合成生物学 ›› 2021, Vol. 2 ›› Issue (6): 982-999.DOI: 10.12211/2096-8280.2021-002
芳香族氨基酸及其衍生物的细胞工厂构建策略
孙薇1,2, 丁冬芹2, 柏丹阳2, 朱亚如2, 解晓彤2,3, 张大伟2
- 1.天津科技大学生物工程学院,天津 300457
2.中国科学院天津工业生物技术研究所,天津 300308
3.大连工业大学生物工程学院,辽宁 大连 116000
-
收稿日期:2021-01-05修回日期:2021-04-16出版日期:2021-12-31发布日期:2021-04-21 -
通讯作者:张大伟 -
作者简介:孙薇 (1998—),女,硕士研究生。研究方向为氨基酸代谢。E-mail:sunw@tib.cas.cn张大伟 (1978—),男,博士,研究员。主要研究方向为采用分子遗传、合成生物技术、生物传感技术等,合成维生素、氨基酸及高附加值化合物;建立蛋白表达平台,实现蛋白质胞内表达或分泌表达,并开发新型表达系统。E-mail:zhang_dw@tib.cas.cn -
基金资助:国家重点研发计划(2018YFA0903700);天津市杰出青年科学基金(17JCJQJC45300);天津市合成生物技术创新能力提升行动项目(TSBICIP-CXRC-029)
Strategies of cell factory construction for the production of aromatic amino acids and their derivatives
SUN Wei1,2, DING Dongqin2, BAI Danyang2, ZHU Yaru2, XIE Xiaotong2,3, ZHANG Dawei2
- 1.Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
2.College of Bioengineering,Tianjin University of Science and Technology,Tianjin 300308,China
3.College of Bioengineering,Dalian University of Technology,Dalian 116000,Liaoning,China
-
Received:2021-01-05Revised:2021-04-16Online:2021-12-31Published:2021-04-21 -
Contact:ZHANG Dawei
摘要:
芳香族氨基酸及其衍生物由于其特定的生理活性,已广泛应用于医药、食品、饲料和化工等行业。利用重组微生物发酵生产芳香族氨基酸及其衍生物是满足全球日益增长需求的有效途径。通过将代谢工程策略与合成生物学、系统生物学和生物工程的发展相结合,在菌株的改造及优化方面取得了显著的进展。然而,合成芳香族氨基酸及其衍生物的代谢途径长且调控机制复杂,通过简单的代谢途径改造难以大幅提高产量,因此,近年来出现了很多相关的改造方法,为克服代谢途径中的限速问题提供了很好的借鉴意义。本文回顾和比较了最近在芳香族氨基酸及其衍生物合成方面应用的成熟技术和策略,包括常用的代谢途径改造策略(如增加前体供给、解除关键酶和阻遏蛋白的反馈抑制和阻遏抑制、改造转运系统、全局调节系统)以及菌株生长与生产产品耦联和菌株构建方法(如基于生物传感器的高通量筛选以及对培养基和培养条件的优化等),未来相关前沿技术如计算机辅助途径酶改造技术和筛选高产菌株的定向进化技术将助力芳香族氨基酸及其衍生物高产菌株的构建。
中图分类号:
引用本文
孙薇, 丁冬芹, 柏丹阳, 朱亚如, 解晓彤, 张大伟. 芳香族氨基酸及其衍生物的细胞工厂构建策略[J]. 合成生物学, 2021, 2(6): 982-999.
SUN Wei, DING Dongqin, BAI Danyang, ZHU Yaru, XIE Xiaotong, ZHANG Dawei. Strategies of cell factory construction for the production of aromatic amino acids and their derivatives[J]. Synthetic Biology Journal, 2021, 2(6): 982-999.
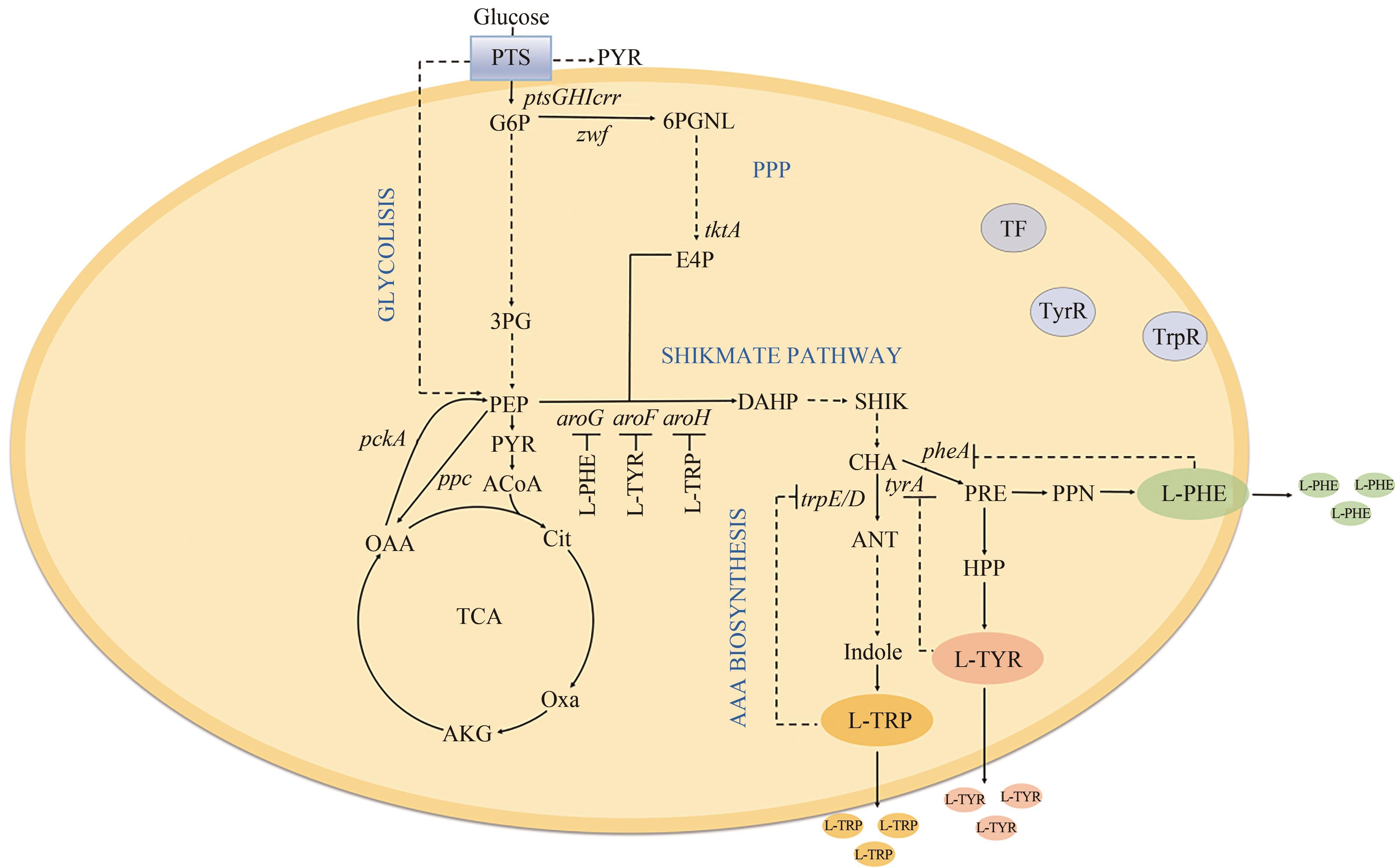
图1 芳香族氨基酸的合成途径PTS—磷酸转移酶系统;G6P—葡萄糖-6-磷酸;3PG—3-磷酸甘油酸;PYR—丙酮酸;ACoA—乙酰辅酶A;Cit—柠檬酸;Oxa—草酰琥珀酸;AKG—α-酮戊二酸;OAA—草酰乙酸;6PGNL—6-磷酸葡萄糖内脂;ANT—邻氨基苯甲酸;Indole—吲哚;PRE—预苯酸;PPN—苯丙酮酸;HPP—4-羟基苯丙酮酸;ptsGHIccr—编码磷酸转移酶基因;ppc—编码PEP羧化酶基因;pckA—编码PEP羧激酶基因;tktA—编码转酮酶基因;aroG/F/H—编码DAHP合酶基因;trpE/D—编码ANT合酶基因,tyrA/pheA—编码CHA变位酶基因
Fig. 1 Synthesis of aromatic amino acidsPTS—phosphotransferase system; G6P—glucose-6-phosphate; 3PG—3-phosphoglyceric acid; PYR—pyruvic acid; ACoA—acetyl coenzyme A; Cit—citrate; Oxa—oxalylsuccinic acid; AKG—α-ketoglutarate; OAA—oxaloacetic acid; 6PGNL—glucose 6-phosphate endolipid; ANT—o-aminobenzoic acid; PRE—prephenic acid; PPN—phenylpyruvic acid; HPP—4-hydroxyphenylpyruvic acidGene: ptsGHccr—ecoding phosphotransferase; ppc—encoding PEP carboxylase; pckA—encoding PEP carboxykinase; tktA—encoding transketolase; aroG/F/H—encoding DAHP synthase; trpE/D—encoding ANTA synthase; tyrA/pheA—encoding CHA mutase
| 生产方法 | 原理 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|---|
| 天然蛋白质水解法 | 利用盐酸使蛋白质发生水解 | 最初的提取方法 | 方法原始、产率极低、产物易被破坏 | [ |
| 化学合成法 | 添加化学试剂,改变碳链结构 | 快速、高效 | 工艺复杂、成本高、副产物多 | [ |
| 酶法 | 利用微生物中的芳香族氨基酸合成酶系 | 产物转化率高、副产物少、操作简便 | 筛选强活力酶困难、反应平衡难操控 | [ |
| 微生物转化法 | 以糖类为碳源,同时添加芳香族氨基酸的前体 | 耗能少、纯化简便 | 前体价格昂贵、改造周期较长 | [ |
| 微生物发酵法 | 以葡萄糖为碳源,利用微生物发酵生产芳香族氨基酸 | 成本低、环保、工艺流程易控制 | 获得工业化高产菌株较难 | [ |
表1 芳香族氨基酸生产方法的比较
Tab. 1 Comparison of production methods of aromatic amino acids
| 生产方法 | 原理 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|---|
| 天然蛋白质水解法 | 利用盐酸使蛋白质发生水解 | 最初的提取方法 | 方法原始、产率极低、产物易被破坏 | [ |
| 化学合成法 | 添加化学试剂,改变碳链结构 | 快速、高效 | 工艺复杂、成本高、副产物多 | [ |
| 酶法 | 利用微生物中的芳香族氨基酸合成酶系 | 产物转化率高、副产物少、操作简便 | 筛选强活力酶困难、反应平衡难操控 | [ |
| 微生物转化法 | 以糖类为碳源,同时添加芳香族氨基酸的前体 | 耗能少、纯化简便 | 前体价格昂贵、改造周期较长 | [ |
| 微生物发酵法 | 以葡萄糖为碳源,利用微生物发酵生产芳香族氨基酸 | 成本低、环保、工艺流程易控制 | 获得工业化高产菌株较难 | [ |
| 影响因素 | 详情 |
|---|---|
| 前体的供给 | PEP,涉及其生成和消耗的系统和酶有:PEP合酶、PEP羧激酶[ E4P,涉及其生成的酶有转酮酶、转醛酶[ |
| 关键酶的反馈抑制 | DAHP合酶、ANT合酶[ |
| 调控因子TyrR和TrpR对合成途径的转录调节 | TyrR调控蛋白调节8个非连续操纵子的表达[ TrpR调控蛋白调节基因aroH、mtr和色氨酸操纵子[ |
| 转运系统 | 分泌系统:转运基因tnaB、mtr、aroP负责芳香族氨基酸的运输[ 膜蛋白YddG[ |
| 其他因素 | L-Trp分支途径:色氨酸酶(tnaA)[ L-Tyr分支途径:SHIK脱氢酶(aroB)、SHIK激酶(aroK)[ L-Phe分支途径:全局调节系统——Csr[ |
表2 芳香族氨基酸合成的影响因素
Tab. 2 Factors affecting synthesis of aromatic amino acids
| 影响因素 | 详情 |
|---|---|
| 前体的供给 | PEP,涉及其生成和消耗的系统和酶有:PEP合酶、PEP羧激酶[ E4P,涉及其生成的酶有转酮酶、转醛酶[ |
| 关键酶的反馈抑制 | DAHP合酶、ANT合酶[ |
| 调控因子TyrR和TrpR对合成途径的转录调节 | TyrR调控蛋白调节8个非连续操纵子的表达[ TrpR调控蛋白调节基因aroH、mtr和色氨酸操纵子[ |
| 转运系统 | 分泌系统:转运基因tnaB、mtr、aroP负责芳香族氨基酸的运输[ 膜蛋白YddG[ |
| 其他因素 | L-Trp分支途径:色氨酸酶(tnaA)[ L-Tyr分支途径:SHIK脱氢酶(aroB)、SHIK激酶(aroK)[ L-Phe分支途径:全局调节系统——Csr[ |
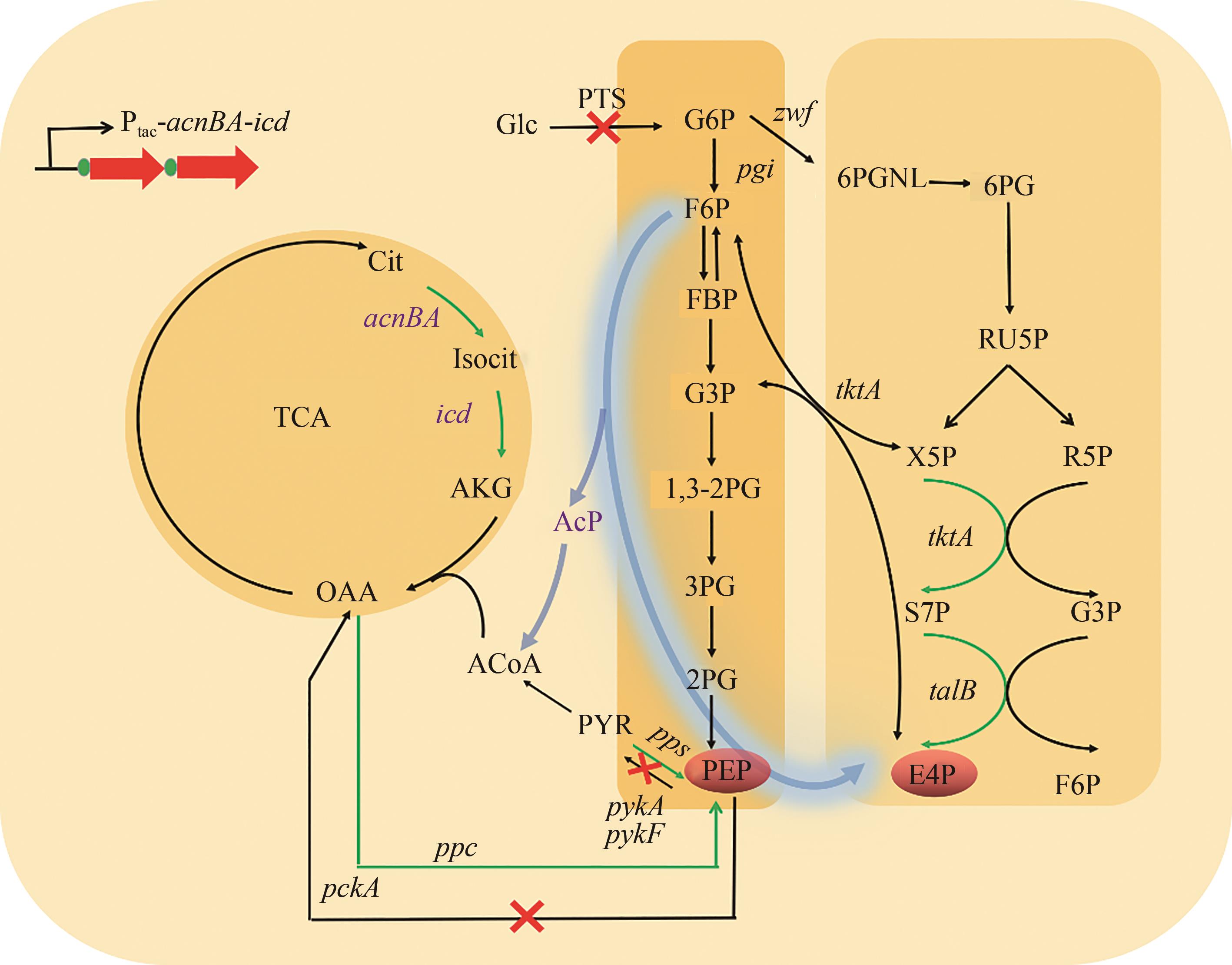
图2 PEP和E4P合成途径的改造(“绿色箭头”表示过表达,“红色叉号”表示敲除)F6P—果糖-6-磷酸;FBP—果糖1,6-二磷酸;G3P—甘油醛-3-磷酸;1,3-2PG—1,3-二磷酸甘油酸;3PG—3-磷酸甘油酸;2PG—2-磷酸甘油酸;Isocit—异柠檬酸;AKG—α-酮戊二酸;6PG—6-磷酸葡萄糖酸;RU5P—核酮糖-5-磷酸;X5P—木酮糖-5-磷酸;R5P—核酮糖-5-磷酸;S7P—景天庚酮糖-7-磷酸;G3P—甘油醛-3-磷酸;acnBA—编码乌头酸酶基因;icd—编码异柠檬酸脱氢酶基因;zwf—编码葡萄糖-6-磷酸脱氢酶基因
Fig. 2 Modification of synthesis route of PEP and E4P(''Green arrow'' indicates overexpression, ''red cross'' indicates knockout) F6P—fructose-6-phosphate; FBP—fructose 1,6-diphosphate; G3P—glyceraldehyde-3-phosphate; 1,3-2PG—1,3-diphosphoglyceride; 3PG—3-phosphoglyceric acid; 2PG—2-phosphoglyceric acid; Isocit—Isocitric acid; AKG—α-ketoglutarate; 6PG—6-phosphogluconic acid; RU5P—ketose-5-phosphate; X5P—xylulose-5-phosphate; R5P—ketose-5-phosphate; S7P—sedum heptanulose-7-phosphate; G3P—glyceraldehyde-3-phosphateGene: acnBA—encoding aconitase; icd—encoding isocitrate dehydrogenase; zwf—encoding glucose-6-phosphate dehydrogenase
| 菌种 | 关键酶 | 编码基因 | 终端产物反馈抑制 | 定点突变 | 参考文献 |
|---|---|---|---|---|---|
| 大肠杆菌 | DAHP合酶 | aroF(占比19%) | L-Tyr | Pro148Leu | [ |
| Gln152Ile | [ | ||||
| Asn8Lys(N末端) | [ | ||||
| aroG(占比80%) | L-Phe | Leu76Val | [ | ||
| Pro150Leu | [ | ||||
| Asp146Asn | [ | ||||
| aroH(占比1%) | L-Trp | [ | |||
| ANT合酶 | trpE | L-Trp | Ser40Phe | [ | |
| Met1293Thr | [ | ||||
| CM-PDT | pheA | L-Phe | Glu39Lys | [ | |
| Gly309Cys | [ | ||||
| tyrA | L-Tyr | 缺失305~386位基因 | [ | ||
| Ala354Val, Met53Ile | [ | ||||
| Gla534Val, Phe357Leu, Tyr263His | [ |
表3 芳香族氨基酸合成途径中抗反馈抑制的关键酶
Tab. 3 Key enzymes in aromatic amino acid synthesis against feedback inhibition
| 菌种 | 关键酶 | 编码基因 | 终端产物反馈抑制 | 定点突变 | 参考文献 |
|---|---|---|---|---|---|
| 大肠杆菌 | DAHP合酶 | aroF(占比19%) | L-Tyr | Pro148Leu | [ |
| Gln152Ile | [ | ||||
| Asn8Lys(N末端) | [ | ||||
| aroG(占比80%) | L-Phe | Leu76Val | [ | ||
| Pro150Leu | [ | ||||
| Asp146Asn | [ | ||||
| aroH(占比1%) | L-Trp | [ | |||
| ANT合酶 | trpE | L-Trp | Ser40Phe | [ | |
| Met1293Thr | [ | ||||
| CM-PDT | pheA | L-Phe | Glu39Lys | [ | |
| Gly309Cys | [ | ||||
| tyrA | L-Tyr | 缺失305~386位基因 | [ | ||
| Ala354Val, Met53Ile | [ | ||||
| Gla534Val, Phe357Leu, Tyr263His | [ |
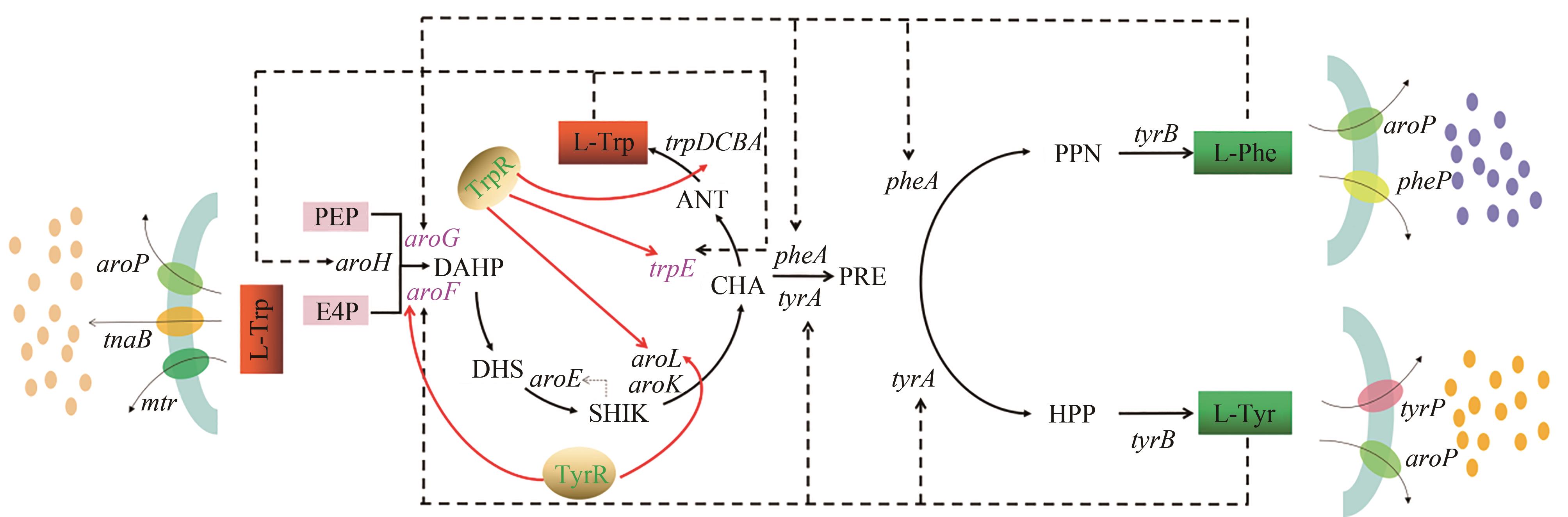
图3 关键酶的反馈抑制和调控蛋白的阻遏作用(“虚线”表示反馈抑制,“红色实线箭头”表示阻遏作用)
Fig. 3 Feedback inhibition of key enzymes and repression of regulatory proteins("Dotted line" indicates feedback inhibition, "red solid line arrow" indicates inhibition)
| 芳香族氨基酸 及其衍生物 | 宿主细胞/ 生产菌株 | 改造策略 | 发酵方式 | 产量 /(g/L) | 参考文献 |
|---|---|---|---|---|---|
| L-Phe | E.coli W3110 突变菌株 | pheAThr326Pro、aroF、galp、glk、aroD、tyrRT495I、ΔptsH | 分批补料 | 72.9 | [ |
| S/R-扁桃酸 | E.coli | 引入A. orientalis的HamS、ΔtyrA和共表达S. coelicolor的hmo、Rhodotorula graminis的dmd | 摇瓶发酵 | 0.74/0.68 | [ |
| 苯乳酸 | E.coli (L-Phe 高产菌株) | ldhA、来自Wickerhamia fluorescens的pprA | 分批发酵 | 29 | [ |
| 肉桂酸/醛 | E.coli | pal/pal、ccl、ccr | 摇瓶发酵 | 0.287/0.075 | [ |
| 肉桂醇 | S.cerevisiae | pal2、acar、entD、Adh6 | 摇瓶发酵 | 0.0278 | [ |
| 苯乙烯 | E.coli | pal2、fdc1 | 摇瓶发酵 | 0.26 | [ |
| L-Trp | E.coli W3110 | 多次随机诱变 | 分批补料 | 54.5 | [ |
| 生长素 | E.coli | aspC、ipdC、iad1 | 催化转化 | 3 | [ |
| 紫色杆菌素 | C.glutamicum | vioA、vioB、vioC、vioD、vioE | 分批补料 | 5.436 | [ |
| 脱氧紫色杆菌素 | E.coli | vioA、vioB、vioC、vioE | 摇瓶发酵 | 0.3241 | [ |
| 血清素 | E.coli | T-5H、共表达来自Catharanthus roseus的TDC | 催化转化 | 24 | [ |
| 靛红、靛蓝 | E.coli | tnaA、引入来自Methylophaga aminisulfidivorans的fmo | 催化转化 | 5/0.92 | [ |
| L-Tyr | E.coli MG1655衍生菌株 | tyrA、ΔpheA、ΔpheL | 分批补料 | 55 | [ |
| 丹参素 | E.coli | d-ldhY52A、hpaBC | 分批补料 | 7.1 | [ |
| 对羟基肉桂酸 | Yeast | TAL、用SET3p、CDC24p和ALD5p替换PFK1、PFK2和PYK1天然启动子 | 分批补料 | 12.5 | [ |
| 4-羟基苯乙烯 | E.coli | pal、pdc | 分批补料 | 0.4 | [ |
| L-多巴 | E.coli | 引入来自Zymomonas mobilis的tyrC、hpaBC | 分批补料 | 1.51 | [ |
表4 芳香族氨基酸及其衍生物生产概况
Tab. 4 General profile of production of aromatic amino acids and their derivatives
| 芳香族氨基酸 及其衍生物 | 宿主细胞/ 生产菌株 | 改造策略 | 发酵方式 | 产量 /(g/L) | 参考文献 |
|---|---|---|---|---|---|
| L-Phe | E.coli W3110 突变菌株 | pheAThr326Pro、aroF、galp、glk、aroD、tyrRT495I、ΔptsH | 分批补料 | 72.9 | [ |
| S/R-扁桃酸 | E.coli | 引入A. orientalis的HamS、ΔtyrA和共表达S. coelicolor的hmo、Rhodotorula graminis的dmd | 摇瓶发酵 | 0.74/0.68 | [ |
| 苯乳酸 | E.coli (L-Phe 高产菌株) | ldhA、来自Wickerhamia fluorescens的pprA | 分批发酵 | 29 | [ |
| 肉桂酸/醛 | E.coli | pal/pal、ccl、ccr | 摇瓶发酵 | 0.287/0.075 | [ |
| 肉桂醇 | S.cerevisiae | pal2、acar、entD、Adh6 | 摇瓶发酵 | 0.0278 | [ |
| 苯乙烯 | E.coli | pal2、fdc1 | 摇瓶发酵 | 0.26 | [ |
| L-Trp | E.coli W3110 | 多次随机诱变 | 分批补料 | 54.5 | [ |
| 生长素 | E.coli | aspC、ipdC、iad1 | 催化转化 | 3 | [ |
| 紫色杆菌素 | C.glutamicum | vioA、vioB、vioC、vioD、vioE | 分批补料 | 5.436 | [ |
| 脱氧紫色杆菌素 | E.coli | vioA、vioB、vioC、vioE | 摇瓶发酵 | 0.3241 | [ |
| 血清素 | E.coli | T-5H、共表达来自Catharanthus roseus的TDC | 催化转化 | 24 | [ |
| 靛红、靛蓝 | E.coli | tnaA、引入来自Methylophaga aminisulfidivorans的fmo | 催化转化 | 5/0.92 | [ |
| L-Tyr | E.coli MG1655衍生菌株 | tyrA、ΔpheA、ΔpheL | 分批补料 | 55 | [ |
| 丹参素 | E.coli | d-ldhY52A、hpaBC | 分批补料 | 7.1 | [ |
| 对羟基肉桂酸 | Yeast | TAL、用SET3p、CDC24p和ALD5p替换PFK1、PFK2和PYK1天然启动子 | 分批补料 | 12.5 | [ |
| 4-羟基苯乙烯 | E.coli | pal、pdc | 分批补料 | 0.4 | [ |
| L-多巴 | E.coli | 引入来自Zymomonas mobilis的tyrC、hpaBC | 分批补料 | 1.51 | [ |
| 影响组分 | 作用 | 不同种类 | 参考文献 |
|---|---|---|---|
| 碳源 | 提供能量和碳链骨架 | 葡萄糖、果糖、蔗糖、乳糖、麦芽糖、淀粉 | [ |
| 氮源 | 合成氨基酸、蛋白质和含氮代谢物 | 酵母粉、牛肉膏、蛋白胨、酵母膏、NH4Cl、NH4NO3、(NH4)2SO4、NH4H2PO4 | [ |
| 无机盐 | 调节pH,维持渗透压 | MgSO4、MgCl、CaCO3、KCl、KH2PO4 | [ |
表5 培养基中各组分的优化
Tab. 5 The components optimization in the medium
| 影响组分 | 作用 | 不同种类 | 参考文献 |
|---|---|---|---|
| 碳源 | 提供能量和碳链骨架 | 葡萄糖、果糖、蔗糖、乳糖、麦芽糖、淀粉 | [ |
| 氮源 | 合成氨基酸、蛋白质和含氮代谢物 | 酵母粉、牛肉膏、蛋白胨、酵母膏、NH4Cl、NH4NO3、(NH4)2SO4、NH4H2PO4 | [ |
| 无机盐 | 调节pH,维持渗透压 | MgSO4、MgCl、CaCO3、KCl、KH2PO4 | [ |
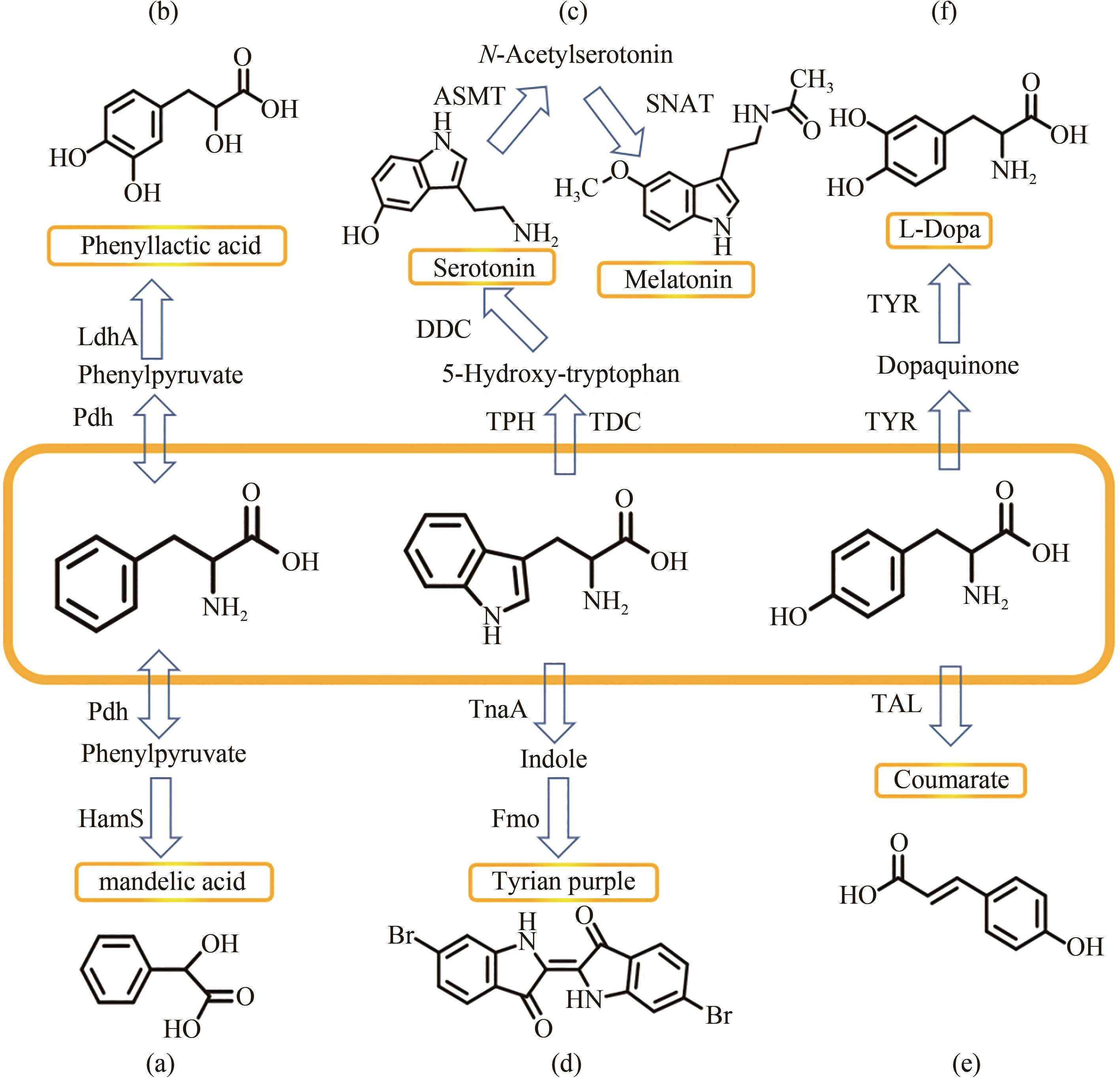
图4 几种芳香族氨基酸衍生物的具体合成途径Pdh—苯丙氨酸脱氢酶;HamS—4-羟基扁桃酸合酶;LdhA—乳酸脱氢酶;TPH—色氨酸5-羟化酶;TDC—色氨酸脱羧酶;DDC—芳香族L-氨基酸脱羧酶;ASMT—乙酰羟色胺O-甲基转移酶;SNAT—芳烷基胺N-乙酰基转移酶;TnaA—色氨酸酶;Fmo—黄素单加氧酶;TAL—苯丙氨酸解氨酶;TYR—酪氨酸酶
Fig. 4 Full synthetic routes of several aromatic amino acid derivativesPdh—phenylalanine dehydrogenase; HamS—4-hydroxymandelate synthase; LdhA—lactate dehydrogenase; TPH—tryptophan 5-hydroxylase; TDC—tryptophan decarboxylase; DDC—aromatic L-amino acid decarboxylase; ASMT—acetyl hydroxytryptamine O-methyltransferase; SNAT—arylalkylamine N-acetyltransferase; TnaA—tryptophan enzyme; Fmo—flavin monooxygenase; TAL—phenylalanine ammonia lyase; TYR—tyrosinase
| 1 | 鄢芳清, 韩亚昆, 李娟, 等. 大肠杆菌芳香族氨基酸代谢工程研究进展[J]. 生物加工过程, 2017, 15(5): 32-39, 85. |
| YAN F Q, HAN K, LI J, et al. Metabolic engineering of aromatic amino acids in Escherichia coli [J]. Chinese Journal of Bioprocess Engineering, 2017, 15(5): 32-39, 85. | |
| 2 | 于金龙. 大肠杆菌芳香族氨基酸生物合成途径的代谢调控研究[D]. 北京: 中国人民解放军军事医学科学院, 2008. |
| YU J L. Metabolic regulation of aromatic amino acid biosynthesis pathway in Escherichia coli [D]. Beijing: Chinese Academy of Military Sciences, 2008. | |
| 3 | 吴凤礼, 彭彦峰, 徐毅诚, 等. 代谢工程改造微生物生产芳香族化合物的研究进展[J]. 生物加工过程, 2017, 15(5): 9-23. |
| WU F L, PENG Y F, XU Y C, et al. Advances in microbial metabolic engineering for producing aromatic chemicals[J]. Chinese Journal of Bioprocess Engineering, 2017, 15(5): 9-23. | |
| 4 | ZHANG R H, LI C Y, WANG J, et al. Microbial production of small medicinal molecules and biologics: From nature to synthetic pathways[J]. Biotechnology Advances, 2018, 36(8): 2219-2231. |
| 5 | 李飞飞, 赵广荣. 大肠杆菌代谢工程生产芳香族化合物研究进展[J]. 食品与发酵工业, 2014, 40(6): 128-134. |
| LI F F, ZHAO G R. Advances in metabolic engineering of Escherichia coli for producing aromatic compounds[J]. Food and Fermentation Industries, 2014, 40(6):128-134. | |
| 6 | PITTARD J, YANG J. Biosynthesis of the aromatic amino acids[J]. EcoSal Plus, 2008, 3(1): 285-356. |
| 7 | JOO J C, OH Y H, YU J H, et al. Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a Miscanthus hydrolysate solution prepared by a newly developed Miscanthus hydrolysis process[J]. Bioresource Technology, 2017, 245(Pt B): 1692-1700. |
| 8 | WILLKE T. Methionine production—a critical review[J]. Applied Microbiology and Biotechnology, 2014, 98(24): 9893-9914. |
| 9 | GRÖGER H. Biocatalytic concepts for synthesizing amine bulk chemicals: recent approaches towards linear and cyclic aliphatic primary amines and ω-substituted derivatives thereof[J]. Applied Microbiology and Biotechnology, 2019, 103(1): 83-95. |
| 10 | WENDISCH V F. Metabolic engineering advances and prospects for amino acid production[J]. Metabolic Engineering, 2020, 58(1): 17-34. |
| 11 | WENDISCH V F. Microbial production of amino acids and derived chemicals: synthetic biology approaches to strain development[J]. Current Opinion in Biotechnology, 2014, 30(3): 51-58. |
| 12 | 申晓林, 袁其朋. 生物合成芳香族氨基酸及其衍生物的研究进展[J]. 生物技术通报, 2017, 33(1): 24-34. |
| SHEN X L, YUAN Q P. Research advance on biosynthesis of aromatic amino acids and their derivatives[J]. Biotechnology Bulletin, 2017,33(1): 24-34. | |
| 13 | 赵志军. L-色氨酸生产菌株的构建及代谢调控研究[D]. 无锡: 江南大学, 2011. |
| ZHAO Z J. Construction and metabolic regulation of L-tryptophan producing strain[D]. Wuxi: Jiangnan University, 2011. | |
| 14 | BONGAERTS J, KRÄMER M, MÜLLER U, et al. Metabolic engineering for microbial production of aromatic amino acids and derived compounds[J]. Metabolic Engineering, 2001, 3(4): 289-300. |
| 15 | 王健, 陈宁. 基于途径分析及代谢流量分析的L-色氨酸发酵条件优化[J]. 云南大学学报(自然科学版), 2004, 26(S2): 68-73. |
| WANG J, CHEN N. Optimization of L-tryptophan fermentation conditions based on pathway analysis and metabolic flux analysis[J]. Journal of Yunnan University (Natural Science Edition), 2004, 26(S2): 68-73. | |
| 16 | FLORES S, GOSSET G, FLORES N, et al. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by 13C labeling and NMR spectroscopy[J]. Metabolic Engineering, 2002, 4(2): 124-137. |
| 17 | 汪多仁. L-苯丙氨酸生产与应用[J]. 发酵科技通讯, 2011, 40(1): 29-36. |
| WANG D R. Production and application of L-phenylalanine[J]. Fermentation Technology Communication, 2011, 40(1): 29-36. | |
| 18 | LIU Q L, YU T, LI X W, et al. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals[J]. Nature Communications, 2019, 10(1): 4976. |
| 19 | PONCE E, MARTÍNEZ A, BOLÍVAR F, et al. Stimulation of glucose catabolism through the pentose pathway by the absence of the two pyruvate kinase isoenzymes in Escherichia coli [J]. Biotechnology and Bioengineering, 1998, 58(2/3): 292-295. |
| 20 | SPRAGGON G, KIM C, NGUYEN-HUU X, et al. The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(11): 6021-6026. |
| 21 | BRAUS G H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway[J]. Microbiological Reviews, 1991, 55(3): 349-370. |
| 22 | WALLACE B J, PITTARD J. Regulator gene controlling enzymes concerned in tyrosine biosynthesis in Escherichia coli [J]. Journal of Bacteriology, 1969, 97(3): 1234-1241. |
| 23 | SINGLETON C K, ROEDER W D, BOGOSIAN G, et al. DNA sequence of the E. coli trpR gene and prediction of the amino acid sequence of Trp repressor[J]. Nucleic Acids Research, 1980, 8(7): 51-60. |
| 24 | YANOFSKY C, HORN V, GOLLNICK P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli [J]. Journal of Bacteriology, 1991, 173(19): 6009-6017. |
| 25 | IKEDA M, KATSUMATA R. Tryptophan production by transport mutants of Corynebacterium glutamicum [J]. Bioscience, Biotechnology, and Biochemistry, 1995, 59(8): 1600-1602. |
| 26 | DOROSHENKO V, AIRICH L, VITUSHKINA M, et al. YddG from Escherichia coli promotes export of aromatic amino acids[J]. FEMS Microbiology Letters, 2007, 275(2): 312-318. |
| 27 | AIBA S, TSUNEKAWA H, IMANAKA T. New approach to tryptophan production by Escherichia coli: genetic manipulation of composite plasmids in vitro [J]. Applied and Environmental Microbiology, 1982, 43(2): 289-297. |
| 28 | SHI S, CHEN T, ZHANG Z, et al. Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production[J]. Metabolic Engineering, 2009, 11(4/5): 243-252. |
| 29 | LÜTKE-EVERSLOH T, STEPHANOPOULOS G. Combinatorial pathway analysis for improved L-tyrosine production in Escherichia coli: identification of enzymatic bottlenecks by systematic gene overexpression[J]. Metabolic Engineering, 2008, 10(2): 69-77. |
| 30 | OH J S, PARK H H, PARK T H. Temperature management strategy for efficient gene expression in a thermally inducible Escherichia coli/bacteriophage system[J]. Biotechnology and Bioprocess Engineering, 2008, 13(4): 470-475. |
| 31 | SUÁSTEGUI M, GUO W H, FENG X Y, et al. Investigating strain dependency in the production of aromatic compounds in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2016, 113(12): 2676-2685. |
| 32 | PATNAIK R, SPITZER R G, LIAO J C. Pathway engineering for production of aromatics in Escherichia coli: confirmation of stoichiometric analysis by independent modulation of AroG, TktA, and Pps activities[J]. Biotechnology and Bioengineering, 1995, 46(4): 361-370. |
| 33 | CARMONA S B, MORENO F, BOLÍVAR F, et al. Inactivation of the PTS as a strategy to engineer the production of aromatic metabolites in Escherichia coli [J]. Journal of Molecular Microbiology & Biotechnology, 2015, 25(2/3): 195-208. |
| 34 | CHEN Y Y, LIU Y F, DING D Q, et al. Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production[J]. Journal of Industrial Microbiology and Biotechnology, 2018, 45(5): 357-367. |
| 35 | CARMONA S B, FLORES N, MARTÍNEZ-ROMERO E, et al. Evolution of an Escherichia coli PTS-strain: a study of reproducibility and dynamics of an adaptive evolutive process[J]. Applied Microbiology and Biotechnology, 2020, 104(21): 9309-9325. |
| 36 | CHEN L, ZENG A P. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration[J]. Applied Microbiology and Biotechnology, 2017, 101(2): 559-568. |
| 37 | 吴涛, 赵津津, 毛贤军. 大肠杆菌磷酸烯醇式丙酮酸-糖磷酸转移酶系统改造对产L-色氨酸的影响[J]. 生物工程学报, 2017, 33(11): 1877-1882. |
| WU T, ZHAO J J, MAO X J. Effect of PTS modifications on L-tryptophan production in Escherichia coli [J]. Chinese Journal of Biotechnology, 2017, 33(11): 1877-1882. | |
| 38 | GOSSET G, YONG-XIAO J, BERRY A. A direct comparison of approaches for increasing carbon flow to aromatic biosynthesis in Escherichia coli [J]. Journal of Industrial Microbiology, 1996, 17(1): 47-52. |
| 39 | BERRY A. Improving production of aromatic compounds in Escherichia coli by metabolic engineering[J]. Trends in Biotechnology, 1996, 14(7): 250-256. |
| 40 | DU L H, ZHANG Z, XU Q Y, et al. Central metabolic pathway modification to improve L-tryptophan production in Escherichia coli [J]. Bioengineered, 2019, 10(1): 59-70. |
| 41 | 吴永庆, 江培翊, 范长胜, 等. 大肠杆菌ppsA, pckA基因的克隆与串联表达[J]. 复旦学报(自然科学版), 2002, 41(1): 31-35. |
| WU Y Q, JIANG P X, FAN C S, et al. Cloning and co-expression of ppsA and pckA genes in Escherichia coli [J]. Journal of Fudan University (Natural Sciences), 2002, 41(1): 31-35. | |
| 42 | TYAGI N, SAINI D, GULERIA R, et al. Designing an Escherichia coli strain for phenylalanine overproduction by metabolic engineering[J]. Molecular Biotechnology, 2017, 59(4/5): 168-178. |
| 43 | MASCARENHAS D, ASHWORTH D J, CHEN C S. Deletion of pgi alters tryptophan biosynthesis in a genetically engineered strain of Escherichia coli [J]. Applied and Environmental Microbiology, 1991, 57(10): 2995-2999. |
| 44 | GUO W, HUANG Q L, FENG Y H, et al. Rewiring central carbon metabolism for tyrosol and salidroside production in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2020, 117(8): 2410-2419. |
| 45 | TRENCHARD I J, SMOLKE C D. Engineering strategies for the fermentative production of plant alkaloids in yeast[J]. Metabolic Engineering, 2015, 30: 96-104. |
| 46 | JOSSEK R, BONGAERTS J, SPRENGER G A. Characterization of a new feedback-resistant 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroF of Escherichia coli [J]. FEMS Microbiology Letters, 2001, 202(1): 145-148. |
| 47 | LIU L N, DUAN X G, WU J. Modulating the direction of carbon flow in Escherichia coli to improve L-tryptophan production by inactivating the global regulator FruR[J]. Journal of Biotechnology, 2016, 231(4): 138-141. |
| 48 | 李剑欣, 郭长江, 刘云, 等. 大肠杆菌邻氨基苯甲酸合成酶编码基因trpED的克隆与表达[J]. 生物技术通讯, 2007, 18(2): 183-185. |
| LI J X, GUO C J, LIU Y, et al. The clone and expression of the anthranilate synthetase coding gene in Escherichia coli [J]. Letters in Biotechnology, 2007, 18(2): 183-185. | |
| 49 | IKEDA M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering[J]. Applied Microbiology and Biotechnology, 2006, 69(6): 615-626. |
| 50 | LÜTKE-EVERSLOH T, STEPHANOPOULOS G. L-tyrosine production by deregulated strains of Escherichia coli [J]. Applied Microbiology and Biotechnology, 2007, 75(1): 103-110. |
| 51 | LIU Y J, LI P P, ZHAO K X, et al. Corynebacterium glutamicum contains 3-deoxy-D-arabino-heptulosonate 7-phosphate synthases that display novel biochemical features[J]. Applied and Environmental Microbiology, 2008, 74(17): 5497-5503. |
| 52 | RODRIGUEZ A, CHEN Y, KHOOMRUNG S, et al. Comparison of the metabolic response to over-production of p-coumaric acid in two yeast strains[J]. Metabolic Engineering, 2017, 44: 265-272. |
| 53 | SCHENCK C A, MAEDA H A. Tyrosine biosynthesis, metabolism, and catabolism in plants[J]. Phytochemistry, 2018, 149: 82-102. |
| 54 | LÜTKE-EVERSLOH T, STEPHANOPOULOS G. Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: generation and characterization of tyrosine-insensitive mutants[J]. Applied and Environmental Microbiology, 2005, 71(11): 7224-7228. |
| 55 | DODGE T C, GERSTNER J M. Optimization of the glucose feed rate profile for the production of tryptophan from recombinant E coli [J]. Journal of Chemical Technology and Biotechnology, 2002, 77(11): 45-123. |
| 56 | LIU S P, LIU R X, XIAO M R, et al. A systems level engineered E. coli capable of efficiently producing L-phenylalanine[J]. Process Biochemistry, 2014, 49(5): 751-757 |
| 57 | FERNSTROM J D. A perspective on the safety of supplemental tryptophan based on its metabolic fates[J]. The Journal of Nutrition, 2016, 146(12): 2601S-2608S. |
| 58 | 古鹏飞. 利用重组大肠杆菌生产L-色氨酸的研究[D]. 济南: 山东大学, 2013. |
| GU P F. Production of L-tryptophan by recombinant Escherichia coli [D]. Jinan: Shandong University, 2013. | |
| 59 | MUÑOZ A J, HERNÁNDEZ-CHÁVEZ G, DE ANDA R, et al. Metabolic engineering of Escherichia coli for improving 1-3,4-dihydroxyphenylalanine (L-Dopa) synthesis from glucose[J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(11): 1845-1852. |
| 60 | KIM B, BINKLEY R, KIM H U, et al. Metabolic engineering of Escherichia coli for the enhanced production of L-tyrosine[J]. Biotechnology and Bioengineering, 2018, 115(10): 2554-2564. |
| 61 | LIU Y F, XU Y R, DING D Q, et al. Genetic engineering of Escherichia coli to improve L-phenylalanine production[J]. BMC Biotechnology, 2018, 18(1): 5. |
| 62 | SUN Z T, NING Y Y, LIU L X, et al. Metabolic engineering of the L-phenylalanine pathway in Escherichia coli for the production of S- or R-mandelic acid[J]. Microbial Cell Factories, 2011, 10(1): 71. |
| 63 | FUJITA T, NGUYEN H D, ITO T, et al. Microbial monomers custom-synthesized to build true bio-derived aromatic polymers[J]. Applied Microbiology and Biotechnology, 2013, 97(20): 8887-8894. |
| 64 | BANG H B, LEE Y H, KIM S C, et al. Metabolic engineering of Escherichia coli for the production of cinnamaldehyde[J]. Microbial Cell Factories, 2016, 15(1): 16. |
| 65 | GOTTARDI M, KNUDSEN J D, PRADO L, et al. De novo biosynthesis of trans-cinnamic acid derivatives in Saccharomyces cerevisiae [J]. Applied Microbiology and Biotechnology, 2017, 101(12): 4883-4893. |
| 66 | MCKENNA R, NIELSEN D R. Styrene biosynthesis from glucose by engineered E. coli [J]. Metabolic Engineering, 2011, 13(5): 544-554. |
| 67 | ROMASI E F, LEE J. Development of indole-3-acetic acid-producing Escherichia coli by functional expression of IpdC, AspC, and Iad1[J]. Journal of microbiology and biotechnology, 2013, 23(12): 26-36. |
| 68 | SUN H, ZHAO D, XIONG B, et al. Engineering Corynebacterium glutamicum for violacein hyper production[J]. Microbial Cell Factories, 2016, 15(1): 148. |
| 69 | RODRIGUES A L, TRACHTMANN N, BECKER J, et al. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein[J]. Metabolic Engineering, 2013, 20: 29-41. |
| 70 | PARK S, KANG K, LEE S W, et al. Production of serotonin by dual expression of tryptophan decarboxylase and tryptamine 5-hydroxylase in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2011, 89(5): 1387-1394. |
| 71 | 韩晓红, 王伟, 肖兴国. 靛蓝及其同类色素的微生物生产与转化[J]. 生物工程学报, 2008, 24(6): 921-926. |
| HAN X H, WANG W, XIAO X G. Microbial biosynthesis and biotransformation of indigo and indigo-like pigments[J]. Chinese Journal of Biotechnology, 2008, 24(6): 921-926. | |
| 72 | YAO Y F, WANG C S, QIAO J, et al. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway[J]. Metabolic Engineering, 2013, 19: 79-87. |
| 73 | QI W W, VANNELLI T, BREINIG S, et al. Functional expression of prokaryotic and eukaryotic genes in Escherichia coli for conversion of glucose to p-hydroxystyrene[J]. Metabolic Engineering, 2007, 9(3): 268-276. |
| 74 | NIU H, LI R R, LIANG Q F, et al. Metabolic engineering for improving L-tryptophan production in Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(1): 55-65. |
| 75 | PÉREZ-GARCÍA F, WENDISCH V F. Transport and metabolic engineering of the cell factory Corynebacterium glutamicum [J]. FEMS Microbiology Letters, 2018, 365(16): fny166. |
| 76 | GU P F, YANG F, LI F F, et al. Knocking out analysis of tryptophan permeases in Escherichia coli for improving L-tryptophan production[J]. Applied Microbiology and Biotechnology, 2013, 97(15): 6677-6683. |
| 77 | TATARKO M, ROMEO T. Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosynthesis in Escherichia coli [J]. Current Microbiology, 2001, 43(1): 26-32. |
| 78 | YAKANDAWALA N, ROMEO T, FRIESEN A D, et al. Metabolic engineering of Escherichia coli to enhance phenylalanine production[J]. Applied Microbiology and Biotechnology, 2008, 78(2): 283-291. |
| 79 | TSOI R, WU F L, ZHANG C, et al. Metabolic division of labor in microbial systems[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2526-2531. |
| 80 | DINH C V, PRATHER K L J. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(51): 25562-25568. |
| 81 | WANG J, ZHANG R H, ZHANG Y, et al. Developing a pyruvate-driven metabolic scenario for growth-coupled microbial production[J]. Metabolic Engineering, 2019, 55(6): 191-200. |
| 82 | YOUNGER A K, DALVIE N C, ROTTINGHAUS A G, et al. Engineering modular biosensors to confer metabolite-responsive regulation of transcription[J]. ACS Synthetic Biology, 2017, 6(2): 311-325. |
| 83 | RAMAN S, ROGERS J K, TAYLOR N D, et al. Evolution-guided optimization of biosynthetic pathways[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(50): 17803-17808. |
| 84 | SHI S B, ANG E L, ZHAO H M. In vivo biosensors: mechanisms, development, and applications[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(7): 491-516. |
| 85 | FANG M Y, WANG T M, ZHANG C, et al. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli [J]. Metabolic Engineering, 2016, 33: 41-51. |
| 86 | LIU Y F, ZHUANG Y Y, DING D Q, et al. Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli [J]. ACS Synthetic Biology, 2017, 6(5): 837-848. |
| 87 | DING D Q, LI J L, BAI D Y, et al. Biosensor-based monitoring of the central metabolic pathway metabolites[J]. Biosensors & Bioelectronics, 2020, 167: 112456. |
| 88 | CHEN M, CHEN L, ZENG A P. CRISPR/Cas9-facilitated engineering with growth-coupled and sensor-guided in vivo screening of enzyme variants for a more efficient chorismate pathway in E. coli [J]. Metabolic Engineering Communications, 2019, 9(2): e00094. |
| 89 | 陈胜杰. L-色氨酸生产菌构建及发酵条件优化[D]. 天津: 天津科技大学, 2016. |
| CHRN S J. Construction of L-tryptophan producing strain and optimization of fermentation conditions[D]. TianJin: Tianjin University of Science and Technology, 2016. | |
| 90 | 黄静, 史建明, 霍文婷, 等. 氮源对L-色氨酸发酵的影响[J]. 食品与发酵工业, 2011, 37(5): 21-25. |
| HUANG J, SHI J M, HUO W T, et al. The effects of nitrogen sources on the fermentation of L-tryptophan[J]. Food and Fermentation Industries, 2011, 37(5): 21-25. | |
| 91 | 杨梦晨. L-色氨酸新型清液发酵工艺研究[D]. 天津: 天津科技大学, 2018. |
| YANG M C. Study on fermentation and fermentation technology of L-tryptophan[D]. Tianjin: Tianjin University of Science and Technology, 2018. | |
| 92 | 娄秀平. Escherichia coli JN8发酵产L-色氨酸的研究[D]. 无锡: 江南大学, 2014. |
| LOU X P. Production of L-tryptophan by Escherichia coli JN8[D]. Wuxi: Jiangnan University, 2014. | |
| 93 | 张婷婷. 微生物法生产L-色氨酸的检测及发酵过程优化[D]. 开封: 河南大学, 2017. |
| ZHANG T T. Determination of L-tryptophan grade in microbial production and optimization of fermentation process[D]. Kaifeng: Henan University, 2017. | |
| 94 | PAREKH S, VINCI V A, STROBEL R J. Improvement of microbial strains and fermentation processes[J]. Applied Microbiology and Biotechnology, 2000, 54(3): 287-301. |
| 95 | JING K J, TANG Y W, YAO C Y, et al. Overproduction of L-tryptophan via simultaneous feed of glucose and anthranilic acid from recombinant Escherichia coli W3110: kinetic modeling and process scale-up[J]. Biotechnology and Bioengineering, 2018, 115(2): 371-381. |
| 96 | REIFENRATH M, BOLES E. Engineering of hydroxymandelate synthases and the aromatic amino acid pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae [J]. Metabolic Engineering, 2018, 45(2): 246-254. |
| 97 | CAO M F, GAO M R, SUÁSTEGUI M, et al. Building microbial factories for the production of aromatic amino acid pathway derivatives: from commodity chemicals to plant-sourced natural products[J]. Metabolic Engineering, 2020, 58: 94-132. |
| 98 | FUJIWARA R, NODA S, TANAKA T, et al. Muconic acid production using gene-level fusion proteins in Escherichia coli [J]. ACS Synthetic Biology, 2018, 7(11): 2698-2705. |
| 99 | RAMAKRISHNA A, GIRIDHAR P, RAVISHANKAR G A. Phytoserotonin: a review[J]. Plant Signaling & Behavior, 2011, 6(6): 80-92. |
| 100 | DU J, YANG D, LUO Z W, et al. Metabolic engineering of Escherichia coli for the production of indirubin from glucose[J]. Journal of Biotechnology, 2018, 267: 19-28. |
| 101 | HAN G H, GIM G H, KIM W, et al. Enhanced indirubin production in recombinant Escherichia coli harboring a flavin-containing monooxygenase gene by cysteine supplementation[J]. Journal of Biotechnology, 2012, 164(2): 179-187. |
| 102 | RODRIGUEZ A, KILDEGAARD K R, LI M, et al. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis[J]. Metabolic Engineering, 2015, 31: 181-188. |
| 103 | WEI T, CHENG B Y, LIU J Z. Genome engineering Escherichia coli for L-Dopa overproduction from glucose[J]. Scientific Reports, 2016, 6: 30080. |
| 104 | VALLIERE M A, KORMAN T P, WOODALL N B, et al. A cell-free platform for the prenylation of natural products and application to cannabinoid production[J]. Nature Communications, 2019, 10(1): 565. |
| 105 | LEE M Y, HUNG W P, TSAI S H. Improvement of shikimic acid production in Escherichia coli with growth phase-dependent regulation in the biosynthetic pathway from glycerol[J]. World Journal of Microbiology and Biotechnology, 2017, 33(2): 1-8. |
| 106 | SILVA D A, YU S, ULGE U Y, et al. De novo design of potent and selective mimics of IL-2 and IL-15[J]. Nature, 2019, 565(7738): 186-191. |
| 107 | YANG X, YUAN Q Q, LUO H, et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways[J]. Metabolic Engineering, 2019, 56(1): 142-153. |
| 108 | D'OELSNITZ S, ELLINGTON A. Continuous directed evolution for strain and protein engineering[J]. Current Opinion in Biotechnology, 2018, 53: 158-163. |
| 109 | BRYSON D I, FAN C, GUO L T, et al. Continuous directed evolution of aminoacyl-tRNA synthetases[J]. Nature Chemical Biology, 2017, 13(12): 1253-1260. |
| 110 | SONG L, ZENG A P. Engineering 'cell robots' for parallel and highly sensitive screening of biomolecules under in vivo conditions[J]. Scientific Reports, 2017, 7(1): 15-45. |
| 111 | BADRAN A H, LIU D R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra[J]. Nature Communications, 2015, 6: 8425. |
| 112 | JAKOČIŪNAS T, PEDERSEN L E, LIS A V, et al. CasPER, a method for directed evolution in genomic contexts using mutagenesis and CRISPR/Cas9[J]. Metabolic Engineering, 2018, 48(2): 88-96. |
| 113 | HALPERIN S O, TOU C J, WONG E B, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560(7717): 248-252. |
| [1] | 吕靖伟, 邓子新, 张琪, 丁伟. 基于深度学习识别RiPPs前体肽及裂解位点[J]. 合成生物学, 2022, 3(6): 1262-1276. |
| [2] | 尤迪, 叶邦策. 从翻译后修饰角度解析人工合成途径与底盘细胞的适配性[J]. 合成生物学, 2020, 1(2): 212-225. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
