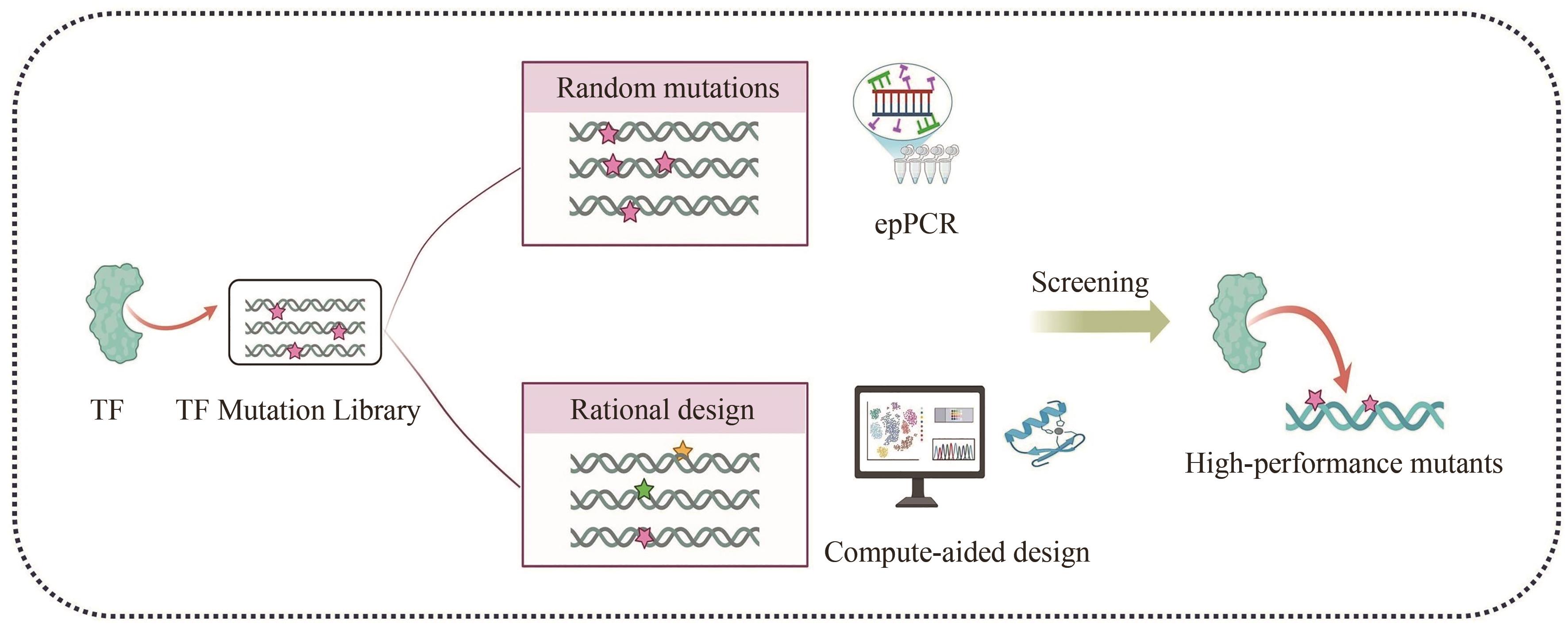
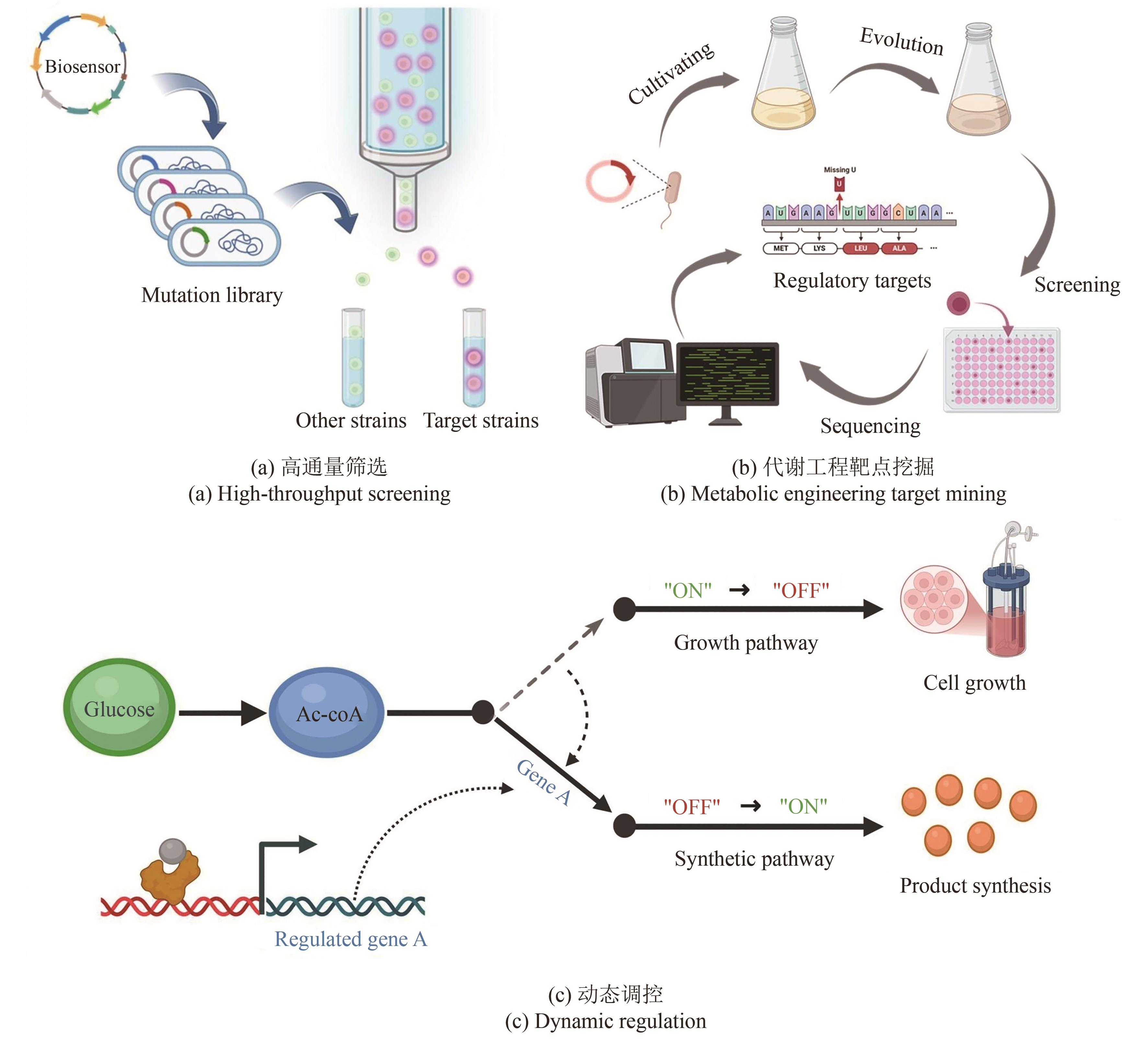
Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (4): 829-845.DOI: 10.12211/2096-8280.2025-030
• Invited Review • Previous Articles Next Articles
Construction and advances in the applications of transcription factor-based biosensors
WANG Hong1, LU Kongyong2, ZHENG Yangyang1, CHEN Tao1, WANG Zhiwen1,2
- 1.School of Chemical Engineering and Technology,Tianjin University,Tianjin 300350,China
2.College of Life Science,Ningxia University,Yinchuan 750021,Ningxia,China
-
Received:2025-03-28Revised:2025-05-29Online:2025-09-03Published:2025-08-31 -
Contact:WANG Zhiwen
基于转录因子生物传感器的构建与应用进展
王宏1, 陆孔泳2, 郑洋洋1, 陈涛1, 王智文1,2
- 1.天津大学化工学院,天津 300350
2.宁夏大学生命科学学院,宁夏 银川 750021
-
通讯作者:王智文 -
作者简介:王宏 (1999—),男,硕士研究生。研究方向为代谢工程与合成生物学。 E-mail:wh17826529977@163.com陆孔泳 (1989—),女,博士,副教授,硕士生导师。研究方向为合成生物学、生物发酵与代谢调控、微藻生物技术、工业微生物技术等。E-mail:lky@nxu.edu.cn王智文 (1981—),男,博士,教授,博士生导师。研究方向为基因组编辑与合成生物学元件开发、基因组尺度网络模型构建与途径模拟设计、合成高附加值生物医药与生物基化学品人工细胞工厂构建、微生物资源挖掘与利用等。E-mail:zww@tju.edu.cn
第一联系人:共同第一作者 -
基金资助:宁夏重点研发基金资助项目(2024BEE02005)
CLC Number:
Cite this article
WANG Hong, LU Kongyong, ZHENG Yangyang, CHEN Tao, WANG Zhiwen. Construction and advances in the applications of transcription factor-based biosensors[J]. Synthetic Biology Journal, 2025, 6(4): 829-845.
王宏, 陆孔泳, 郑洋洋, 陈涛, 王智文. 基于转录因子生物传感器的构建与应用进展[J]. 合成生物学, 2025, 6(4): 829-845.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-030
| 转录因子 | 宿主 | 响应物质 | 主要构建策略 | 传感器性能 | 参考文献 |
|---|---|---|---|---|---|
| EryD | 解脂耶氏酵母 | 赤藓糖醇 | 启动子筛选替换 | 检测范围扩展至0~400 mmol/L,500 mmol/L时信号饱和 | [ |
| CouR | 酿酒酵母 | 对香豆酰辅酶 A | 启动子元件重构 | 动态范围高达21.4 倍,具有高度底物特异性 | [ |
| TrpR | 大肠杆菌 | 色氨酸 | TrpR及启动子的定向进化 | 检测范围从125 mg/L扩展到500 mg/L | [ |
| BsNadR | 大肠杆菌 | 烟酸 | BsNadR的定向进化 | 检测范围扩展至0~50 mmol/L | [ |
| RamR | 大肠杆菌 | 苄基异喹啉生物碱 | RamR的定向进化 | 对5种BIA具有高特异性和灵敏度 | [ |
| AlkS | 大肠杆菌 | 短链氯代脂肪烃 | AlkS的定向进化 | 荧光输出增加150倍,检测限降低至0.03 mg/kg | [ |
| BenM | 大肠杆菌 | 己二酸 | BenM的理性设计 | 配体特异性改变,灵敏度提高约3倍 | [ |
| MarR | 大肠杆菌 | 阿司匹林 | MarR的理性设计 | 配体特异性改变,检测限低至0.01 mmol/L | [ |
| FeaR | 大肠杆菌 | 芳香胺 | FeaR的理性设计 | 动态范围高达580倍,对苯乙胺特异性响应 | [ |
| MyrR | 大肠杆菌 | β-蒎烯 | 启动子元件重构 | 动态范围提高54倍,检测范围扩展至0~160 mg/L | [ |
| TtgV | 大肠杆菌 | 3-甲基吲哚 | 启动子及质粒拷贝数优化 | 检测限低至10 μmol/L,检测范围扩展至10~1750 μmol/L | [ |
| LldR | 大肠杆菌 | 乳酸 | 启动子元件重构 | 检测限低至2.34 mmol/L,动态范围提高14倍 | [ |
| BreR | 大肠杆菌 | 胆汁酸 | 启动子元件重构 | 动态范围提高至470倍,检测限低至0.61 μmol/L | [ |
| MphR | 大肠杆菌 | 红霉素 | RBS替换优化MphR表达 | 获得了灵敏度差异超过10倍的传感器变体 | [ |
| CdaR | 大肠杆菌 | 葡萄糖酸 | 交叉RBS组合文库筛选 | 动态范围从最初的9倍提升到最高247倍 | [ |
| FapR | 大肠杆菌 | 丙二酰辅酶A | 启动子与RBS替换 | 低浓度mCoA下表现出更高的荧光输出 | [ |
| CamR | 恶臭假单胞菌 | 丁醇类 | CamR的定向进化 | 特异性响应正丁醇,展现出显著底物区分能力 | [ |
| LysG | 谷氨酸棒状杆菌 | γ-氨基丁酸 | LysG的定向进化 | 检测限降低至0.2 μmol/L,动态范围扩展至350倍 | [ |
| PdhR | 枯草芽孢杆菌 | 丙酮酸 | 启动子元件重构 | 动态范围从0.6倍提升至30.7倍 | [ |
Table 1 Biosensors based on transcription factors
| 转录因子 | 宿主 | 响应物质 | 主要构建策略 | 传感器性能 | 参考文献 |
|---|---|---|---|---|---|
| EryD | 解脂耶氏酵母 | 赤藓糖醇 | 启动子筛选替换 | 检测范围扩展至0~400 mmol/L,500 mmol/L时信号饱和 | [ |
| CouR | 酿酒酵母 | 对香豆酰辅酶 A | 启动子元件重构 | 动态范围高达21.4 倍,具有高度底物特异性 | [ |
| TrpR | 大肠杆菌 | 色氨酸 | TrpR及启动子的定向进化 | 检测范围从125 mg/L扩展到500 mg/L | [ |
| BsNadR | 大肠杆菌 | 烟酸 | BsNadR的定向进化 | 检测范围扩展至0~50 mmol/L | [ |
| RamR | 大肠杆菌 | 苄基异喹啉生物碱 | RamR的定向进化 | 对5种BIA具有高特异性和灵敏度 | [ |
| AlkS | 大肠杆菌 | 短链氯代脂肪烃 | AlkS的定向进化 | 荧光输出增加150倍,检测限降低至0.03 mg/kg | [ |
| BenM | 大肠杆菌 | 己二酸 | BenM的理性设计 | 配体特异性改变,灵敏度提高约3倍 | [ |
| MarR | 大肠杆菌 | 阿司匹林 | MarR的理性设计 | 配体特异性改变,检测限低至0.01 mmol/L | [ |
| FeaR | 大肠杆菌 | 芳香胺 | FeaR的理性设计 | 动态范围高达580倍,对苯乙胺特异性响应 | [ |
| MyrR | 大肠杆菌 | β-蒎烯 | 启动子元件重构 | 动态范围提高54倍,检测范围扩展至0~160 mg/L | [ |
| TtgV | 大肠杆菌 | 3-甲基吲哚 | 启动子及质粒拷贝数优化 | 检测限低至10 μmol/L,检测范围扩展至10~1750 μmol/L | [ |
| LldR | 大肠杆菌 | 乳酸 | 启动子元件重构 | 检测限低至2.34 mmol/L,动态范围提高14倍 | [ |
| BreR | 大肠杆菌 | 胆汁酸 | 启动子元件重构 | 动态范围提高至470倍,检测限低至0.61 μmol/L | [ |
| MphR | 大肠杆菌 | 红霉素 | RBS替换优化MphR表达 | 获得了灵敏度差异超过10倍的传感器变体 | [ |
| CdaR | 大肠杆菌 | 葡萄糖酸 | 交叉RBS组合文库筛选 | 动态范围从最初的9倍提升到最高247倍 | [ |
| FapR | 大肠杆菌 | 丙二酰辅酶A | 启动子与RBS替换 | 低浓度mCoA下表现出更高的荧光输出 | [ |
| CamR | 恶臭假单胞菌 | 丁醇类 | CamR的定向进化 | 特异性响应正丁醇,展现出显著底物区分能力 | [ |
| LysG | 谷氨酸棒状杆菌 | γ-氨基丁酸 | LysG的定向进化 | 检测限降低至0.2 μmol/L,动态范围扩展至350倍 | [ |
| PdhR | 枯草芽孢杆菌 | 丙酮酸 | 启动子元件重构 | 动态范围从0.6倍提升至30.7倍 | [ |
| 转录因子 | 宿主 | 响应物质 | 应用领域 | 应用结果 | 参考文献 |
|---|---|---|---|---|---|
| TtgR | 大肠杆菌 | 2S-柚皮素 | 高通量筛选 | 筛选出催化活性提高2.34倍的查尔酮合酶突变体 | [ |
| BmoR | 大肠杆菌 | 乙二醇 | 高通量筛选 | 筛选获得的SMM3F突变体催化活性提高1.52倍 | [ |
| YqhC | 大肠杆菌 | 香兰素 | 高通量筛选 | 筛选获得的Mu176突变体催化活性提高7倍 | [ |
| XylS | 大肠杆菌 | 3-甲基水杨酸 | 高通量筛选 | 筛选出催化效率比野生型提高15倍的突变酶 | [ |
| AlkS | 大肠杆菌 | 异戊醇 | 高通量筛选 | 筛选出的异戊醇产量提高45倍的突变菌株 | [ |
| LysG | 谷氨酸棒状杆菌 | L-组氨酸 | 高通量筛选 | 筛选出100个独立的L-组氨酸高产菌株 | [ |
| Leu3p | 酿酒酵母 | α-异丙基苹果酸 | 高通量筛选 | 筛选出异丁醇产量高达725 mg/L的突变菌株 | [ |
| EryD | 解脂耶氏酵母 | 赤藓糖醇 | 高通量筛选 | 筛选出赤藓糖醇产量较原始菌株提升4.4倍的突变菌株 | [ |
| PadR | 大肠杆菌 | 对香豆酸 | 代谢工程靶点挖掘 | 挖掘到与对香豆酸生产相关的靶点pfkA和ptsI | [ |
| LldR | 运动发酵单胞菌 | D-乳酸 | 代谢工程靶点挖掘 | 挖掘到与D-乳酸生产相关的靶点ZMO1323和ZMO1530 | [ |
| Lrp | 谷氨酸棒状杆菌 | 支链氨基酸 | 代谢工程靶点挖掘 | 挖掘到与支链氨基酸合成相关的靶点AHAS | [ |
| CouR | 酿酒酵母 | 对香豆酰辅酶A | 动态调控 | 柚皮素产量达47.3 mg/L,与未调控相比提高15倍 | [ |
| Mlc | 大肠杆菌 | 葡萄糖 | 动态调控 | 动态调控大肠杆菌葡萄糖摄取速率 | [ |
| GlcC | 大肠杆菌 | 乙醇酸 | 动态调控 | 动态调控gltA、ycdW和aceA的表达水平,乙醇酸产量达到52.2 g/L | [ |
| ivbL、BmoR | 大肠杆菌 | 氨基酸、高级醇 | 动态调控 | 动态平衡氨基酸向高级醇转化,异丁醇产量达40.4 g/L | [ |
| PadR | 大肠杆菌 | 对香豆酸 | 动态调控 | 动态调控丙二酰辅酶A合成,覆盆子酮产量提高32.4倍 | [ |
| LacI | 枯草芽孢杆菌 | 乳糖 | 动态调控 | 动态调控glcK表达,2'-岩藻糖基乳糖产量达到30.1 g/L | [ |
| Rex | 希瓦氏菌 | NADH/NAD⁺ | 动态调控 | 动态调控异丁醇合成途径,异丁醇产量提高10.8倍 | [ |
| ChnR | 谷氨酸棒状杆菌 | 戊内酰胺 | 动态调控 | 动态上调Act的表达水平,戊内酰胺产量提高10倍以上 | [ |
Table 2 Applications of transcription factor-based biosensors in microbial cell factory
| 转录因子 | 宿主 | 响应物质 | 应用领域 | 应用结果 | 参考文献 |
|---|---|---|---|---|---|
| TtgR | 大肠杆菌 | 2S-柚皮素 | 高通量筛选 | 筛选出催化活性提高2.34倍的查尔酮合酶突变体 | [ |
| BmoR | 大肠杆菌 | 乙二醇 | 高通量筛选 | 筛选获得的SMM3F突变体催化活性提高1.52倍 | [ |
| YqhC | 大肠杆菌 | 香兰素 | 高通量筛选 | 筛选获得的Mu176突变体催化活性提高7倍 | [ |
| XylS | 大肠杆菌 | 3-甲基水杨酸 | 高通量筛选 | 筛选出催化效率比野生型提高15倍的突变酶 | [ |
| AlkS | 大肠杆菌 | 异戊醇 | 高通量筛选 | 筛选出的异戊醇产量提高45倍的突变菌株 | [ |
| LysG | 谷氨酸棒状杆菌 | L-组氨酸 | 高通量筛选 | 筛选出100个独立的L-组氨酸高产菌株 | [ |
| Leu3p | 酿酒酵母 | α-异丙基苹果酸 | 高通量筛选 | 筛选出异丁醇产量高达725 mg/L的突变菌株 | [ |
| EryD | 解脂耶氏酵母 | 赤藓糖醇 | 高通量筛选 | 筛选出赤藓糖醇产量较原始菌株提升4.4倍的突变菌株 | [ |
| PadR | 大肠杆菌 | 对香豆酸 | 代谢工程靶点挖掘 | 挖掘到与对香豆酸生产相关的靶点pfkA和ptsI | [ |
| LldR | 运动发酵单胞菌 | D-乳酸 | 代谢工程靶点挖掘 | 挖掘到与D-乳酸生产相关的靶点ZMO1323和ZMO1530 | [ |
| Lrp | 谷氨酸棒状杆菌 | 支链氨基酸 | 代谢工程靶点挖掘 | 挖掘到与支链氨基酸合成相关的靶点AHAS | [ |
| CouR | 酿酒酵母 | 对香豆酰辅酶A | 动态调控 | 柚皮素产量达47.3 mg/L,与未调控相比提高15倍 | [ |
| Mlc | 大肠杆菌 | 葡萄糖 | 动态调控 | 动态调控大肠杆菌葡萄糖摄取速率 | [ |
| GlcC | 大肠杆菌 | 乙醇酸 | 动态调控 | 动态调控gltA、ycdW和aceA的表达水平,乙醇酸产量达到52.2 g/L | [ |
| ivbL、BmoR | 大肠杆菌 | 氨基酸、高级醇 | 动态调控 | 动态平衡氨基酸向高级醇转化,异丁醇产量达40.4 g/L | [ |
| PadR | 大肠杆菌 | 对香豆酸 | 动态调控 | 动态调控丙二酰辅酶A合成,覆盆子酮产量提高32.4倍 | [ |
| LacI | 枯草芽孢杆菌 | 乳糖 | 动态调控 | 动态调控glcK表达,2'-岩藻糖基乳糖产量达到30.1 g/L | [ |
| Rex | 希瓦氏菌 | NADH/NAD⁺ | 动态调控 | 动态调控异丁醇合成途径,异丁醇产量提高10.8倍 | [ |
| ChnR | 谷氨酸棒状杆菌 | 戊内酰胺 | 动态调控 | 动态上调Act的表达水平,戊内酰胺产量提高10倍以上 | [ |
| [1] | SHI A Q, ZHU X N, LU J, et al. Activating transhydrogenase and NAD kinase in combination for improving isobutanol production[J]. Metabolic Engineering, 2013, 16: 1-10. |
| [2] | LUCKIE B A, KASHYAP M, PEARSON A N, et al. Development of Corynebacterium glutamicum as a monoterpene production platform[J]. Metabolic Engineering, 2024, 81: 110-122. |
| [3] | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| [4] | 于政, 申晓林, 孙新晓, 等. 动态调控策略在代谢工程中的应用研究进展[J]. 合成生物学, 2020, 1(4): 440-453. |
| YU Z, SHEN X L, SUN X X, et al. Application of dynamic regulation strategies in metabolic engineering[J]. Synthetic Biology Journal, 2020, 1(4): 440-453. | |
| [5] | MACHADO D, COSTA R S, FERREIRA E C, et al. Exploring the gap between dynamic and constraint-based models of metabolism[J]. Metabolic Engineering, 2012, 14(2): 112-119. |
| [6] | 程术, 邓子新, 卞光凯, 等. 萜类高效合成平台的搭建与萜类产物批量挖掘[J]. 生命科学, 2019, 31(5): 449-457. |
| CHENG S, DENG Z X, BIAN G K, et al. Construction of high-efficient terpenoid platform and the application in terpenoid discovery[J]. Chinese Bulletin of Life Sciences, 2019, 31(5): 449-457. | |
| [7] | XU X H, LV X Q, BI X Y, et al. Genetic circuits for metabolic flux optimization[J]. Trends in Microbiology, 2024, 32(8): 791-806. |
| [8] | 洪霞, 田开仁, 乔建军, 等. 基因编码型生物传感器在微生物细胞工厂中的应用进展[J]. 中国生物工程杂志, 2023, 43(9): 62-76. |
| HONG X, TIAN K R, QIAO J J, et al. Application progress of genetically encoded biosensors in microbial cell factory[J]. China Biotechnology, 2023, 43(9): 62-76. | |
| [9] | YU W W, XU X H, JIN K, et al. Genetically encoded biosensors for microbial synthetic biology: from conceptual frameworks to practical applications[J]. Biotechnology Advances, 2023, 62: 108077. |
| [10] | VERMA A K, NOUMANI A, YADAV A K, et al. FRET based biosensor: principle applications recent advances and challenges[J]. Diagnostics, 2023, 13(8): 1375. |
| [11] | FERREIRA S S, ANTUNES M S. Re-engineering plant phenylpropanoid metabolism with the aid of synthetic biosensors[J]. Frontiers in Plant Science, 2021, 12: 701385. |
| [12] | CARPENTER A C, PAULSEN I T, WILLIAMS T C. Blueprints for biosensors: design, limitations, and applications[J]. Genes, 2018, 9(8): 375. |
| [13] | LI C F, WANG C, ZHU J, et al. Advances and prospects of transcription-factor-based biosensors in high-throughput screening for cell factories construction[J]. Food Bioengineering, 2022, 1(2): 135-147. |
| [14] | TU R, ZHANG Y, HUA E B, et al. Droplet-based microfluidic platform for high-throughput screening of Streptomyces [J]. Communications Biology, 2021, 4: 647. |
| [15] | YI D, BAYER T, BADENHORST C P S, et al. Recent trends in biocatalysis[J]. Chemical Society Reviews, 2021, 50(14): 8003-8049. |
| [16] | KIM H, JU J, LEE H N, et al. Genetically encoded biosensors based on fluorescent proteins[J]. Sensors, 2021, 21(3): 795. |
| [17] | 赵静宇, 张健, 祁庆生, 等. 基于细菌双组分系统的生物传感器的研究进展[J]. 合成生物学, 2024, 5(1): 38-52. |
| ZHAO J Y, ZHANG J, QI Q S, et al. Research progress in biosensors based on bacterial two-component systems[J]. Synthetic Biology Journal, 2024, 5(1): 38-52. | |
| [18] | CHEN L, ZHANG Z H, LI Z H, et al. Learning protein fitness landscapes with deep mutational scanning data from multiple sources[J]. Cell Systems, 2023, 14(8): 706-721.e5. |
| [19] | LI J W, QIN Z Q, ZHANG B H, et al. Development of transcriptional factor-based whole-cell biosensors to monitor and degrade antibiotics using mutant cells obtained via adaptive laboratory evolution[J]. Journal of Hazardous Materials, 2024, 473: 134536. |
| [20] | KANG Z Q, ZHANG M M, GAO K Y, et al. An l-2-hydroxyglutarate biosensor based on specific transcriptional regulator LhgR[J]. Nature Communications, 2021, 12: 3619. |
| [21] | 周子莹, 宋晓东, 刘洋儿, 等. 变构转录因子生物传感器构建策略及在食品安全中的应用进展[J]. 生物技术通报, 2024, 40(12): 20-33. |
| ZHOU Z Y, SONG X D, LIU Y E, et al. Construction strategies of allosteric transcription factor biosensors and their application advances in food safety[J]. Biotechnology Bulletin, 2024, 40(12): 20-33. | |
| [22] | LI M, CHEN Z Y, HUO Y X. Application evaluation and performance-directed improvement of the native and engineered biosensors[J]. ACS Sensors, 2024, 9(10): 5002-5024. |
| [23] | XIAO C F, PAN Y Y, HUANG M T. Advances in the dynamic control of metabolic pathways in Saccharomyces cerevisiae [J]. Engineering Microbiology, 2023, 3(4): 100103. |
| [24] | MITCHLER M M, GARCIA J M, MONTERO N E, et al. Transcription factor-based biosensors: a molecular-guided approach for natural product engineering[J]. Current Opinion in Biotechnology, 2021, 69: 172-181. |
| [25] | LIU Y E, ZHOU Z Y, WU Y F, et al. Engineered transcription factor-binding diversed functional nucleic acid-based synthetic biosensor[J]. Biotechnology Advances, 2024, 77: 108463. |
| [26] | DE PAEPE B, DE MEY M. Biological switches: past and future milestones of transcription factor-based biosensors[J]. ACS Synthetic Biology, 2025, 14(1): 72-86. |
| [27] | CHAISUPA P, WRIGHT R C. State-of-the-art in engineering small molecule biosensors and their applications in metabolic engineering[J]. SLAS Technology, 2024, 29(2): 100113. |
| [28] | HUTTANUS H M, TRIOLA E H, VELASQUEZ-GUZMAN J C, et al. Targeted mutagenesis and high-throughput screening of diversified gene and promoter libraries for isolating gain-of-function mutations[J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1202388. |
| [29] | ZHANG Y F, CORTEZ J D, HAMMER S K, et al. Biosensor for branched-chain amino acid metabolism in yeast and applications in isobutanol and isopentanol production[J]. Nature Communications, 2022, 13: 270. |
| [30] | LI H M, ZHANG W, HAN Y Y, et al. Programming a bacterial biosensor for directed evolution of tryptophan hydroxylase via high-throughput droplet sorting[J]. Biosensors & Bioelectronics, 2025, 271: 117072. |
| [31] | 刘静, 李龙, 王云霞, 等. 细菌DeoR家族转录调控因子的研究进展[J]. 微生物学报, 2022, 62(3): 906-917. |
| LIU J, LI L, WANG Y X, et al. Progress on the DeoR family transcriptional regulators in bacteria[J]. Acta Microbiologica Sinica, 2022, 62(3): 906-917. | |
| [32] | YEOM S J, KIM M, KWON K K, et al. A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts[J]. Nature Communications, 2018, 9: 5053. |
| [33] | PHAM C, STOGIOS P J, SAVCHENKO A, et al. Advances in engineering and optimization of transcription factor-based biosensors for plug-and-play small molecule detection[J]. Current Opinion in Biotechnology, 2022, 76: 102753. |
| [34] | SHEN Y P, PAN Y Y, NIU F X, et al. Biosensor-assisted evolution for high-level production of 4-hydroxyphenylacetic acid in Escherichia coli [J]. Metabolic Engineering, 2022, 70: 1-11. |
| [35] | SU H F, CHEN S J, CHEN X L, et al. Utilizing a high-throughput visualization screening technology to develop a genetically encoded biosensor for monitoring 5-aminolevulinic acid production in engineered Escherichia coli [J]. Biosensors and Bioelectronics, 2025, 267: 116806. |
| [36] | CHEN D D, XU S M, LI S L, et al. Directly evolved AlkS-based biosensor platform for monitoring and high-throughput screening of alkane production[J]. ACS Synthetic Biology, 2023, 12(3): 832-841. |
| [37] | PU W, CHEN J Z, LIU P, et al. Directed evolution of linker helix as an efficient strategy for engineering LysR-type transcriptional regulators as whole-cell biosensors[J]. Biosensors and Bioelectronics, 2023, 222: 115004. |
| [38] | TENG Y X, GONG X Y, ZHANG J L, et al. Investigating and engineering an 1, 2-propanediol-responsive transcription factor-based biosensor[J]. ACS Synthetic Biology, 2024, 13(7): 2177-2187. |
| [39] | ROTTINGHAUS A G, XI C G, AMROFELL M B, et al. Engineering ligand-specific biosensors for aromatic amino acids and neurochemicals[J]. Cell Systems, 2022, 13(3): 204-214.e4. |
| [40] | COULSON T J D, PATTEN C L. The TyrR transcription factor regulates the divergent akr-ipdC operons of Enterobacter cloacae UW5[J]. PLoS One, 2015, 10(3): e0121241. |
| [41] | D’OELSNITZ S, NGUYEN V, ALPER H S, et al. Evolving a generalist biosensor for bicyclic monoterpenes[J]. ACS Synthetic Biology, 2022, 11(1): 265-272. |
| [42] | DABIRIAN Y, LI X W, CHEN Y, et al. Expanding the dynamic range of a transcription factor-based biosensor in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2019, 8(9): 1968-1975. |
| [43] | CHEN C, LIU J J, YAO G, et al. A novel, genetically encoded whole-cell biosensor for directed evolution of myrcene synthase in Escherichia coli [J]. Biosensors and Bioelectronics, 2023, 228: 115176. |
| [44] | LEBOVICH M, ANDREWS L B. Surveying the genetic design space for transcription factor-based metabolite biosensors: synthetic gamma-aminobutyric acid and propionate biosensors in E. coli Nissle 1917[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 938056. |
| [45] | WEI W P, SHANG Y Z, ZHANG P, et al. Engineering prokaryotic transcriptional activator XylR as a xylose-inducible biosensor for transcription activation in yeast[J]. ACS Synthetic Biology, 2020, 9(5): 1022-1029. |
| [46] | BEABOUT K, EHRENWORTH BREEDON A M, BLUM S M, et al. Detection of bile acids in complex matrices using a transcription factor-based biosensor[J]. ACS Biomaterials Science & Engineering, 2023, 9(9): 5151-5162. |
| [47] | DING N N, YUAN Z Q, ZHANG X J, et al. Programmable cross-ribosome-binding sites to fine-tune the dynamic range of transcription factor-based biosensor[J]. Nucleic Acids Research, 2020, 48(18): 10602-10613. |
| [48] | LI C Y, ZHOU Y Y, ZOU Y S, et al. Identifying, characterizing, and engineering a phenolic acid-responsive transcriptional factor from Bacillus amyloliquefaciens [J]. ACS Synthetic Biology, 2023, 12(8): 2382-2392. |
| [49] | ZHAO J Y, SUN H H, WANG G G, et al. Engineering chimeric chemoreceptors and two-component systems for orthogonal and leakless biosensing of extracellular γ-aminobutyric acid[J]. Journal of Agricultural and Food Chemistry, 2024, 72(25): 14216-14228. |
| [50] | LI L, ZHANG Q Q, SHI R R, et al. Multidimensional combinatorial screening for high-level production of erythritol in Yarrowia lipolytica [J]. Bioresource Technology, 2024, 406: 131035. |
| [51] | LIU D, SICA M S, MAO J W, et al. A p-coumaroyl-CoA biosensor for dynamic regulation of naringenin biosynthesis in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2022, 11(10): 3228-3238. |
| [52] | GONG X Y, ZHANG R H, WANG J, et al. Engineering of a TrpR-based biosensor for altered dynamic range and ligand preference[J]. ACS Synthetic Biology, 2022, 11(6): 2175-2183. |
| [53] | HAN L C, LIU X Y, CHENG Z Y, et al. Construction and application of a high-throughput in vivo screening platform for the evolution of nitrile metabolism-related enzymes based on a desensitized repressive biosensor[J]. ACS Synthetic Biology, 2022, 11(4): 1577-1587. |
| [54] | D’OELSNITZ S, KIM W, BURKHOLDER N T, et al. Using fungible biosensors to evolve improved alkaloid biosyntheses[J]. Nature Chemical Biology, 2022, 18(9): 981-989. |
| [55] | CHEN D D, ZHAO J D, XU S M, et al. Detection of short-chain chlorinated aliphatic hydrocarbons through an engineered biosensor with tailored ligand specificity[J]. Analytical Chemistry, 2024, 96(39): 15614-15623. |
| [56] | PHAM C, STOGIOS P J, SAVCHENKO A, et al. Computation-guided transcription factor biosensor specificity engineering for adipic acid detection[J]. Computational and Structural Biotechnology Journal, 2024, 23: 2211-2219. |
| [57] | KIM Y, JEON Y, SONG K, et al. Development of an Escherichia coli cell-based biosensor for aspirin monitoring by genetic engineering of MarR[J]. Biosensors, 2024, 14(11): 547. |
| [58] | PHAM C, STOGIOS P J, SAVCHENKO A, et al. Design and characterization of a generalist biosensor for indole derivatives[J]. ACS Synthetic Biology, 2024, 13(7): 2246-2252. |
| [59] | XIAO D, HU C X, XU X Z, et al. A D, L-lactate biosensor based on allosteric transcription factor LldR and amplified luminescent proximity homogeneous assay[J]. Biosensors and Bioelectronics, 2022, 211: 114378. |
| [60] | WANG Y, LI S X, XUE N, et al. Modulating sensitivity of an erythromycin biosensor for precise high-throughput screening of strains with different characteristics[J]. ACS Synthetic Biology, 2023, 12(6): 1761-1771. |
| [61] | KALKREUTER E, KEELER A M, MALICO A A, et al. Development of a genetically encoded biosensor for detection of polyketide synthase extender units in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8(6): 1391-1400. |
| [62] | XU X H, LI X L, LIU Y F, et al. Pyruvate-responsive genetic circuits for dynamic control of central metabolism[J]. Nature Chemical Biology, 2020, 16(11): 1261-1268. |
| [63] | 杨璐, 吴楠, 白茸茸, 等. 基因回路型全细胞微生物传感器的设计、优化与应用[J]. 合成生物学, 2022, 3(6): 1061-1080. |
| YANG L, WU N, BAI R R, et al. Design, optimization and application of whole-cell microbial biosensors with engineered genetic circuits[J]. Synthetic Biology Journal, 2022, 3(6): 1061-1080. | |
| [64] | DING N N, YUAN Z N, MA Z, et al. AI-assisted rational design and activity prediction of biological elements for optimizing transcription-factor-based biosensors[J]. Molecules, 2024, 29(15): 3512. |
| [65] | 赵梅, 罗佳璐, 王震, 等. 基于转录因子的生物传感器研究进展[J]. 食品与发酵工业, 2024, 50(12): 362-369. |
| ZHAO M, LUO J L, WANG Z, et al. Research progress on transcription-factor-based biosensors[J]. Food and Fermentation Industries, 2024, 50(12): 362-369. | |
| [66] | ZHEN Z, XIANG L, LI S Z, et al. Designing a whole-cell biosensor applicable for S-adenosyl-L-methionine-dependent methyltransferases[J]. Biosensors and Bioelectronics, 2025, 268: 116904. |
| [67] | LI C, GAO X, QI H B, et al. Substantial improvement of an epimerase for the synthesis of D-allulose by biosensor-based high-throughput microdroplet screening[J]. Angewandte Chemie International Edition, 2023, 62(10): e202216721. |
| [68] | QIU X L, XU P, ZHAO X R, et al. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica [J]. Metabolic Engineering, 2020, 60: 66-76. |
| [69] | GAO J S, DU M H, ZHAO J H, et al. Design of a genetically encoded biosensor to establish a high-throughput screening platform for L-cysteine overproduction[J]. Metabolic Engineering, 2022, 73: 144-157. |
| [70] | TRIVEDI V D, MOHAN K, CHAPPELL T C, et al. Cheating the cheater: suppressing false-positive enrichment during biosensor-guided biocatalyst engineering[J]. ACS Synthetic Biology, 2022, 11(1): 420-429. |
| [71] | LI S S, LI Z L, TAN G Y, et al. In vitro allosteric transcription factor-based biosensing[J]. Trends in Biotechnology, 2023, 41(8): 1080-1095. |
| [72] | MORASKIE M, ROSHID M H O, O’CONNOR G, et al. Microbial whole-cell biosensors: current applications, challenges, and future perspectives[J]. Biosensors and Bioelectronics, 2021, 191: 113359. |
| [73] | 贾男, 臧国伟, 李春, 等. 辅因子在微生物细胞工厂中的代谢调控与应用[J]. 中国生物工程杂志, 2022, 42(7): 79-89. |
| JIA N, ZANG G W, LI C, et al. Metabolic regulations and applications of cofactors in microbial cell factories[J]. China Biotechnology, 2022, 42(7): 79-89. | |
| [74] | DONG C, SCHULTZ J C, LIU W, et al. Identification of novel metabolic engineering targets for S-adenosyl-L-methionine production in Saccharomyces cerevisiae via genome-scale engineering[J]. Metabolic Engineering, 2021, 66: 319-327. |
| [75] | LI X Y, ZHOU M H, ZENG D W, et al. Membrane transport engineering for efficient yeast biomanufacturing[J]. Bioresource Technology, 2025, 418: 131890. |
| [76] | WANG J, LI C Y, JIANG T, et al. Biosensor-assisted titratable CRISPRi high-throughput (BATCH) screening for over-production phenotypes[J]. Metabolic Engineering, 2023, 75: 58-67. |
| [77] | PENG Q Q, BAO W W, GENG B N, et al. Biosensor-assisted CRISPRi high-throughput screening to identify genetic targets in Zymomonas mobilis for high d-lactate production[J]. Synthetic and Systems Biotechnology, 2024, 9(2): 242-249. |
| [78] | BAUMANN L, BRUDER S, KABISCH J, et al. High-throughput screening of an octanoic acid producer strain library enables detection of new targets for increasing titers in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2021, 10(5): 1077-1086. |
| [79] | STELLA R G, GERTZEN C G W, SMITS S H J, et al. Biosensor-based growth-coupling and spatial separation as an evolution strategy to improve small molecule production of Corynebacterium glutamicum [J]. Metabolic Engineering, 2021, 68: 162-173. |
| [80] | KRÜGER A, GÖDDECKE J, OSTHEGE M, et al. Biosensor-based growth-coupling as an evolutionary strategy to improve heme export in Corynebacterium glutamicum [J]. Microbial Cell Factories, 2024, 23(1): 276. |
| [81] | GEORGE K W, THOMPSON M G, KIM J, et al. Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli [J]. Metabolic Engineering, 2018, 47: 60-72. |
| [82] | LU L Y, WANG X L, WANG T, et al. A bacterial platform for producing aromatic esters from glycerol[J]. Nature Chemical Engineering, 2024, 1(12): 751-764. |
| [83] | ZHANG Q W, XU X H, ZHANG W, et al. De novo 2'-fucosyllactose biosynthesis using glucose as the sole carbon source by multiple engineered Bacillus subtilis [J]. Metabolic Engineering, 2025, 88: 85-93. |
| [84] | YU F, LI C Y, ZHANG T, et al. Developing a novel heme biosensor to produce high-active hemoproteins in Pichia pastoris through comparative transcriptomics[J]. Metabolic Engineering, 2024, 84: 59-68. |
| [85] | DING D Q, ZHU Y R, BAI D Y, et al. Monitoring and dynamically controlling glucose uptake rate and central metabolism[J]. Nature Chemical Engineering, 2025, 2(1): 50-62. |
| [86] | TONG Y J, LI N, ZHOU S H, et al. Improvement of Chalcone synthase activity and high-efficiency fermentative production of (2S)-naringenin via in vivo biosensor-guided directed evolution[J]. ACS Synthetic Biology, 2024, 13(5): 1454-1466. |
| [87] | LI M, CHEN Z Y, ZHANG W Y, et al. Customization of ethylene glycol (EG)-induced BmoR-based biosensor for the directed evolution of PET degrading enzymes[J]. Advanced Science, 2025, 12(13): e2413205. |
| [88] | DONG P Y, FAN Y J, HUO Y X, et al. Pathway-adapted biosensor for high-throughput screening of O-methyltransferase and its application in vanillin synthesis[J]. ACS Synthetic Biology, 2024, 13(9): 2873-2886. |
| [89] | OGAWA Y, SAITO Y, YAMAGUCHI H, et al. Engineering the substrate specificity of toluene degrading enzyme XylM using biosensor XylS and machine learning[J]. ACS Synthetic Biology, 2023, 12(2): 572-582. |
| [90] | BAHLS M O, PLATZ L, MORGADO G, et al. Directed evolution of biofuel-responsive biosensors for automated optimization of branched-chain alcohol biosynthesis[J]. Metabolic Engineering, 2022, 69: 98-111. |
| [91] | BAUMANN P T, MOLIN M DAL, ARING H, et al. Beyond rational-biosensor-guided isolation of 100 independently evolved bacterial strain variants and comparative analysis of their genomes[J]. BMC Biology, 2023, 21(1): 183. |
| [92] | YANG H N, HE Y C, ZHOU S H, et al. Dynamic regulation and cofactor engineering of Escherichia coli to enhance production of glycolate from corn stover hydrolysate[J]. Bioresource Technology, 2024, 398: 130531. |
| [93] | CHEN Z Y, YU S Z, LIU J, et al. Concentration recognition-based auto-dynamic regulation system (CRUISE) enabling efficient production of higher alcohols[J]. Advanced Science, 2024, 11(23): e2310215. |
| [94] | ZHOU S H, ZHANG Q Y, YUAN M W, et al. Static and dynamic regulation of precursor supply pathways to enhance raspberry ketone synthesis from glucose in Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2024, 72(42): 23411-23421. |
| [95] | YU H, LI F, WANG Y X, et al. Electro-controlled distribution of reducing equivalents to boost isobutanol biosynthesis in microbial electro-fermentation of S. oneidensis[J]. Joule, 2025, 9(1): 101773. |
| [96] | ZHAO X X, WU Y L, FENG T Y, et al. Dynamic upregulation of the rate-limiting enzyme for valerolactam biosynthesis in Corynebacterium glutamicum [J]. Metabolic Engineering, 2023, 77: 89-99. |
| [97] | SUN H H, ZHAO H M, ANG E L. A new biosensor for stilbenes and a cannabinoid enabled by genome mining of a transcriptional regulator[J]. ACS Synthetic Biology, 2020, 9(4): 698-705. |
| [98] | HANKO E K R, JOOSAB NOOR MAHOMED T A, STONEY R A, et al. TFBMiner: a user-friendly command line tool for the rapid mining of transcription factor-based biosensors[J]. ACS Synthetic Biology, 2023, 12(5): 1497-1507. |
| [99] | EKAS H M, WANG B, SILVERMAN A D, et al. An automated cell-free workflow for transcription factor engineering[J]. ACS Synthetic Biology, 2024, 13(10): 3389-3399. |
| [100] | LIU K, ZHANG Y S, LIU K, et al. De novo design of a transcription factor for a progesterone biosensor[J]. Biosensors and Bioelectronics, 2022, 203: 113897. |
| [101] | ZHAO M, HU M K, HAN R M, et al. Dynamics design of a non-natural transcription factor responding to androst-4-ene-3, 17-dione[J]. Synthetic and Systems Biotechnology, 2024, 9(3): 436-444. |
| [102] | RICHTER M F, ZHAO K T, ETON E, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity[J]. Nature Biotechnology, 2020, 38(7): 883-891. |
| [103] | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| [104] | RUBE H T, RASTOGI C, FENG S Q, et al. Prediction of protein-ligand binding affinity from sequencing data with interpretable machine learning[J]. Nature Biotechnology, 2022, 40(10): 1520-1527. |
| [105] | ZHANG P C, WANG H C, XU H W, et al. Deep flanking sequence engineering for efficient promoter design using DeepSEED[J]. Nature Communications, 2023, 14: 6309. |
| [106] | BAYER T, HÄNEL L, HUSARCIKOVA J, et al. In vivo detection of low molecular weight platform chemicals and environmental contaminants by genetically encoded biosensors[J]. ACS Omega, 2023, 8(26): 23227-23239. |
| [107] | TELLECHEA-LUZARDO J, STIEBRITZ M T, CARBONELL P. Transcription factor-based biosensors for screening and dynamic regulation[J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1118702. |
| [108] | NEUBAUER P, JUNNE S. Scale-down simulators for metabolic analysis of large-scale bioprocesses[J]. Current Opinion in Biotechnology, 2010, 21(1): 114-121. |
| [109] | 王晟, 王泽琛, 陈威华, 等. 基于人工智能和计算生物学的合成生物学元件设计[J]. 合成生物学, 2023, 4(3): 422-443. |
| WANG S, WANG Z C, CHEN W H, et al. Design of synthetic biology components based on artificial intelligence and computational biology[J]. Synthetic Biology Journal, 2023, 4(3): 422-443. | |
| [110] | ZHANG C, LIU H, LI X J, et al. Modularized synthetic biology enabled intelligent biosensors[J]. Trends in Biotechnology, 2023, 41(8): 1055-1065. |
| [111] | BOADA Y, VIGNONI A, PICÓ J, et al. Extended metabolic biosensor design for dynamic pathway regulation of cell factories[J]. iScience, 2020, 23(7): 101305. |
| [112] | ABRAMSON J, ADLER J, DUNGER J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 630(8016): 493-500. |
| [113] | CHAN C T Y, KENNEDY V, KINSHUK S. A domain swapping strategy to create modular transcriptional regulators for novel topology in genetic network[J]. Biotechnology Advances, 2024, 72: 108345. |
| [114] | DEMEESTER W, DE BAETS J, DUCHI D, et al. MoBioS: modular platform technology for high-throughput construction and characterization of tunable transcriptional biological sensors[J]. Biosensors, 2023, 13(6): 590. |
| [115] | LEE H, XIE T, KANG B, et al. Plug-and-play protein biosensors using aptamer-regulated in vitro transcription[J]. Nature Communications, 2024, 15: 7973. |
| [1] | LIU Huan, CUI Qiu. Advances and applications of ambient ionization mass spectrometry in screening of microbial strains [J]. Synthetic Biology Journal, 2023, 4(5): 980-999. |
| [2] | WU Yujie, LIU Xinxin, LIU Jianhui, Yang Kaiguang, SUI Zhigang, ZHANG Lihua, ZHANG Yukui. Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry [J]. Synthetic Biology Journal, 2023, 4(5): 1000-1019. |
| [3] | QIN Weitong, YANG Guangyu. Research and application progress of microdroplets high throughput screening methods [J]. Synthetic Biology Journal, 2023, 4(5): 966-979. |
| [4] | ZHAO Guomiao, YANG Xin, ZHANG Yuan, WANG Jing, TAN Jian, WEI Chao, ZHOU Nana, LI Fan, WANG Xiaoyan. Biofoundry and its industrial application [J]. Synthetic Biology Journal, 2023, 4(5): 892-903. |
| [5] | GAO Xianyun, NIU Lingxue, JIAN Ni, GUAN Ningzi. Applications of microbial synthetic biology in the diagnosis and treatment of diseases [J]. Synthetic Biology Journal, 2023, 4(2): 263-282. |
| [6] | TU Ran, LI Shixin, LI Haoni, WANG Meng. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains [J]. Synthetic Biology Journal, 2023, 4(1): 165-184. |
| [7] | REN Shichao, SUN Qiuyan, FENG Xudong, LI Chun. Biosynthesis of pentacyclic triterpenoid saponins in microbial cell factories [J]. Synthetic Biology Journal, 2022, 3(1): 168-183. |
| [8] | CHEN Jiuzhou, WANG Yu, PU Wei, ZHENG Ping, SUN Jibin. Advances and perspective on bioproduction of 5-aminolevulinic acid [J]. Synthetic Biology Journal, 2021, 2(6): 1000-1016. |
| [9] | XU Peng. In memory of Prof. Daniel I.C. Wang: Engineering Yarrowia lipolytica for the production of plant-based lipids: technical constraints and perspectives for a sustainable cellular agriculture economy [J]. Synthetic Biology Journal, 2021, 2(4): 509-527. |
| [10] | XIA Siyang, JIANG Lihong, CAI Jin, HUANG Lei, XU Zhinan, LIAN Jiazhang. Advances in genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2020, 1(5): 556-569. |
| [11] | XU Ke, WANG Jingnan, LI Chun. Intelligent microbial cell factory with tolerance for green biological manufacturing [J]. Synthetic Biology Journal, 2020, 1(4): 427-439. |
| [12] | Yu Zheng, SHEN Xiaolin, Sun Xinxiao, Wang Jia, Yuan Qipeng. Application of dynamic regulation strategies in metabolic engineering [J]. Synthetic Biology Journal, 2020, 1(4): 440-453. |
| [13] | SHI Shuobo, MENG Qiongyu, QIAO Weibo, ZHAO Huimin. Establishing carbon dioxide-based third-generation biorefinery for a sustainable low-carbon economy [J]. Synthetic Biology Journal, 2020, 1(1): 44-59. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||