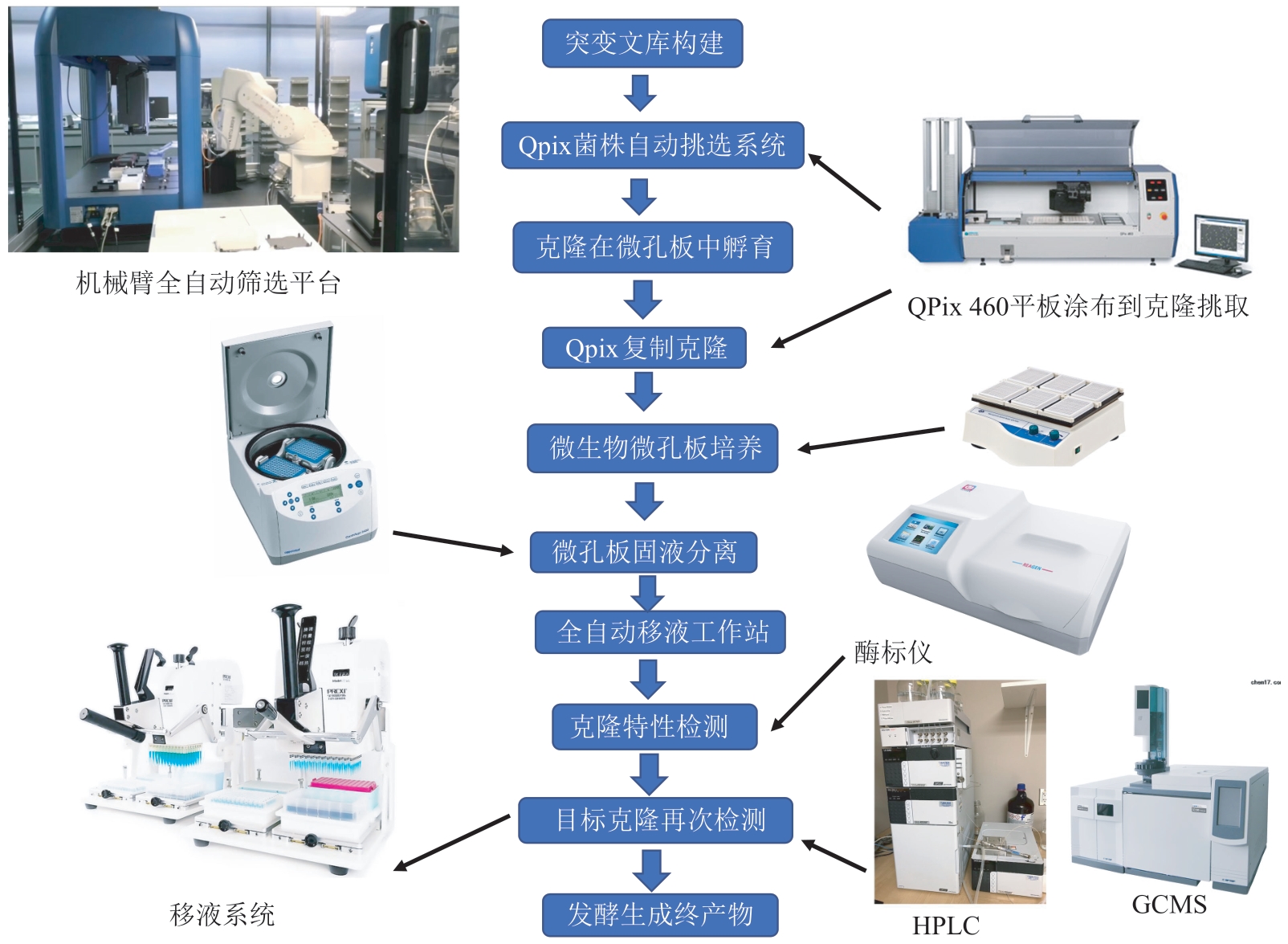
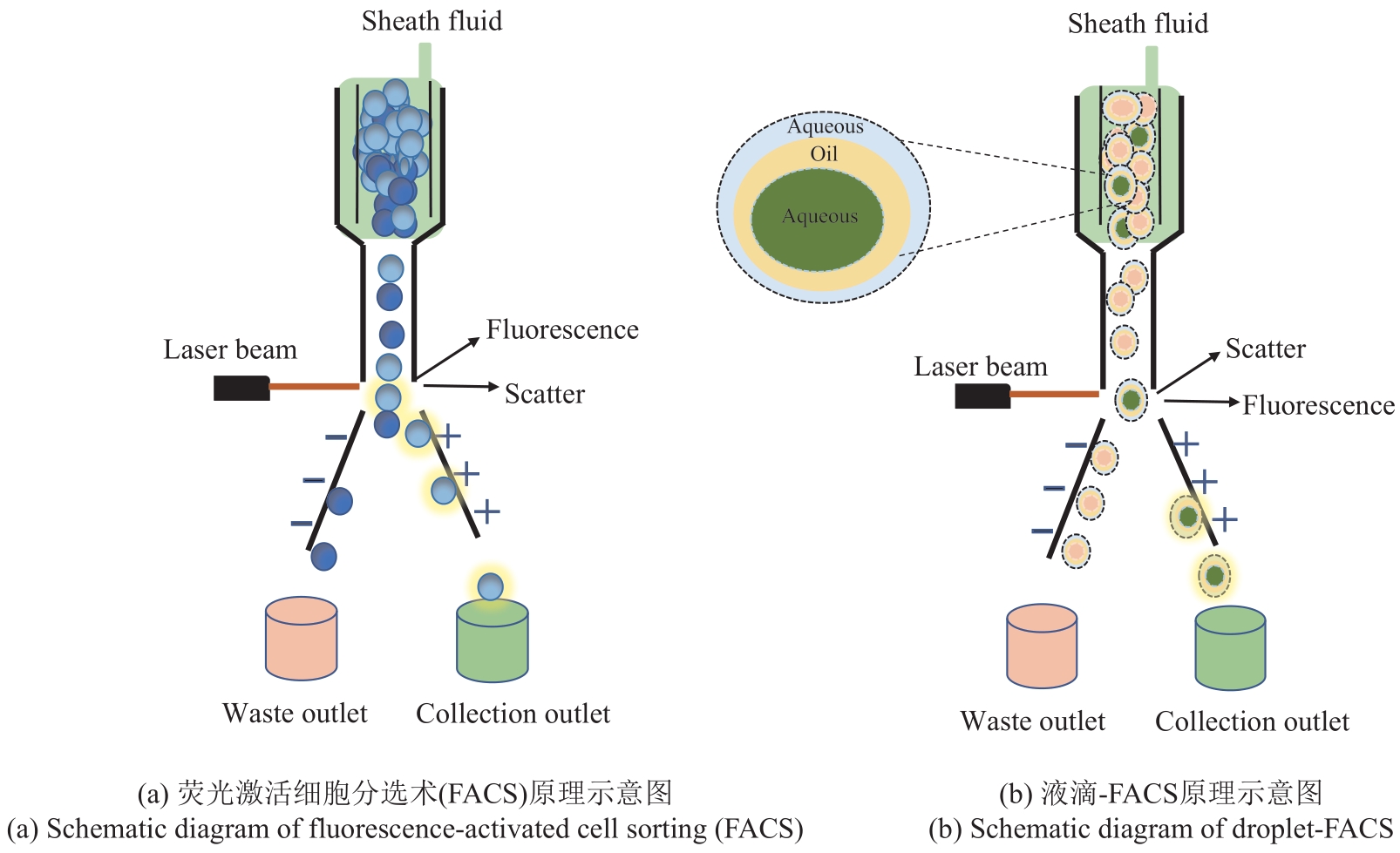
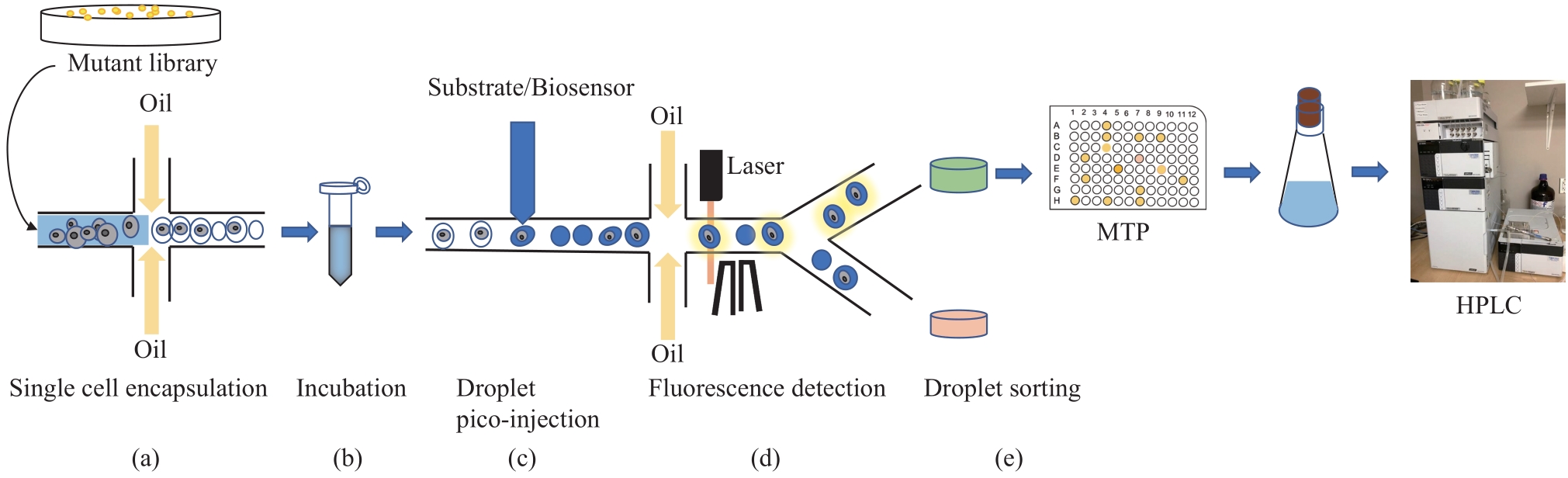
合成生物学 ›› 2023, Vol. 4 ›› Issue (5): 947-965.DOI: 10.12211/2096-8280.2023-017
基于荧光检测的高通量筛选技术和装备助力细胞工厂构建
孙梦楚, 陆亮宇, 申晓林, 孙新晓, 王佳, 袁其朋
- 北京化工大学化工资源有效利用国家重点实验室,北京软物质科学与工程高精尖创新中心,北京 100029
-
收稿日期:2023-02-28修回日期:2023-04-17出版日期:2023-10-31发布日期:2023-11-15 -
通讯作者:王佳,袁其朋 -
作者简介:孙梦楚 (1992—),女,博士研究生。研究方向为代谢工程及合成生物学。E-mail:xcs624816@163.com王佳 (1989—),女,博士,副教授。研究方向为代谢工程及合成生物学。E-mail:wangjia@mail.buct.edu.cn袁其朋 (1969—),男,博士,教授。研究方向为生物化工,代谢工程及微生物合成生物学。E-mail:yuanqp@mail.buct.edu.cn -
基金资助:国家自然科学基金(21908003);国家重点研发计划(2018YFA0903000)
Fluorescence detection-based high-throughput screening systems and devices facilitate cell factories construction
SUN Mengchu, LU Liangyu, SHEN Xiaolin, SUN Xinxiao, WANG Jia, YUAN Qipeng
- State Key Laboratory of Chemical Resource Engineering,Beijing Advanced Innovation Center for Soft Matter Science and Engineering,Beijing University of Chemical Technology,Beijing 100029,China
-
Received:2023-02-28Revised:2023-04-17Online:2023-10-31Published:2023-11-15 -
Contact:WANG Jia, YUAN Qipeng
摘要:
微生物工业制造是以微生物细胞工厂为核心,利用低成本、可再生资源为原料,实现高附加值化合物的绿色生产。依赖于“设计-构建-测试-学习”(DBTL)循环的微生物细胞工厂开发过程中“测试”阶段已成为制约合成生物学和代谢工程发展的瓶颈之一。基于微量滴定板(MTP)高通量自动化筛选平台极大降低了高通量筛选过程的劳动强度,流式细胞术和液滴微流控技术的发展大幅度提高了筛选通量。尤其是荧光激活液滴分选(FADS)高通量筛选技术的开发为自动化、高通量和低消耗筛选工作提供了新的解决方案。本文综述了不同高通量筛选技术在合成生物学和代谢工程领域应用的主要进展,重点介绍了近几年荧光激活细胞分选技术(FACS)和FADS在微生物细胞工厂和酶定向进化方面的应用实例,关注了待测分子与荧光信号偶联的常用策略,并简单介绍目前国内外基于液滴微流控技术高通量筛选装备的研发情况。
中图分类号:
引用本文
孙梦楚, 陆亮宇, 申晓林, 孙新晓, 王佳, 袁其朋. 基于荧光检测的高通量筛选技术和装备助力细胞工厂构建[J]. 合成生物学, 2023, 4(5): 947-965.
SUN Mengchu, LU Liangyu, SHEN Xiaolin, SUN Xinxiao, WANG Jia, YUAN Qipeng. Fluorescence detection-based high-throughput screening systems and devices facilitate cell factories construction[J]. Synthetic Biology Journal, 2023, 4(5): 947-965.
| 菌种 | 目标产物 | 成果 | 技术 | 信号检测 | 文献 |
|---|---|---|---|---|---|
| 酿酒酵母 | 左旋多巴 | 左旋多巴(L-DOPA)产量增加 | FACS | 左旋多巴的酶偶联生物传感器 | [ |
| 芽孢杆菌 | 抗生素Ami | 通过基因挖掘鉴定抗生素Ami | FACS (液滴) | 生长偶联 | [ |
| 大肠杆菌 | L-半胱氨酸 | 诱导半胱氨酸产生增加约2.67倍 | FACS | L-半胱氨酸响应性转录因子生物传感器 | [ |
| 解脂耶氏酵母 | β-胡萝卜素 | 产生9.4 g/L β-胡萝卜素的菌株 | FACS | β-胡萝卜素的菌株自身荧光 | [ |
| 里氏木霉 | 十六烷醇 | 构建首个丝状真菌脂肪醇生产细胞工厂 | FACS | GFP-融合耦合荧光 | [ |
| 凝结芽孢杆菌 | 乳酸 | 乳酸滴度增加52% | FACS (液滴) | 荧光染料(探针1) | [ |
| 大肠杆菌 | 丙二酰-CoA | 显著提高了柚皮素的产量,达到(523.7±51.8)mg/L | FACS | 基于柚皮素和对香豆酸生物传感器 | [ |
| 毕赤酵母 | 木聚糖酶 | 木聚糖酶活性提高1.3倍 | FACS | 荧光标记底物 | [ |
| 光滑念珠菌 | 丙酮酸 | 丙酮酸产量增加73.6% | FACS (液滴) | pH敏感荧光蛋白 | [ |
| 谷氨酸棒状杆菌 | L-赖氨酸 | 获得高产L-赖氨酸的丙酮酸羧化酶突变体 | FACS | 基于赖氨酸转录因子生物传感器 | [ |
| 大肠杆菌 | 引入非天然氨基酸 | 成功筛选到可将非天然氨基酸插入到蛋白质特定位点的功能性PylRS突变体 | FACS | 荧光蛋白融合表达 | [ |
| 大肠杆菌 | α-1,3-岩藻糖基转移酶 | 成功筛选到活性提高14倍的α-1,3-岩藻糖基转移酶突变 | FACS | 荧光标记底物及利用细胞膜的选择性转运蛋白实现细胞捕获荧光产物 | [ |
| 大肠杆菌 | 单胺氧化酶 | 获得对仲胺底物具有催化活性的突变体 | FACS | 多级酶偶联反应,过氧化氢与荧光探针反应释放荧光基团 | [ |
| 自来水样本 | 酯酶 | 挖掘具有高催化效率新型酯酶EstWY | FACS (液滴) | 荧光标记底物 | [ |
表1 FACS应用工业微生物菌株构建和酶分子定向进化及实例
Table 1 Summary of recent FACS applications for microbial cell factories and directed evolution
| 菌种 | 目标产物 | 成果 | 技术 | 信号检测 | 文献 |
|---|---|---|---|---|---|
| 酿酒酵母 | 左旋多巴 | 左旋多巴(L-DOPA)产量增加 | FACS | 左旋多巴的酶偶联生物传感器 | [ |
| 芽孢杆菌 | 抗生素Ami | 通过基因挖掘鉴定抗生素Ami | FACS (液滴) | 生长偶联 | [ |
| 大肠杆菌 | L-半胱氨酸 | 诱导半胱氨酸产生增加约2.67倍 | FACS | L-半胱氨酸响应性转录因子生物传感器 | [ |
| 解脂耶氏酵母 | β-胡萝卜素 | 产生9.4 g/L β-胡萝卜素的菌株 | FACS | β-胡萝卜素的菌株自身荧光 | [ |
| 里氏木霉 | 十六烷醇 | 构建首个丝状真菌脂肪醇生产细胞工厂 | FACS | GFP-融合耦合荧光 | [ |
| 凝结芽孢杆菌 | 乳酸 | 乳酸滴度增加52% | FACS (液滴) | 荧光染料(探针1) | [ |
| 大肠杆菌 | 丙二酰-CoA | 显著提高了柚皮素的产量,达到(523.7±51.8)mg/L | FACS | 基于柚皮素和对香豆酸生物传感器 | [ |
| 毕赤酵母 | 木聚糖酶 | 木聚糖酶活性提高1.3倍 | FACS | 荧光标记底物 | [ |
| 光滑念珠菌 | 丙酮酸 | 丙酮酸产量增加73.6% | FACS (液滴) | pH敏感荧光蛋白 | [ |
| 谷氨酸棒状杆菌 | L-赖氨酸 | 获得高产L-赖氨酸的丙酮酸羧化酶突变体 | FACS | 基于赖氨酸转录因子生物传感器 | [ |
| 大肠杆菌 | 引入非天然氨基酸 | 成功筛选到可将非天然氨基酸插入到蛋白质特定位点的功能性PylRS突变体 | FACS | 荧光蛋白融合表达 | [ |
| 大肠杆菌 | α-1,3-岩藻糖基转移酶 | 成功筛选到活性提高14倍的α-1,3-岩藻糖基转移酶突变 | FACS | 荧光标记底物及利用细胞膜的选择性转运蛋白实现细胞捕获荧光产物 | [ |
| 大肠杆菌 | 单胺氧化酶 | 获得对仲胺底物具有催化活性的突变体 | FACS | 多级酶偶联反应,过氧化氢与荧光探针反应释放荧光基团 | [ |
| 自来水样本 | 酯酶 | 挖掘具有高催化效率新型酯酶EstWY | FACS (液滴) | 荧光标记底物 | [ |
| 化合物/酶分子 | 表达菌株 | 成果 | 信号检测 | 文献 |
|---|---|---|---|---|
| 微生物菌群 | 筛选PET降解菌株,挖掘并验证两种PET降解酶 | 荧光探针 | [ | |
| L-色氨酸 | 大肠杆菌 | 筛选色氨酸增加165.9%突变菌株 | 核糖体开关生物传感器 | [ |
| α-L-苏糖核酸 | 大肠杆菌 | α-L-苏糖核酸合成提高10倍 | 荧光标记底物 | [ |
| 纤维素 | 里氏木霉 | 纤维素产量增加46% | 荧光标记底物 | [ |
| 3-脱氢莽草酸 | 大肠杆菌 | 3-脱氢莽草酸产量增加30% | 荧光生物传感器 | [ |
| 纳米抗体VHH | 谷氨酸棒状杆菌 | 挖掘蛋白分泌关联基因位点并构建了VHH产量提高2.78倍的底盘菌株 | 氧化还原反应转录因子生物传感器 | [ |
| 硫酸酯酶 | 大肠杆菌 | 获得的酶变体对4-硝基苯基硫酸盐催化活性提高6.2倍,对二硫酸荧光素催化活性提高30倍 | 荧光标记底物 | [ |
| L-色氨酸 | 大肠杆菌 | 获得产量提高145%的变体菌株 | 核糖开关生物传感器 | [ |
| 醛缩酶 | 大肠杆菌 | 单轮筛选中醛缩酶RA95.0催化活性大幅跃升高达80倍 | 荧光标记底物 | [ |
| 谷氨酸 | 谷氨酸棒杆菌 | 获得谷氨酸产量分别提升了25.8%和19.1%的变体菌 | 谷氨酸感应荧光报告器 | [ |
| 谷氨酰胺酶 | 解淀粉芽孢杆菌 | 筛选产生了谷氨酰胺酶产量增加47%的菌株 | 响应谷氨酸iGluSnFR生物传感器 | [ |
| 环己胺氧化酶 | 大肠杆菌 | 环己胺氧化酶在一轮定向进化后催化效率提高了960倍 | 多酶级联反应辅助荧光底物释放荧光基团 | [ |
| 丙酮酸 | 光滑球拟酵母 | 最终获得了1株高产突变菌株4H2,摇瓶产量达48.6 g/L,相比出发菌株提高了73.6% | pH敏感传感器 | [ |
| D-阿洛酮糖 | 大肠杆菌将 | 获得催化效率提高了17倍的突变体 | D-阿洛酮糖响应转录因子生物传感器 | [ |
| 抗生素红霉素 | 放线菌 | 筛选出高产红霉素放线菌 | 全细胞转录因子生物传感器 | [ |
| α-淀粉酶 | 地衣芽孢杆菌 | 成功鉴定出α-淀粉酶生产能力较高的突变体 | 荧光标记底物 | [ |
表2 FADS应用工业微生物菌株构建和酶分子定向进化及实例
Table 2 Summary of recent FADS applications for microbial cell factories and directed evolution
| 化合物/酶分子 | 表达菌株 | 成果 | 信号检测 | 文献 |
|---|---|---|---|---|
| 微生物菌群 | 筛选PET降解菌株,挖掘并验证两种PET降解酶 | 荧光探针 | [ | |
| L-色氨酸 | 大肠杆菌 | 筛选色氨酸增加165.9%突变菌株 | 核糖体开关生物传感器 | [ |
| α-L-苏糖核酸 | 大肠杆菌 | α-L-苏糖核酸合成提高10倍 | 荧光标记底物 | [ |
| 纤维素 | 里氏木霉 | 纤维素产量增加46% | 荧光标记底物 | [ |
| 3-脱氢莽草酸 | 大肠杆菌 | 3-脱氢莽草酸产量增加30% | 荧光生物传感器 | [ |
| 纳米抗体VHH | 谷氨酸棒状杆菌 | 挖掘蛋白分泌关联基因位点并构建了VHH产量提高2.78倍的底盘菌株 | 氧化还原反应转录因子生物传感器 | [ |
| 硫酸酯酶 | 大肠杆菌 | 获得的酶变体对4-硝基苯基硫酸盐催化活性提高6.2倍,对二硫酸荧光素催化活性提高30倍 | 荧光标记底物 | [ |
| L-色氨酸 | 大肠杆菌 | 获得产量提高145%的变体菌株 | 核糖开关生物传感器 | [ |
| 醛缩酶 | 大肠杆菌 | 单轮筛选中醛缩酶RA95.0催化活性大幅跃升高达80倍 | 荧光标记底物 | [ |
| 谷氨酸 | 谷氨酸棒杆菌 | 获得谷氨酸产量分别提升了25.8%和19.1%的变体菌 | 谷氨酸感应荧光报告器 | [ |
| 谷氨酰胺酶 | 解淀粉芽孢杆菌 | 筛选产生了谷氨酰胺酶产量增加47%的菌株 | 响应谷氨酸iGluSnFR生物传感器 | [ |
| 环己胺氧化酶 | 大肠杆菌 | 环己胺氧化酶在一轮定向进化后催化效率提高了960倍 | 多酶级联反应辅助荧光底物释放荧光基团 | [ |
| 丙酮酸 | 光滑球拟酵母 | 最终获得了1株高产突变菌株4H2,摇瓶产量达48.6 g/L,相比出发菌株提高了73.6% | pH敏感传感器 | [ |
| D-阿洛酮糖 | 大肠杆菌将 | 获得催化效率提高了17倍的突变体 | D-阿洛酮糖响应转录因子生物传感器 | [ |
| 抗生素红霉素 | 放线菌 | 筛选出高产红霉素放线菌 | 全细胞转录因子生物传感器 | [ |
| α-淀粉酶 | 地衣芽孢杆菌 | 成功鉴定出α-淀粉酶生产能力较高的突变体 | 荧光标记底物 | [ |
| 1 | LEGGIERI P A, LIU Y Y, HAYES M, et al. Integrating systems and synthetic biology to understand and engineer microbiomes[J]. Annual Review of Biomedical Engineering, 2021, 23: 169-201. |
| 2 | NAKAMURA C E, WHITED G M. Metabolic engineering for the microbial production of 1,3-propanediol[J]. Current Opinion in Biotechnology, 2003, 14(5): 454-459. |
| 3 | LUO X Z, REITER M A, D'ESPAUX L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast[J]. Nature, 2019, 567(7746): 123-126. |
| 4 | LIU Y Z, CRUZ-MORALES P, ZARGAR A, et al. Biofuels for a sustainable future[J]. Cell, 2021, 184(6): 1636-1647. |
| 5 | ZARGAR A, BAILEY C B, HAUSHALTER R W, et al. Leveraging microbial biosynthetic pathways for the generation of 'drop-in' biofuels[J]. Current Opinion in Biotechnology, 2017, 45: 156-163. |
| 6 | ZHANG J, HANSEN L G, GUDICH O, et al. A microbial supply chain for production of the anti-cancer drug vinblastine[J]. Nature, 2022, 609(7926): 341-347. |
| 7 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12): 725-741. |
| 8 | LOK C. Mining the microbial dark matter[J]. Nature, 2015, 522(7556): 270-273. |
| 9 | CHOI K R, JANG W D, YANG D S, et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering[J]. Trends in Biotechnology, 2019, 37(8): 817-837. |
| 10 | SALESKI T E, KERNER A R, CHUNG M T, et al. Syntrophic co-culture amplification of production phenotype for high-throughput screening of microbial strain libraries[J]. Metabolic Engineering, 2019, 54: 232-243. |
| 11 | MICHAEL S, AULD D, KLUMPP C, et al. A robotic platform for quantitative high-throughput screening[J]. ASSAY and Drug Development Technologies, 2008, 6(5): 637-657. |
| 12 | LONG Q, LIU X X, YANG Y K, et al. The development and application of high throughput cultivation technology in bioprocess development[J]. Journal of Biotechnology, 2014, 192 Pt B: 323-338. |
| 13 | MAYR L M, FUERST P. The future of high-throughput screening[J]. Journal of Biomolecular Screening, 2008, 13(6): 443-448. |
| 14 | MAYR L M, BOJANIC D. Novel trends in high-throughput screening[J]. Current Opinion in Pharmacology, 2009, 9(5): 580-588. |
| 15 | WANG B L, GHADERI A, ZHOU H, et al. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption[J]. Nature Biotechnology, 2014, 32(5): 473-478. |
| 16 | WAGNER J M, LIU L Q, YUAN S F, et al. A comparative analysis of single cell and droplet-based FACS for improving production phenotypes: Riboflavin overproduction in Yarrowia lipolytica [J]. Metabolic Engineering, 2018, 47: 346-356. |
| 17 | JOENSSON H N, ANDERSSON SVAHN H. Droplet microfluidics—a tool for single-cell analysis[J]. Angewandte Chemie International Edition, 2012, 51(49): 12176-12192. |
| 18 | STUCKI A, VALLAPURACKAL J, WARD T R, et al. Droplet microfluidics and directed evolution of enzymes: an intertwined journey[J]. Angewandte Chemie International Edition, 2021, 60(46): 24368-24387. |
| 19 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 20 | SUN G Y, QU L S, AZI F, et al. Recent progress in high-throughput droplet screening and sorting for bioanalysis[J]. Biosensors and Bioelectronics, 2023, 225: 115107. |
| 21 | CIRINO P C, QIAN S. Chapter 2 - Protein engineering as an enabling tool for synthetic biology [M/OL]//Synthetic biology. Boston: Academic Press, 2013: 23-42 [2023-02-01]. . |
| 22 | ZENG W Z, DU G C, CHEN J, et al. A high-throughput screening procedure for enhancing α-ketoglutaric acid production in Yarrowia lipolytica by random mutagenesis[J]. Process Biochemistry, 2015, 50(10): 1516-1522. |
| 23 | SUN L, ZHANG H, YUAN H, et al. A double-enzyme-coupled assay for high-throughput screening of succinic acid-producing strains[J]. Journal of Applied Microbiology, 2013, 114(6): 1696-1701. |
| 24 | ZHANG P P, HU S, MEI L H, et al. Improving the activity of cytochrome P450 BM-3 catalyzing indole hydroxylation by directed evolution[J]. Applied Biochemistry and Biotechnology, 2013, 171(1): 93-103. |
| 25 | TU R, LV T, SUN L, et al. Development of a simple colorimetric assay for determination of the isoamyl alcohol-producing strain[J].Applied Biochemistry and Biotechnology, 2020, 192(2): 632-642. |
| 26 | ADAN A, ALIZADA G, KIRAZ Y, et al. Flow cytometry: basic principles and applications[J]. Critical Reviews in Biotechnology, 2017, 37(2): 163-176. |
| 27 | MCKINNON K M. Flow cytometry: an overview[J]. Current Protocols in Immunology, 2018, 120: 5.1.-5.1.11. |
| 28 | RADCLIFF G, JAROSZESKI M J. Basics of flow cytometry[M/OL]//Flow Cytometry Protocols- Methods in molecular biology. New Jersey: Humana Press, 2003: 1-24 [2023-02-01]. . |
| 29 | BARET J C, MILLER O J, TALY V, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity[J]. Lab on a Chip, 2009, 9(13): 1850-1858. |
| 30 | MARKEL U, ESSANI K D, BESIRLIOGLU V, et al. Advances in ultrahigh-throughput screening for directed enzyme evolution[J]. Chemical Society Reviews, 2020, 49(1): 233-262. |
| 31 | GAO J S, DU M H, ZHAO J H, et al. Design of a genetically encoded biosensor to establish a high-throughput screening platform for L-cysteine overproduction[J]. Metabolic Engineering, 2022, 73: 144-157. |
| 32 | SAVITSKAYA J, PROTZKO R J, LI F Z, et al. Iterative screening methodology enables isolation of strains with improved properties for a FACS-based screen and increased L-DOPA production[J]. Scientific Reports, 2019, 9: 5815. |
| 33 | TEREKHOV S S, SMIRNOV I V, MALAKHOVA M V, et al. Ultrahigh-throughput functional profiling of microbiota communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(38): 9551-9556. |
| 34 | LIU M M, ZHANG J, LIU X Q, et al. Rapid gene target tracking for enhancing β-carotene production using flow cytometry-based high-throughput screening in Yarrowia lipolytica [J]. Applied and Environmental Microbiology, 2022, 88(19): e0114922. |
| 35 | WANG G K, JIA W D, CHEN N, et al. A GFP-fusion coupling FACS platform for advancing the metabolic engineering of filamentous fungi[J].Biotechnology for Biofuels, 2018, 11: 232. |
| 36 | ZHU X D, SHI X, WANG S W, et al. High-throughput screening of high lactic acid-producing Bacillus coagulans by droplet microfluidic based flow cytometry with fluorescence activated cell sorting[J]. RSC Advances, 2019, 9(8): 4507-4513. |
| 37 | ZHOU S H, YUAN S F, NAIR P H, et al. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli [J]. Metabolic Engineering, 2021, 67: 41-52. |
| 38 | MA C X, TAN Z L, LIN Y, et al. Gel microdroplet-based high-throughput screening for directed evolution of xylanase-producing Pichia pastoris [J]. Journal of Bioscience and Bioengineering, 2019, 128(6): 662-668. |
| 39 | GUO L K, ZENG W Z, XU S, et al. Fluorescence-activated droplet sorting for enhanced pyruvic acid accumulation by Candida glabrata [J]. Bioresource Technology, 2020, 318: 124258. |
| 40 | KORTMANN M, MACK C, BAUMGART M, et al. Pyruvate carboxylase variants enabling improved lysine production from glucose identified by biosensor-based high-throughput fluorescence-activated cell sorting screening[J]. ACS Synthetic Biology, 2019, 8(2): 274-281. |
| 41 | LIN A E, LIN Q. Rapid identification of functional pyrrolysyl-tRNA synthetases via fluorescence-activated cell sorting[J]. International Journal of Molecular Sciences, 2018, 20(1): 29. |
| 42 | TAN Y M, ZHANG Y, HAN Y B, et al. Directed evolution of an α1, 3-fucosyltransferase using a single-cell ultrahigh-throughput screening method[J]. Science Advances, 2019, 5(10): eaaw8451. |
| 43 | SADLER J C, CURRIN A, KELL D B. Ultra-high throughput functional enrichment of large monoamine oxidase (MAO-N) libraries by fluorescence activated cell sorting[J]. Analyst, 2018, 143(19): 4747-4755. |
| 44 | MA F Q, GUO T J, ZHANG Y F, et al. An ultrahigh-throughput screening platform based on flow cytometric droplet sorting for mining novel enzymes from metagenomic libraries[J]. Environmental Microbiology, 2021, 23(2): 996-1008. |
| 45 | LIU H, HOU Y H, WANG Y, et al. Enhancement of sulfur conversion rate in the production of l-cysteine by engineered Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2020, 68(1): 250-257. |
| 46 | WENDISCH V F. Metabolic engineering advances and prospects for amino acid production[J]. Metabolic Engineering, 2020, 58: 17-34. |
| 47 | WOOLSTON B M, ROTH T, KOHALE I, et al. Development of a formaldehyde biosensor with application to synthetic methylotrophy[J]. Biotechnology and Bioengineering, 2018, 115(1): 206-215. |
| 48 | TU R, ZHANG Y, HUA E B, et al. Droplet-based microfluidic platform for high-throughput screening of Streptomyces [J]. Communications Biology, 2021, 4: 647. |
| 49 | WHITESIDES G M. The origins and the future of microfluidics[J]. Nature, 2006, 442(7101): 368-373. |
| 50 | SEEMANN R, BRINKMANN M, PFOHL T, et al. Droplet based microfluidics[J]. Reports on Progress in Physics, 2012, 75(1): 016601. |
| 51 | GIELEN F, HOURS R, EMOND S, et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS)[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(47): E7383-E7389. |
| 52 | MAZUTIS L, GILBERT J, UNG W L, et al. Single-cell analysis and sorting using droplet-based microfluidics[J]. Nature Protocols, 2013, 8(5): 870-891. |
| 53 | ALKAYYALI T, CAMERON T, HALTLI B, et al. Microfluidic and cross-linking methods for encapsulation of living cells and bacteria-a review[J]. Analytica Chimica Acta, 2019, 1053: 1-21. |
| 54 | MASHAGHI S, ABBASPOURRAD A, WEITZ D A, et al. Droplet microfluidics: a tool for biology, chemistry and nanotechnology[J]. TrAC Trends in Analytical Chemistry, 2016, 82: 118-125. |
| 55 | MARUO S, NAKAMURA O, KAWATA S. Three-dimensional microfabrication with two-photon-absorbed photopolymerization[J]. Optics Letters, 1997, 22(2): 132-134. |
| 56 | CHURSKI K, NOWACKI M, KORCZYK P M, et al. Simple modular systems for generation of droplets on demand[J]. Lab on a Chip, 2013, 13(18): 3689-3697. |
| 57 | ANNA S L, BONTOUX N, STONE H A. Formation of dispersions using "flow focusing" in microchannels[J]. Applied Physics Letters, 2003, 82(3): 364-366. |
| 58 | LIAN Z, CHAN Y, LUO Y, et al. Microfluidic formation of highly monodispersed multiple cored droplets using needle-based system in parallel mode[J]. Electrophoresis, 2020, 41(10/11): 891-901. |
| 59 | ZHU P G, WANG L Q. Passive and active droplet generation with microfluidics: a review[J]. Lab on a Chip, 2017, 17(1): 34-75. |
| 60 | BOITARD L, COTTINET D, KLEINSCHMITT C, et al. Monitoring single-cell bioenergetics via the coarsening of emulsion droplets[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(19): 7181-7186. |
| 61 | ZIMMERMANN H F, ANDERLEI T, BÜCHS J, et al. Oxygen limitation is a pitfall during screening for industrial strains[J]. Applied Microbiology and Biotechnology, 2006, 72(6): 1157-1160. |
| 62 | SUEA-NGAM A, HOWES P D, SRISA-ART M, et al. Droplet microfluidics: from proof-of-concept to real-world utility?[J]. Chemical Communications, 2019, 55(67): 9895-9903. |
| 63 | HOLTZE C, ROWAT A C, AGRESTI J J, et al. Biocompatible surfactants for water-in-fluorocarbon emulsions[J]. Lab on a Chip, 2008, 8(10): 1632-1639. |
| 64 | THEBERGE A B, COURTOIS F, SCHAERLI Y, et al. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology[J]. Angewandte Chemie International Edition, 2010, 49(34): 5846-5868. |
| 65 | CHUNG M T, NÚñEZ D, CAI D W, et al. Deterministic droplet-based co-encapsulation and pairing of microparticles via active sorting and downstream merging[J]. Lab on a Chip, 2017, 17(21): 3664-3671. |
| 66 | BRAGHERI F, MARTÍNEZ VÁZQUEZ R, OSELLAME R. Microfluidics[M/OL]//Three-Dimensional Microfabrication Using Two-Photon Polymerization. Amsterdam: Elsevier, 2020: 493-526 [2023-02-01]. . |
| 67 | DEBON A, POTT M, OBEXER R, et al. Ultrahigh-throughput screening enables efficient single-round oxidase remodelling[J]. Nature Catalysis, 2019, 2(9): 740-747. |
| 68 | HASAN S, BLAHA M E, PIENDL S K, et al. Two-photon fluorescence lifetime for label-free microfluidic droplet sorting[J].Analytical and Bioanalytical Chemistry, 2022, 414(1): 721-730. |
| 69 | DUNCOMBE T A, PONTI A, SEEBECK F P, et al. UV-vis spectra-activated droplet sorting for label-free chemical identification and collection of droplets[J]. Analytical Chemistry, 2021, 93(38): 13008-13013. |
| 70 | WANG X X, XIN Y, REN L H, et al. Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo [J]. Science Advances, 2020, 6(32): eabb3521. |
| 71 | HOLLAND-MORITZ D A, WISMER M K, MANN B F, et al. Mass activated droplet sorting (MADS) enables high-throughput screening of enzymatic reactions at nanoliter scale[J]. Angewandte Chemie International Edition, 2020, 59(11): 4470-4477. |
| 72 | GIRAULT M, KIM H, ARAKAWA H, et al. An on-chip imaging droplet-sorting system: a real-time shape recognition method to screen target cells in droplets with single cell resolution[J]. Scientific Reports, 2017, 7: 40072. |
| 73 | SCIAMBI A, ABATE A R. Accurate microfluidic sorting of droplets at 30 kHz[J]. Lab on a Chip, 2015, 15(1): 47-51. |
| 74 | CLARK I C, THAKUR R, ABATE A R. Concentric electrodes improve microfluidic droplet sorting[J]. Lab on a Chip, 2018, 18(5): 710-713. |
| 75 | QIAO Y X, HU R, CHEN D W, et al. Fluorescence-activated droplet sorting of PET degrading microorganisms[J]. Journal of Hazardous Materials, 2022, 424 Pt B: 127417. |
| 76 | LIU Y F, YUAN H L, DING D Q, et al. Establishment of a biosensor-based high-throughput screening platform for tryptophan overproduction[J]. ACS Synthetic Biology, 2021, 10(6): 1373-1383. |
| 77 | NIKOOMANZAR A, VALLEJO D, CHAPUT J C. Elucidating the determinants of polymerase specificity by microfluidic-based deep mutational scanning[J]. ACS Synthetic Biology, 2019, 8(6): 1421-1429. |
| 78 | HE R L, DING R H, HEYMAN J A, et al. Ultra-high-throughput picoliter-droplet microfluidics screening of the industrial cellulase-producing filamentous fungus Trichoderma reesei [J]. Journal of Industrial Microbiology and Biotechnology, 2019, 46(11): 1603-1610. |
| 79 | TU R, LI L P, YUAN H L, et al. Biosensor-enabled droplet microfluidic system for the rapid screening of 3-dehydroshikimic acid produced in Escherichia coli [J]. Journal of Industrial Microbiology and Biotechnology, 2020, 47(12): 1155-1160. |
| 80 | YU X Y, LI S, FENG H B, et al. CRISPRi-microfluidics screening enables genome-scale target identification for high-titer protein production and secretion[J]. Metabolic Engineering, 2023, 75: 192-204. |
| 81 | VAN LOO B, HEBERLEIN M, MAIR P, et al. High-throughput, lysis-free screening for sulfatase activity using Escherichia coli autodisplay in microdroplets[J]. ACS Synthetic Biology, 2019, 8(12): 2690-2700. |
| 82 | JANG S, LEE B, JEONG H H, et al. On-chip analysis, indexing and screening for chemical producing bacteria in a microfluidic static droplet array[J]. Lab on a Chip, 2016, 16(10): 1909-1916. |
| 83 | OBEXER R, POTT M, ZEYMER C, et al. Efficient laboratory evolution of computationally designed enzymes with low starting activities using fluorescence-activated droplet sorting[J]. Protein Engineering, Design and Selection, 2016, 29(9): 355-366. |
| 84 | JIAN X J, GUO X J, CAI Z S, et al. Single-cell microliter-droplet screening system (MISS Cell): an integrated platform for automated high-throughput microbial monoclonal cultivation and picking[J]. Biotechnology and Bioengineering, 2023, 120(3): 778-792. |
| 85 | LI S, LIAO X H, YU X Y, et al. Combining genetically encoded biosensors with droplet microfluidic system for enhanced glutaminase production by Bacillus amyloliquefaciens [J]. Biochemical Engineering Journal, 2022, 186: 108586. |
| 86 | LI C, GAO X, QI H B, et al. Substantial improvement of an epimerase for the synthesis of D-allulose by biosensor-based high-throughput microdroplet screening[J]. Angewandte Chemie International Edition, 2023, 62(10): e202216721. |
| 87 | HUA E B, ZHANG Y, YUN K Y, et al. Whole-cell biosensor and producer Co-cultivation-based microfludic platform for screening Saccharopolyspora erythraea with hyper erythromycin production[J]. ACS Synthetic Biology, 2022, 11(8): 2697-2708. |
| 88 | BEST R J, LYCZAKOWSKI J J, ABALDE-CELA S, et al. Label-free analysis and sorting of microalgae and cyanobacteria in microdroplets by intrinsic chlorophyll fluorescence for the identification of fast growing strains[J]. Analytical Chemistry, 2016, 88(21): 10445-10451. |
| 89 | YUAN H L, TU R, TONG X W, et al. Ultrahigh-throughput screening of industrial enzyme-producing strains by droplet-based microfluidic system[J]. Journal of Industrial Microbiology and Biotechnology, 2022, 49(3): kuac007. |
| 90 | CHEN J, VESTERGAARD M, JENSEN T G, et al. Finding the needle in the haystack—the use of microfluidic droplet technology to identify vitamin-secreting lactic acid bacteria[J]. mBio, 2017, 8(3): e00526-17. |
| 91 | AHARONI A, THIEME K, CHIU C P C, et al. High-throughput screening methodology for the directed evolution of glycosyltransferases[J]. Nature Methods, 2006, 3(8): 609-614. |
| 92 | KALIDASAN K, SU Y, WU X Y, et al. Fluorescence-activated cell sorting and directed evolution of α-N-acetylgalactosaminidases using a quenched activity-based probe (qABP)[J]. Chemical Communications, 2013, 49(65): 7237-7239. |
| 93 | DENNIG A, MARIENHAGEN J, RUFF A J, et al. Directed evolution of P450 BM 3 into a p-xylene hydroxylase[J]. ChemCatChem, 2012, 4(6): 771-773. |
| 94 | BJERREGAARD S, PEDERSEN H, VEDSTESEN H, et al. Parenteral water/oil emulsions containing hydrophilic compounds with enhanced in vivo retention: formulation, rheological characterisation and study of in vivo fate using whole body gamma-scintigraphy[J]. International Journal of Pharmaceutics, 2001, 215(1/2): 13-27. |
| 95 | PRODANOVIC R, OSTAFE R, BLANUSA M, et al. Vanadium bromoperoxidase-coupled fluorescent assay for flow cytometry sorting of glucose oxidase gene libraries in double emulsions[J]. Analytical and Bioanalytical Chemistry, 2012, 404(5): 1439-1447. |
| 96 | WORONOFF G, HARRAK A EL, MAYOT E, et al. New generation of amino coumarin methyl sulfonate-based fluorogenic substrates for amidase assays in droplet-based microfluidic applications[J]. Analytical Chemistry, 2011, 83(8): 2852-2857. |
| 97 | CHENG F, KARDASHLIEV T, PITZLER C, et al. A competitive flow cytometry screening system for directed evolution of therapeutic enzyme[J]. ACS Synthetic Biology, 2015, 4(7): 768-775. |
| 98 | BARAHONA E, ISIDRO E S, SIERRA-HERAS L, et al. A directed genome evolution method to enhance hydrogen production in Rhodobacter capsulatus [J]. Frontiers in Microbiology, 2022, 13: 991123. |
| 99 | BARAHONA E, JIMÉNEZ-VICENTE E, RUBIO L M. Hydrogen overproducing nitrogenases obtained by random mutagenesis and high-throughput screening[J]. Scientific Reports, 2016, 6: 38291. |
| 100 | KASEY C M, ZERRAD M, LI Y W, et al. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology[J]. ACS Synthetic Biology, 2018, 7(1): 227-239. |
| 101 | ZHENG J T, SAGAR V, SMOLINSKY A, et al. Structure and function of the macrolide biosensor protein, MphR(a), with and without erythromycin[J]. Journal of Molecular Biology, 2009, 387(5): 1250-1260. |
| 102 | QIU X L, XU P, ZHAO X R, et al. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica [J]. Metabolic Engineering, 2020, 60: 66-76. |
| 103 | MEYER A, PELLAUX R, POTOT S, et al. Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors[J]. Nature Chemistry, 2015, 7(8): 673-678. |
| 104 | JOSEPHIDES D, DAVOLI S, WHITLEY W, et al. Cyto-mine: an integrated, picodroplet system for high-throughput single-cell analysis, sorting, dispensing, and monoclonality assurance[J]. SLAS Technology, 2020, 25(2): 177-189. |
| 105 | GAA R, MENANG-NDI E, PRATAPA S, et al. Versatile and rapid microfluidics-assisted antibody discovery[J]. mAbs, 2021, 13(1): 1978130. |
| [1] | 晏雄鹰, 王振, 娄吉芸, 张皓瑜, 黄星宇, 王霞, 杨世辉. 生物燃料高效生产微生物细胞工厂构建研究进展[J]. 合成生物学, 2023, 4(6): 1082-1121. |
| [2] | 吴玉洁, 刘欣欣, 刘健慧, 杨开广, 随志刚, 张丽华, 张玉奎. 基于高通量液相色谱质谱技术的菌株筛选与关键分子定量分析研究进展[J]. 合成生物学, 2023, 4(5): 1000-1019. |
| [3] | 高纤云, 牛灵雪, 见妮, 管宁子. 微生物合成生物学在疾病诊疗上的应用进展[J]. 合成生物学, 2023, 4(2): 263-282. |
| [4] | 任师超, 孙秋艳, 冯旭东, 李春. 微生物细胞工厂合成五环三萜皂苷类化合物[J]. 合成生物学, 2022, 3(1): 168-183. |
| [5] | 熊亮斌, 宋璐, 赵云秋, 刘坤, 刘勇军, 王风清, 魏东芝. 甾体化合物绿色生物制造:从生物转化到微生物从头合成[J]. 合成生物学, 2021, 2(6): 942-963. |
| [6] | 郭亮, 高聪, 柳亚迪, 陈修来, 刘立明. 大肠杆菌生产饲用氨基酸的研究进展[J]. 合成生物学, 2021, 2(6): 964-981. |
| [7] | 陈久洲, 王钰, 蒲伟, 郑平, 孙际宾. 5-氨基乙酰丙酸生物合成技术的发展及展望[J]. 合成生物学, 2021, 2(6): 1000-1016. |
| [8] | 袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673. |
| [9] | 夏思杨, 江丽红, 蔡谨, 黄磊, 徐志南, 连佳长. 酿酒酵母基因组进化的研究进展[J]. 合成生物学, 2020, 1(5): 556-569. |
| [10] | 许可, 王靖楠, 李春. 智能抗逆微生物细胞工厂与绿色生物制造[J]. 合成生物学, 2020, 1(4): 427-439. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||