合成生物学 ›› 2023, Vol. 4 ›› Issue (5): 932-946.DOI: 10.12211/2096-8280.2023-035
自动化高通量技术在天然产物生物合成中的应用
胡哲辉1,2, 徐娟2, 卞光凯1
- 1.中国科学院深圳先进技术研究院合成生物学研究所,广东 深圳 518055
2.华中农业大学园艺林学学院,果蔬园艺作物种质创新与利用全国重点实验室,湖北 武汉 430071
-
收稿日期:2023-05-06修回日期:2023-07-24出版日期:2023-10-31发布日期:2023-11-15 -
通讯作者:卞光凯 -
作者简介:胡哲辉 (1998—),男,博士研究生。研究方向为植物来源萜类天然产物生物合成。E-mail:zh.hu2@siat.ac.cn卞光凯 (1986—),男,研究员,博士生导师。研究方向为天然产物生物合成及高值天然产物高效合成;利用合成生物学手段改造丝状真菌生产高性能生物材料。E-mail:gk.bian@siat.ac.cn -
基金资助:国家重点研发计划“绿色生物制造”重点专项(2022YFC2106100);国家自然科学基金面上项目(32070063)
Application of automated high-throughput technology in natural product biosynthesis
HU Zhehui1,2, XU Juan2, BIAN Guangkai1
- 1.Insititute of Syntheitc Biology,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
2.National Key Laboeratory for Germplasm Innovation & Utilization of Horticultural Crops,College of Horticulture and Forestry,Huazhong Agricultural University,Wuhan 430071,Hubei,China
-
Received:2023-05-06Revised:2023-07-24Online:2023-10-31Published:2023-11-15 -
Contact:BIAN Guangkai
摘要:
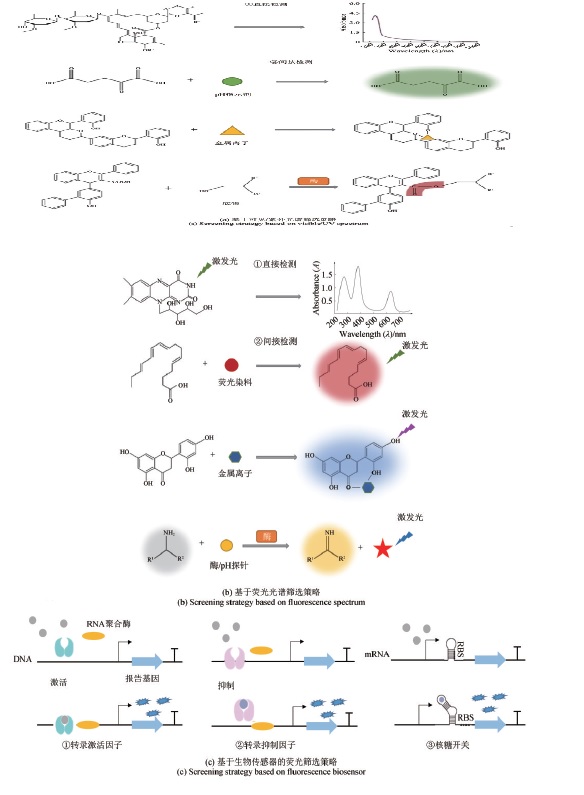
天然产物与人们的日常生活息息相关,是药物、食品、营养添加剂、色素、化妆品等产品的重要来源。发掘新的天然产物并实现其高效合成对于提升人们的生活水平至关重要。然而,研究通量低和产物产量低已成为限制该领域快速发展的两大瓶颈。为了克服这些问题,自动化高通量技术的引入在天然产物生物合成领域具有重要意义。该技术可以将高度依赖人力资源的低通量和随机性手工操作的实验过程转变为基于先进设备的自动化、标准化和高效的研究过程。因此,将自动化高通量技术引入天然产物生物合成领域可以有效解决研究瓶颈,加速研发进程。本文综述了自动化高通量技术在天然产物生物合成领域的应用,阐述了其在天然产物挖掘、高效生物合成和快速检测等方面的优势。最后,讨论了该技术现存缺陷,包括仪器设备造价高昂、操作复杂,并且一些装置尚未得到推广。相信在合成生物、信息技术、自动化等多领域的共同协作下,自动化高通量技术具备的独特优势将为新型天然产物功能产品的开发提供可持续来源。
中图分类号:
引用本文
胡哲辉, 徐娟, 卞光凯. 自动化高通量技术在天然产物生物合成中的应用[J]. 合成生物学, 2023, 4(5): 932-946.
HU Zhehui, XU Juan, BIAN Guangkai. Application of automated high-throughput technology in natural product biosynthesis[J]. Synthetic Biology Journal, 2023, 4(5): 932-946.
| 应用领域 | 传统方法 | 自动化高通量方法 | ||
|---|---|---|---|---|
| 方式 | 特点 | 方式 | 特点 | |
| 天然产物的挖掘 | 直接分离 同源激活 异源表达 | 生物样本单一 局限于单个基因的异源表达 产物丰度低、筛选通量低 | 打造特定微生物高产底盘 基于海量数据库批量挖掘 高通量工作站自动化验证 | 不受限于特定的生物样本 充分释放酶的产物合成潜力 规模化、自动化 |
| 天然产物高效合成 | 突变体文库平板筛选 关键酶的定点诱变 人工改造代谢通路 | 突变体间微小差异分辨率低 试错过程需要消耗大量劳动力 无法完全释放酶的催化潜力 | 基于auto-HTP对突变体文库高效筛选 自动化改造底盘细胞 限速酶定向进化 微生物适应性进化 | 自动化实现海量试错 多维度改造底盘细胞 缩短菌株改造研发周期 |
| 天然产物的快速检测 | 气相色谱、质谱 液相色谱、质谱 核磁共振 基于紫外/可见光谱检测 | 耗时长、通量低 对目标产物的产量和纯度要求高 | 基于荧光光谱检测 基于生物传感器的荧光检测 基于先进光谱技术的检测 | 便捷、高效、省时 对目标产物的动态、实时、自动化检测 |
表1 传统方法与自动化高通量方法在天然产物生物合成中的比较
Table 1 A comparison of automated high-throughput and traditional methods in natural product biosynthesis
| 应用领域 | 传统方法 | 自动化高通量方法 | ||
|---|---|---|---|---|
| 方式 | 特点 | 方式 | 特点 | |
| 天然产物的挖掘 | 直接分离 同源激活 异源表达 | 生物样本单一 局限于单个基因的异源表达 产物丰度低、筛选通量低 | 打造特定微生物高产底盘 基于海量数据库批量挖掘 高通量工作站自动化验证 | 不受限于特定的生物样本 充分释放酶的产物合成潜力 规模化、自动化 |
| 天然产物高效合成 | 突变体文库平板筛选 关键酶的定点诱变 人工改造代谢通路 | 突变体间微小差异分辨率低 试错过程需要消耗大量劳动力 无法完全释放酶的催化潜力 | 基于auto-HTP对突变体文库高效筛选 自动化改造底盘细胞 限速酶定向进化 微生物适应性进化 | 自动化实现海量试错 多维度改造底盘细胞 缩短菌株改造研发周期 |
| 天然产物的快速检测 | 气相色谱、质谱 液相色谱、质谱 核磁共振 基于紫外/可见光谱检测 | 耗时长、通量低 对目标产物的产量和纯度要求高 | 基于荧光光谱检测 基于生物传感器的荧光检测 基于先进光谱技术的检测 | 便捷、高效、省时 对目标产物的动态、实时、自动化检测 |
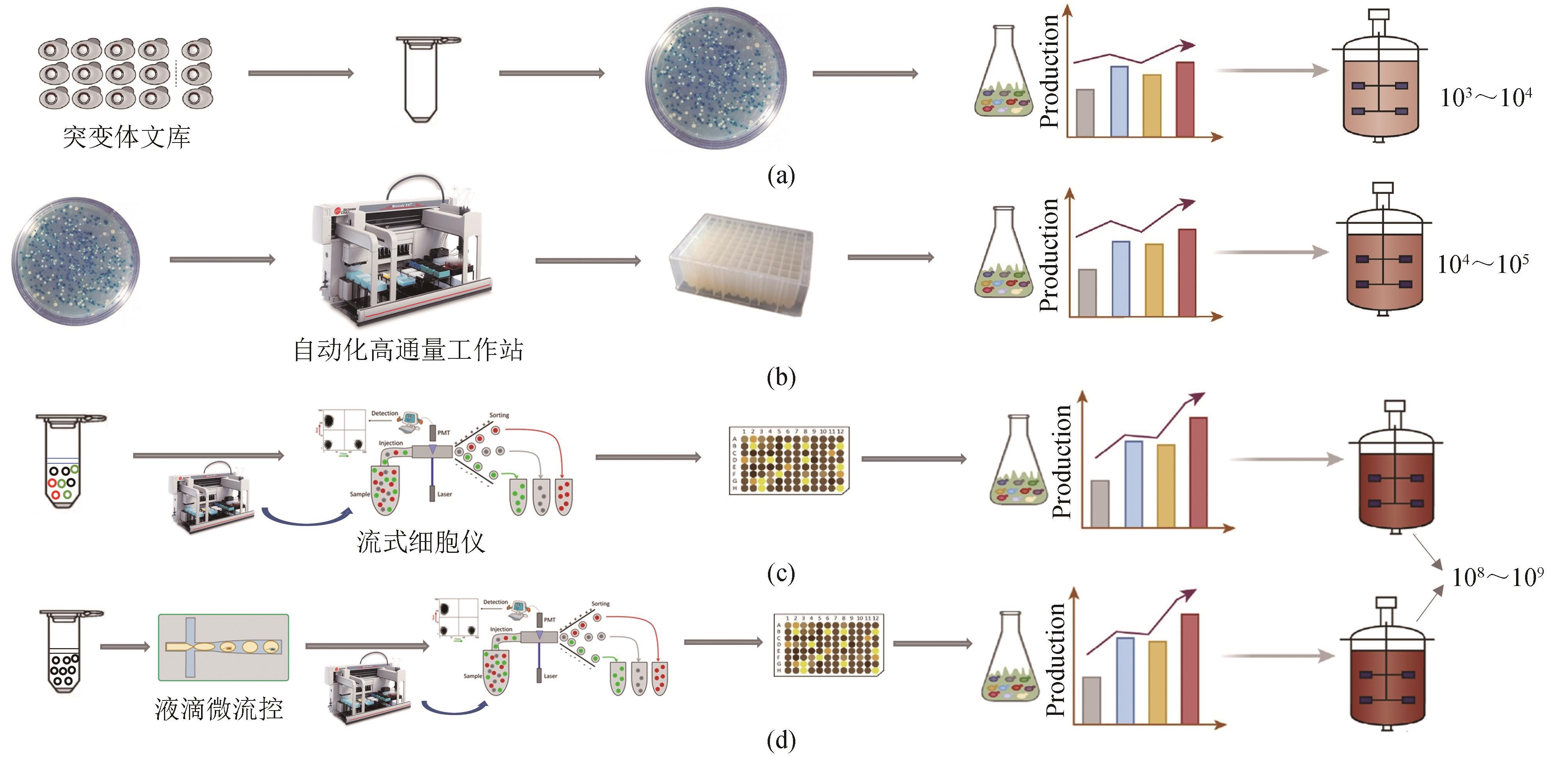
图1 不同通量筛选方法的比较[41-42](a) 传统平板筛选法,筛选通量为103~104;(b) 基于自动化设备的微孔板筛选法,筛选通量为104~105;(c) 基于流式细胞仪的荧光激活细胞分选,筛选通量为108~109;(d) 基于微流控芯片和分选设备的液滴微流控分选,筛选通量为108~109
Fig. 1 A comparison of different screening methods[41-42](a) Traditional screening method based on agar plates, with a screening throughput of 103~104; (b) Microplate screening method based on automation equipment, with a screening throughput of 104~105; (c) Fluorescence activated cell sorting, with a screening throughput of 108~109; (d) Droplet-based microfluidic sorting, with a screening throughput of 108~109
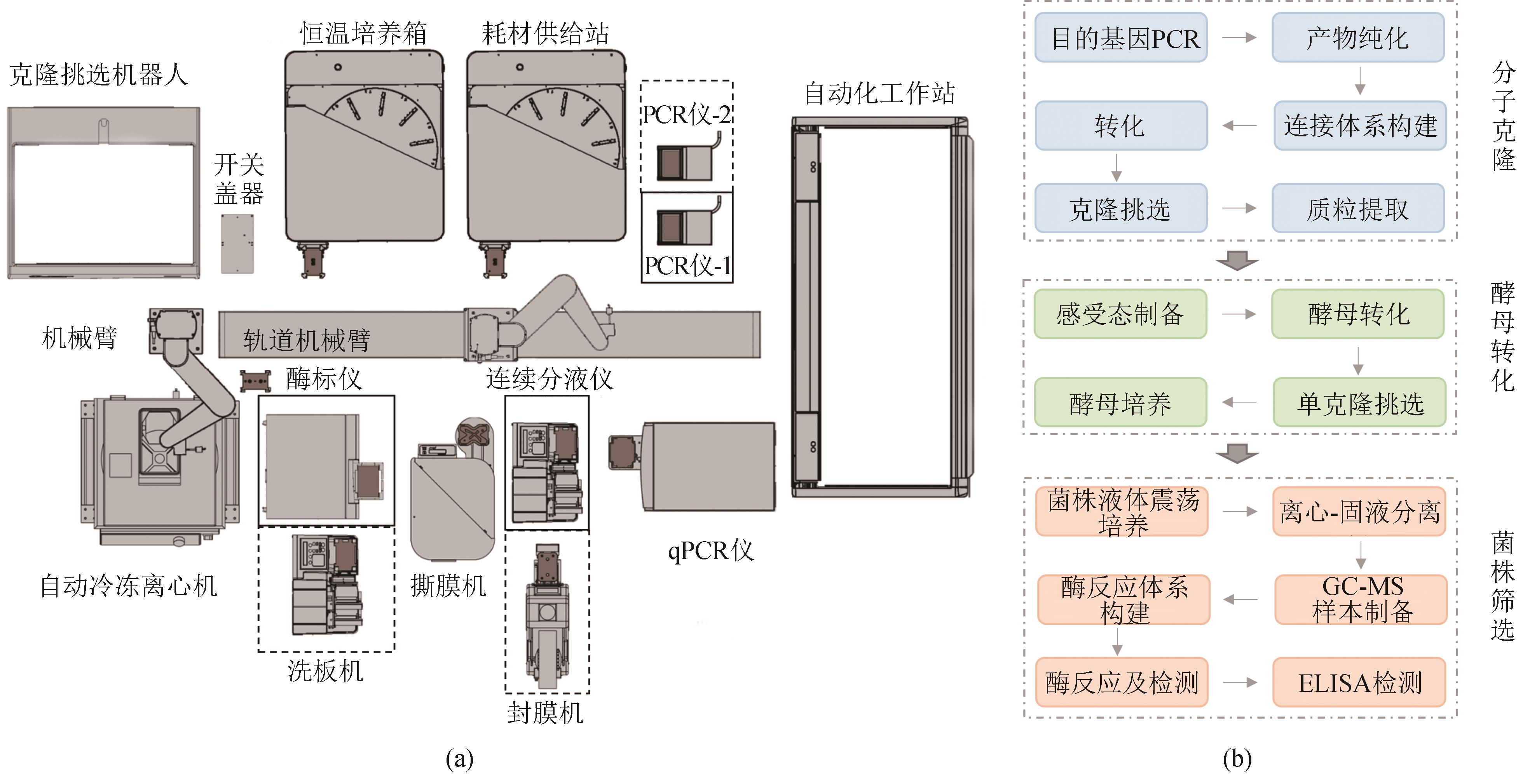
图2 自动化高通量工作站(a)自动化高通量工作站平面示意图;(b)自动化高通量工作流程(以酵母为例)
Fig. 2 Automated high-throughput workstation(a) Schematic diagram of the automated high-throughput workstation; (b) A representative workflow of automated high-throughput process, with yeast engineering shown as an example
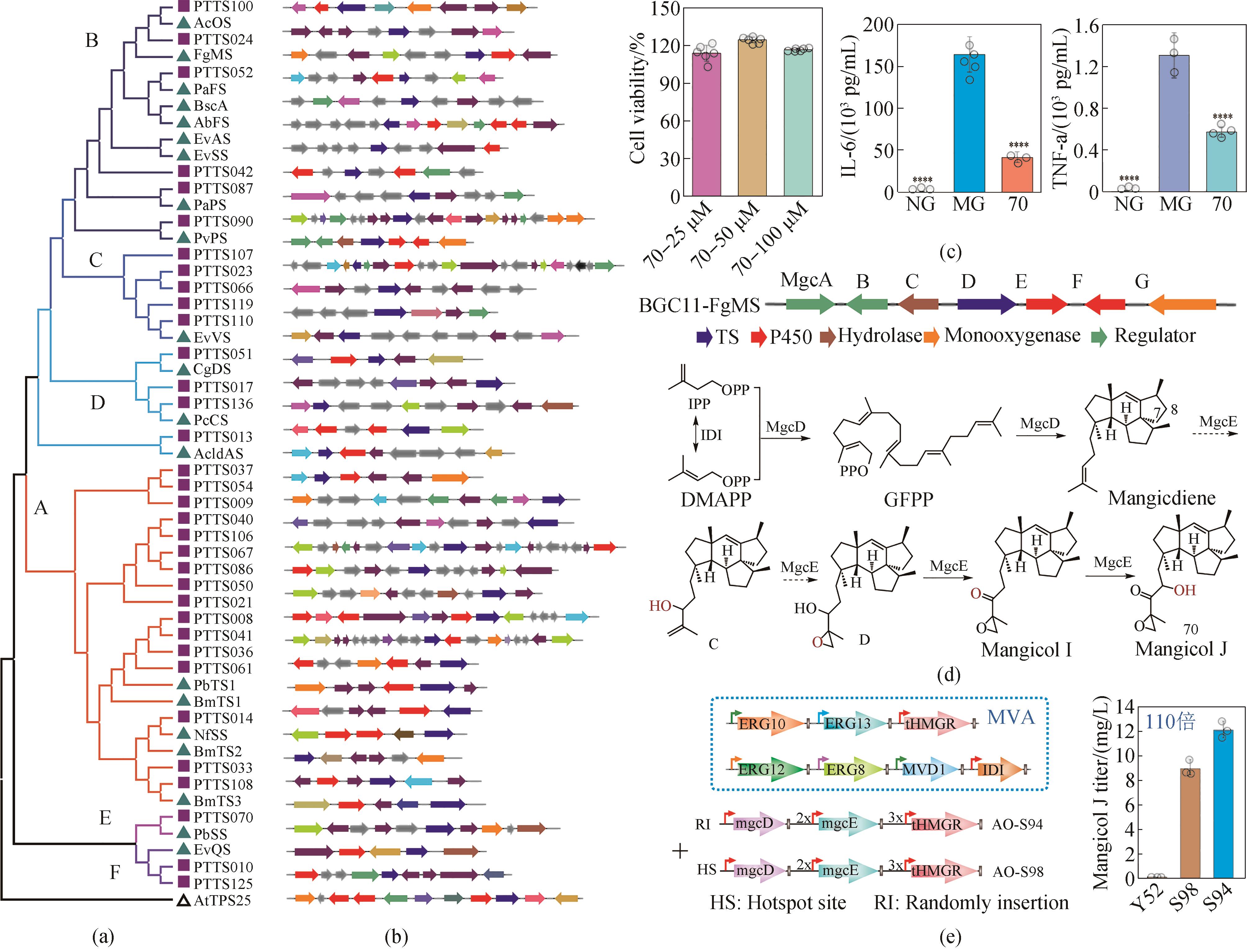
图3 基于生物铸造平台对新功能萜类基因(簇)的高效挖掘及抗炎新分子mangicol J的高效合成[51, 56](a) 34个新嵌合萜类合酶(PTTC)的批量挖掘;(b) 39个萜类基因簇重建为208个突变株;(c) 通过高通量筛选从185个萜类产物中筛选到高抗炎活性70号(mangicol J)化合物;(d) mangicol J生物合成途径解析;(e) 构建通用米曲霉底盘实现mangicol J高效合成
Fig. 3 The application of biocasting in efficient mining of new functional terpenoid genes (gene clusters) and the biosynthesis of mangicol J, a new anti-inflammatory molecule[51, 56](a) The batch mining of 34 new chimeric terpenoid synthases (PTTC); (b) The reconstruction of 39 terpenoid gene clusters into 208 mutants; (c) High-throughput screening of 185 terpenoids leading to the isolation of compound 70 (mangicol J) with high anti-inflammatory activity; (d) The elucidation of the biosynthetic pathway of mangicol J; (e) The construction of a general Aspergillus oryzae chassis for efficient biosynthesis of mangicol J
| 1 | RODRIGUES T, REKER D, SCHNEIDER P, et al. Counting on natural products for drug design[J]. Nature Chemistry, 2016, 8(6): 531-541. |
| 2 | ATANASOV A G, WALTENBERGER B, PFERSCHY-WENZIG E M, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review[J]. Biotechnology Advances, 2015, 33(8): 1582-1614. |
| 3 | HARVEY A L, EDRADA-EBEL R, QUINN R J. The re-emergence of natural products for drug discovery in the genomics era[J]. Nature Reviews Drug Discovery, 2015, 14(2): 111-129. |
| 4 | BIAN G K, HAN Y C, HOU A W, et al. Releasing the potential power of terpene synthases by a robust precursor supply platform[J]. Metabolic Engineering, 2017, 42: 1-8. |
| 5 | BIAN G K, DENG Z X, LIU T G. Strategies for terpenoid overproduction and new terpenoid discovery[J]. Current Opinion in Biotechnology, 2017, 48: 234-241. |
| 6 | GAO J C, JIANG L H, LIAN J Z. Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products[J]. Synthetic and Systems Biotechnology, 2021, 6(2): 110-119. |
| 7 | MORRISON K C, HERGENROTHER P J. Natural products as starting points for the synthesis of complex and diverse compounds[J]. Natural Product Reports, 2014, 31(1): 6-14. |
| 8 | SMANSKI M J, ZHOU H, CLAESEN J, et al. Synthetic biology to access and expand nature's chemical diversity[J]. Nature Reviews Microbiology, 2016, 14(3): 135-149. |
| 9 | PYNE M E, NARCROSS L, MARTIN V J J. Engineering plant secondary metabolism in microbial systems[J]. Plant Physiology, 2019, 179(3): 844-861. |
| 10 | ZHU F Y, ZHONG X F, HU M Z, et al. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli [J]. Biotechnology and Bioengineering, 2014, 111(7): 1396-1405. |
| 11 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 12 | RO D K, PARADISE E M, OUELLET M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast[J]. Nature, 2006, 440(7086): 940-943. |
| 13 | BIAN G K, YUAN Y J, TAO H, et al. Production of taxadiene by engineering of mevalonate pathway in Escherichia coli and endophytic fungus Alternaria alternata TPF6 [J]. Biotechnology Journal, 2017, 12(4): 1600697. |
| 14 | SAGWAN-BARKDOLL L, ANTEROLA A M. Taxadiene-5α-ol is a minor product of CYP725A4 when expressed in Escherichia coli [J]. Biotechnology and Applied Biochemistry, 2018, 65(3): 294-305. |
| 15 | ZHU C Y, YOU X, WU T, et al. Efficient utilization of carbon to produce aromatic valencene in Saccharomyces cerevisiae using mannitol as the substrate[J]. Green Chemistry, 2022, 24(11): 4614-4627. |
| 16 | YE Z L, HUANG Y L, SHI B, et al. Coupling cell growth and biochemical pathway induction in Saccharomyces cerevisiae for production of (+)-valencene and its chemical conversion to (+)-nootkatone[J]. Metabolic Engineering, 2022, 72: 107-115. |
| 17 | VELÁZQUEZ J E, SADAÑOSKI M A, ZAPATA P D, et al. Bioproduction of α-terpineol and R-(+)-limonene derivatives by terpene-tolerant ascomycete fungus as a potential contribution to the citrus value chain[J]. Journal of Applied Microbiology, 2021, 130(1): 76-89. |
| 18 | TAO H, LAUTERBACH L, BIAN G K, et al. Discovery of non-squalene triterpenes[J]. Nature, 2022, 606(7913): 414-419. |
| 19 | ZHU X X, LIU X N, LIU T, et al. Synthetic biology of plant natural products: from pathway elucidation to engineered biosynthesis in plant cells[J]. Plant Communications, 2021, 2(5): 100229. |
| 20 | LV X M, GU J L, WANG F, et al. Combinatorial pathway optimization in Escherichia coli by directed co-evolution of rate-limiting enzymes and modular pathway engineering[J]. Biotechnology and Bioengineering, 2016, 113(12): 2661-2669. |
| 21 | MARKEL U, ESSANI K D, BESIRLIOGLU V, et al. Advances in ultrahigh-throughput screening for directed enzyme evolution[J]. Chemical Society Reviews, 2020, 49(1): 233-262. |
| 22 | SILVERMAN L, CAMPBELL R, BROACH J R. New assay technologies for high-throughput screening[J]. Current Opinion in Chemical Biology, 1998, 2(3): 397-403. |
| 23 | SHAO F C, LEE P W, LI H, et al. Emerging platforms for high-throughput enzymatic bioassays[J]. Trends in Biotechnology, 2023, 41(1): 120-133. |
| 24 | CLARKE L J, KITNEY R I. Synthetic biology in the UK - an outline of plans and progress[J]. Synthetic and Systems Biotechnology, 2016, 1(4): 243-257. |
| 25 | APPLETON E, MADSEN C, ROEHNER N, et al. Design automation in synthetic biology[J]. Cold Spring Harbor Perspectives in Biology, 2017, 9(4): a023978. |
| 26 | 饶聪, 云轩, 虞沂, 等. 微生物药物的合成生物学研究进展[J]. 合成生物学, 2020, 1(1): 92-102. |
| RAO C, YUN X, YU Y, et al. Research progress in synthetic biology of microbial drugs [J]. Synthetic Biology, 2020, 1(1): 92-102. | |
| 27 | HERTZBERG R P, POPE A J. High-throughput screening: new technology for the 21st century[J]. Current Opinion in Chemical Biology, 2000, 4(4): 445-451. |
| 28 | DÖRR M, FIBINGER M P C, LAST D, et al. Fully automatized high-throughput enzyme library screening using a robotic platform[J]. Biotechnology and Bioengineering, 2016, 113(7): 1421-1432. |
| 29 | DU G S, FANG Q, DEN TOONDER J M J. Microfluidics for cell-based high throughput screening platforms—a review[J]. Analytica Chimica Acta, 2016, 903: 36-50. |
| 30 | LUO Z S, ZENG W Z, DU G C, et al. A high-throughput screening procedure for enhancing pyruvate production in Candida glabrata by random mutagenesis[J]. Bioprocess and Biosystems Engineering, 2017, 40(5): 693-701. |
| 31 | MACHILLOT P, QUINTAL C, DALONNEAU F, et al. Automated buildup of biomimetic films in cell culture microplates for high-throughput screening of cellular behaviors[J]. Advanced Materials, 2018, 30(27): 1801097. |
| 32 | CAO X M, LUO Z S, ZENG W Z, et al. Enhanced avermectin production by Streptomyces avermitilis ATCC 31267 using high-throughput screening aided by fluorescence-activated cell sorting[J]. Applied Microbiology and Biotechnology, 2018, 102(2): 703-712. |
| 33 | FENG W Q, UEDA E, LEVKIN P A. Droplet microarrays: from surface patterning to high-throughput applications[J]. Advanced Materials, 2018, 30(20): 1706111. |
| 34 | POPOVIC A, TCHIGVINTSEV A, TRAN H, et al. Metagenomics as a tool for enzyme discovery: hydrolytic enzymes from marine-related metagenomes[M/OL]//Advances in experimental medicine and biology: prokaryotic systems biology. Cham: Springer, 2015, 883: 1-20 [2023-04-10]. . |
| 35 | 杨建花, 苏晓岚, 朱蕾蕾. 高通量筛选系统在定向改造中的新进展[J]. 生物工程学报, 2021, 37(7): 2197-2210. |
| YANG J H, SU X L, ZHU L L. Advances of high-throughput screening system in reengineering of biological entities[J]. Journal of Biotechnology, 2021, 37(7): 2197-2210. | |
| 36 | LONGWELL C K, LABANIEH L, COCHRAN J R. High-throughput screening technologies for enzyme engineering[J]. Current Opinion in Biotechnology, 2017, 48: 196-202. |
| 37 | LEEMHUIS H, KELLY R M, DIJKHUIZEN L. Directed evolution of enzymes: library screening strategies[J]. IUBMB Life, 2009, 61(3): 222-228. |
| 38 | XIAO H, BAO Z H, ZHAO H M. High throughput screening and selection methods for directed enzyme evolution[J]. Industrial & Engineering Chemistry Research, 2015, 54(16): 4011-4020. |
| 39 | YANG G Y, WITHERS S G. Ultrahigh-throughput FACS-based screening for directed enzyme evolution[J]. ChemBioChem, 2009, 10(17): 2704-2715. |
| 40 | MEWIS K, TAUPP M, HALLAM S J. A high throughput screen for biomining cellulase activity from metagenomic libraries[J]. Journal of Visualized Experiments, 2011(48): e2461. |
| 41 | QIN Y L, WU L, WANG J G, et al. A fluorescence-activated single-droplet dispenser for high accuracy single-droplet and single-cell sorting and dispensing[J]. Analytical Chemistry, 2019, 91(10): 6815-6819. |
| 42 | ZENG W Z, GUO L K, XU S, et al. High-throughput screening technology in industrial biotechnology[J]. Trends in Biotechnology, 2020, 38(8): 888-906. |
| 43 | 唐婷, 付立豪, 郭二鹏, 等. 自动化合成生物技术与工程化设施平台[J]. 科学通报, 2021, 66(3): 300-309. |
| TANG T, FU L H, GUO E P, et al. Automation in synthetic biology using biological foundries[J]. Chinese Science Bulletin, 2021, 66(3): 300-309. | |
| 44 | HILLSON N, CADDICK M, CAI Y Z, et al. Building a global alliance of biofoundries[J]. Nature Communications, 2019, 10: 2040. |
| 45 | 崔金明, 张炳照, 马迎飞, 等. 合成生物学研究的工程化平台[J]. 中国科学院院刊, 2018, 33(11): 1249-1257. |
| CUI J M, ZHANG B Z, MA Y F, et al. Engineering platforms for synthetic biology research[J]. Bulletin of Chinese Academy of Sciences, 2018, 33(11): 1249-1257. | |
| 46 | YEOH J W, SWAINSTON N, VEGH P, et al. SynBiopython: an open-source software library for Synthetic Biology[J]. Synthetic Biology, 2021, 6(1): ysab001. |
| 47 | 张亭, 冷梦甜, 金帆, 等. 合成生物研究重大科技基础设施概述[J]. 合成生物学, 2022(1): 184-194. |
| ZHANG T, LENG M T, JIN F, et al. Overview on platform for synthetic biology research at Shenzhen[J]. Synthetic Biology Journal, 2022(1): 184-194. | |
| 48 | CHAO R, LIANG J, TASAN I, et al. Fully automated one-step synthesis of single-transcript TALEN pairs using a biological foundry[J]. ACS Synthetic Biology, 2017, 6(4): 678-685. |
| 49 | SI T, CHAO R, MIN Y H, et al. Automated multiplex genome-scale engineering in yeast[J]. Nature Communications, 2017, 8: 15187. |
| 50 | WANI M C, TAYLOR H L, WALL M E, et al. Plant antitumor agents. Ⅵ. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia [J]. Journal of the American Chemical Society, 1971, 93(9): 2325-2327. |
| 51 | YUAN Y J, CHENG S, BIAN G K, et al. Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi[J]. Nature Catalysis, 2022, 5(4): 277-287. |
| 52 | CHEVRETTE M G, GUTIÉRREZ-GARCÍA K, SELEM-MOJICA N, et al. Evolutionary dynamics of natural product biosynthesis in bacteria[J]. Natural Product Reports, 2020, 37(4): 566-599. |
| 53 | CHEVRETTE M G, GAVRILIDOU A, MANTRI S, et al. The confluence of big data and evolutionary genome mining for the discovery of natural products[J]. Natural Product Reports, 2021, 38(11): 2024-2040. |
| 54 | CHEN S C, ZHANG C, ZHANG L H. Investigation of the molecular landscape of bacterial aromatic polyketides by global analysis of type Ⅱ polyketide synthases[J]. Angewandte Chemie International Edition, 2022, 61(24): e202202286. |
| 55 | ZIEMERT N, ALANJARY M, WEBER T. The evolution of genome mining in microbes - a review[J]. Natural Product Reports, 2016, 33(8): 988-1005. |
| 56 | CHEN R, JIA Q D, MU X, et al. Systematic mining of fungal chimeric terpene synthases using an efficient precursor-providing yeast chassis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(29): e2023247118. |
| 57 | NARITA K, SATO H, MINAMI A, et al. Focused genome mining of structurally related sesterterpenes: enzymatic formation of enantiomeric and diastereomeric products[J]. Organic Letters, 2017, 19(24): 6696-6699. |
| 58 | 范震, 潘海学, 唐功利. 工程酵母助力真菌嵌合萜类合酶的快速系统挖掘[J]. 合成生物学, 2021, 2(5): 666-673. |
| FAN Z, PAN H X, TANG G L. Engineered yeast facilitates rapid and systematic mining of fungal chimeric terpene synthases[J]. Synthetic Biology Journal, 2021, 2(5): 666-673. | |
| 59 | 涂然, 李世新, 李昊霓, 等. 液滴微流控技术在微生物工程菌株选育中的应用进展[J]. 合成生物学, 2023, 4(1): 165-184. |
| TU R, LI S X, LI H N, et al. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains[J]. Synthetic Biology Journal, 2023, 4(1): 165-184. | |
| 60 | MA F Q, CHUNG M T, YAO Y, et al. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform[J]. Nature Communications, 2018, 9: 1030. |
| 61 | LEAVELL M D, SINGH A H, KAUFMANN-MALAGA B B. High-throughput screening for improved microbial cell factories, perspective and promise[J]. Current Opinion in Biotechnology, 2020, 62: 22-28. |
| 62 | MEADOWS A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production[J]. Nature, 2016, 537(7622): 694-697. |
| 63 | LEVISSON M, ARAYA-CLOUTIER C, DE BRUIJN W J C, et al. Toward developing a yeast cell factory for the production of prenylated flavonoids[J]. Journal of Agricultural and Food Chemistry, 2019, 67(49): 13478-13486. |
| 64 | LAUCHLI R, RABE K S, KALBARCZYK K Z, et al. High-throughput screening for terpene-synthase-cyclization activity and directed evolution of a terpene synthase[J]. Angewandte Chemie International Edition, 2013, 52(21): 5571-5574. |
| 65 | FURUBAYASHI M, IKEZUMI M, KAJIWARA J, et al. A high-throughput colorimetric screening assay for terpene synthase activity based on substrate consumption[J]. PLoS One, 2014, 9(3): e93317. |
| 66 | ZHOU S H, LIU P R, CHEN J, et al. Characterization of mutants of a tyrosine ammonia-lyase from Rhodotorula glutinis [J]. Applied Microbiology and Biotechnology, 2016, 100(24): 10443-10452. |
| 67 | HEMMERICH J, NOACK S, WIECHERT W, et al. Microbioreactor systems for accelerated bioprocess development[J]. Biotechnology Journal, 2018, 13(4): 1700141. |
| 68 | WONG B G, MANCUSO C P, KIRIAKOV S, et al. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER[J]. Nature Biotechnology, 2018, 36(7): 614-623. |
| 69 | JIAN X J, GUO X J, WANG J, et al. Microbial microdroplet culture system (MMC): an integrated platform for automated, high-throughput microbial cultivation and adaptive evolution[J]. Biotechnology and Bioengineering, 2020, 117(6): 1724-1737. |
| 70 | OWENS R J. Structural proteomics: high-throughput methods. preface[J]. Methods in Molecular Biology, 2015, 1261: v. |
| 71 | JIAN X J, GUO X J, WANG J, et al. Automated microbial cultivation and adaptive evolution using microbial microdroplet culture system (MMC)[J]. Journal of Visualized Experiments, 2022(180): e62800. |
| 72 | LEE K S, BOCCAZZI P, SINSKEY A J, et al. Microfluidic chemostat and turbidostat with flow rate, oxygen, and temperature control for dynamic continuous culture[J]. Lab on a Chip, 2011, 11(10): 1730-1739. |
| 73 | BENEYTON T, THOMAS S, GRIFFITHS A D, et al. Droplet-based microfluidic high-throughput screening of heterologous enzymes secreted by the yeast Yarrowia lipolytica [J].Microbial Cell Factories, 2017, 16(1): 18. |
| 74 | GRÜNBERGER A, WIECHERT W, KOHLHEYER D. Single-cell microfluidics: opportunity for bioprocess development[J]. Current Opinion in Biotechnology, 2014, 29: 15-23. |
| 75 | PICARD L P, PROSSER R S. Advances in the study of GPCRs by 19F NMR[J]. Current Opinion in Structural Biology, 2021, 69: 169-176. |
| 76 | WANG Q, FENG L R, WEI L, et al. Mutation breeding of lycopene-producing strain Blakeslea trispora by a novel atmospheric and room temperature plasma (ARTP)[J]. Applied Biochemistry and Biotechnology, 2014, 174(1): 452-460. |
| 77 | TAN J, CHU J, HAO Y Y, et al. High-throughput system for screening of cephalosporin C high-yield strain by 48-deep-well microtiter plates[J]. Applied Biochemistry and Biotechnology, 2013, 169(5): 1683-1695. |
| 78 | ZENG W Z, DU G C, CHEN J, et al. A high-throughput screening procedure for enhancing α-ketoglutaric acid production in Yarrowia lipolytica by random mutagenesis[J]. Process Biochemistry, 2015, 50(10): 1516-1522. |
| 79 | GUO C X, HU Y L, YANG C Y, et al. Developing a colorimetric assay for Fe(Ⅱ)/2-oxoglutarate-dependent dioxygenase[J]. Analytical Biochemistry, 2018, 548: 109-114. |
| 80 | HAMEDIRAD M, CHAO R, WEISBERG S, et al. Towards a fully automated algorithm driven platform for biosystems design[J]. Nature Communications, 2019, 10: 5150. |
| 81 | GAO F, HAO Z Z, SUN X H, et al. A versatile system for fast screening and isolation of Trichoderma reesei cellulase hyperproducers based on DsRed and fluorescence-assisted cell sorting[J].Biotechnology for Biofuels, 2018, 11(1): 261. |
| 82 | LI H Q, LI T F, ZUO H, et al. A novel rhodamine-based fluorescent pH probe for high-throughput screening of high-yield polymalic acid strains from random mutant libraries[J]. RSC Advances, 2016, 6(97): 94756-94762. |
| 83 | LIU Y, XUE Z L, CHEN S P, et al. A high-throughput screening strategy for accurate quantification of menaquinone based on fluorescence-activated cell sorting[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(6): 751-760. |
| 84 | ZHOU S H, LYU Y B, LI H Z, et al. Fine-tuning the (2 S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy[J]. Biotechnology and Bioengineering, 2019, 116(6): 1392-1404. |
| 85 | DEBON A, POTT M, OBEXER R, et al. Ultrahigh-throughput screening enables efficient single-round oxidase remodelling[J]. Nature Catalysis, 2019, 2(9): 740-747. |
| 86 | ZHOU S H, LYU Y B, LI H Z, et al. Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy[J]. Biotechnology and Bioengineering, 2019, 116(6): 1392-1404. |
| 87 | EGGELING L, BOTT M, MARIENHAGEN J. Novel screening methods—biosensors[J]. Current Opinion in Biotechnology, 2015, 35: 30-36. |
| 88 | DEKKER L, POLIZZI K M. Sense and sensitivity in bioprocessing—detecting cellular metabolites with biosensors[J]. Current Opinion in Chemical Biology, 2017, 40: 31-36. |
| 89 | JUÁREZ J F, LECUBE-AZPEITIA B, BROWN S L, et al. Biosensor libraries harness large classes of binding domains for construction of allosteric transcriptional regulators[J]. Nature Communications, 2018, 9: 3101. |
| 90 | LIM H G, JANG S H, JANG S Y, et al. Design and optimization of genetically encoded biosensors for high-throughput screening of chemicals[J]. Current Opinion in Biotechnology, 2018, 54: 18-25. |
| 91 | SUN H H, ZHAO H M, ANG E L. A new biosensor for stilbenes and a cannabinoid enabled by genome mining of a transcriptional regulator[J]. ACS Synthetic Biology, 2020, 9(4): 698-705. |
| 92 | CALERO P, VOLKE D C, LOWE P T, et al. A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida [J]. Nature Communications, 2020, 11: 5045. |
| 93 | XIU Y, JANG S, JONES J A, et al. Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures[J]. Biotechnology and Bioengineering, 2017, 114(10): 2235-2244. |
| 94 | SARNAIK A, LIU A, NIELSEN D, et al. High-throughput screening for efficient microbial biotechnology[J]. Current Opinion in Biotechnology, 2020, 64: 141-150. |
| 95 | SHI S Y, XIE Y H, WANG G L, et al. Metabolite-based biosensors for natural product discovery and overproduction[J]. Current Opinion in Biotechnology, 2022, 75: 102699. |
| 96 | WANG X X, REN L H, SU Y T, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells[J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 97 | 张凡忠, 相长君, 张骊駻. 进化与大数据导向生物信息学在天然产物研究中的发展及应用 [J]. 合成生物学, 2023, 4(4): |
| ZHANG F Z, XIANG C J, ZHANG L X. Advances and applications of evolutionary analysis and big-data guided bioinformatics in natural product research[J]. Synthetic Biology Journal, 2023, 4(4): | |
| 98 | GAVRIILIDOU A, KAUTSAR S A, ZABURANNYI N, et al. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes[J]. Nature Microbiology, 2022, 7(5): 726-735. |
| 99 | SMOLKE C D, SILVER P A. Informing biological design by integration of systems and synthetic biology[J]. Cell, 2011, 144(6): 855-859. |
| 100 | KHAMBHATI K, BHATTACHARJEE G, GOHIL N, et al. Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 248. |
| 101 | YANG J H, TU R, YUAN H L, et al. Recent advances in droplet microfluidics for enzyme and cell factory engineering[J]. Critical Reviews in Biotechnology, 2021, 41(7): 1023-1045. |
| 102 | BOUZETOS E, GANAR K A, MASTROBATTISTA E, et al. (R)evolution-on-a-chip[J]. Trends in Biotechnology, 2022, 40(1): 60-76. |
| 103 | GIELEN F, HOURS R, EMOND S, et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS)[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(47): E7383-E7389. |
| 104 | LIN G M, WARDEN-ROTHMAN R, VOIGT C A. Retrosynthetic design of metabolic pathways to chemicals not found in nature[J]. Current Opinion in Systems Biology, 2019, 14: 82-107. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 程中玉, 李付琸. 基于P450选择性氧化的天然产物化学-酶法合成进展[J]. 合成生物学, 2024, 5(5): 960-980. |
| [13] | 张守祺, 王涛, 孔尧, 邹家胜, 刘元宁, 徐正仁. 天然产物的化学-酶法合成:方法与策略的演进[J]. 合成生物学, 2024, 5(5): 913-940. |
| [14] | 谢向前, 郭雯, 王欢, 李进. 含氨基乙烯半胱氨酸核糖体肽的生物合成与化学合成[J]. 合成生物学, 2024, 5(5): 981-996. |
| [15] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||