合成生物学 ›› 2021, Vol. 2 ›› Issue (6): 902-919.DOI: 10.12211/2096-8280.2021-049
人工代谢途径合成有机醇有机酸的研究进展
曹晨凯, 李佳隆, 张科春
- 西湖大学工学院,浙江省海岸带环境与资源研究重点实验室,浙江 杭州 310024
-
收稿日期:2021-04-23修回日期:2021-11-19出版日期:2021-12-31发布日期:2022-01-21 -
通讯作者:张科春 -
作者简介:曹晨凯 (1996—),男,博士研究生。研究方向为代谢工程。E-mail:caochenkai@westlake.edu.cn李佳隆 (1996—),男,博士研究生。研究方向为代谢工程。E-mail:lijialong@westlake.edu.cn张科春 (1978—),特聘研究员,博士生导师。研究方向为应用合成生物、绿色化学、材料科学和工程优化的方法,设计绿色新化工生产路线和开发环保新材料,为循环经济向前发展提供新的解决方案。E-mail:zhangkechun@westlake.edu.cn -
基金资助:国家自然科学基金(22078267)
Progress in artificial metabolic pathways for biosynthesis of organic alcohols & acids
CAO Chenkai, LI Jialong, ZHANG Kechun
- Key Laboratory of Coastal Environment and Resources of Zhejiang Province,School of Engineering,Westlake University,Hangzhou 310024,Zhejiang,China
-
Received:2021-04-23Revised:2021-11-19Online:2021-12-31Published:2022-01-21 -
Contact:ZHANG Kechun
摘要:
有机酸有机醇不仅是食品、化工、医药领域的原料,更是一类重要的生物基燃料,相比于传统的石油基燃料,具有原料可再生,生产过程清洁的特点,是应对能源危机、环境污染问题的有效对策。传统的代谢工程方法是通过在微生物中过表达产物相关的特异性酶实现的,应用时常有发酵原料单一,代谢效率不高,产物种类受限等问题。筛选并重新组合不同来源的酶来构建人工代谢途径是一个有效的解决方案,同时也是代谢工程方法发展的一个重要趋势。本文综述了近年来构建人工代谢途径生产有机酸有机醇取得的突破与进展,具体阐述了5种创新生物合成途径(一碳化合物同化途径、非磷酸化途径、酮酸途径、β-氧化逆循环途径和聚酮化合物途径)的相关机理,为未来构建低成本、高效、产物多样化的有机酸有机醇生物合成平台提供了可能。
中图分类号:
引用本文
曹晨凯, 李佳隆, 张科春. 人工代谢途径合成有机醇有机酸的研究进展[J]. 合成生物学, 2021, 2(6): 902-919.
CAO Chenkai, LI Jialong, ZHANG Kechun. Progress in artificial metabolic pathways for biosynthesis of organic alcohols & acids[J]. Synthetic Biology Journal, 2021, 2(6): 902-919.
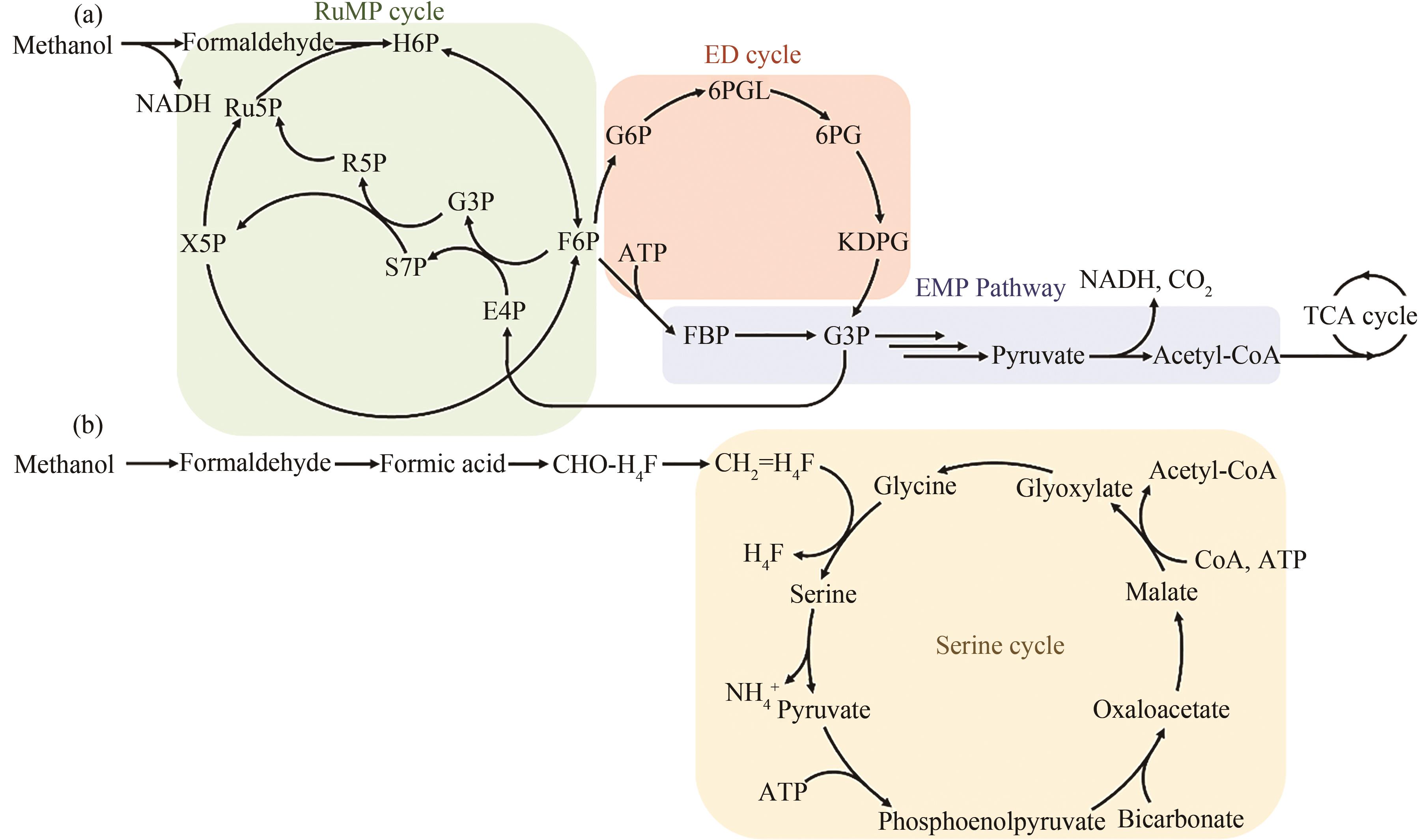
图1 RuMP途径、ED途径、EMP途径(a)和丝氨酸循环(b)[19]H6P—Hexulose-6-phosphate; Ru5P—ribulose-5-phosphate; X5P—xylulose 5-phosphate; F6P—fructose-6-phosphate; R5P—ribose-5-phosphate; G3P—glyceraldehyde-3-phosphate; S7P—sedoheptulose-7-phosphate; E4P—erythrose 4-phosphate; G6P—glucose-6-phosphate; 6PGL—6-phospho-D-glucono-1,5-lactone; 6PG—6-phospho-D-glonate; KDPG—2-dehydro-3-deoxy-D-gluconate-6-phosphate; FBP—fructose-1,6-bisphosphate; TCA—tricarboxylic acid
Fig. 1 C1 compound assimilation through ribulose-5-phosphate (RuMP), ED (Entner-Doudoroff), EMP (Embden-Meyerhof-Parnas) pathway (a) and serine cycle (b)[19]
| 菌株 | 方法 | 培养基 | 产物 | 产量/(g/L) | 时间 | 文献 |
|---|---|---|---|---|---|---|
| 甲醇芽孢杆菌 | 补料分批发酵 | 含200 mmol/L甲醇的MVcM培养基 | γ-氨基丁酸 | 9.0 | 2016 | [ |
| α-变形杆菌甲基杆菌AM1 | 摇瓶发酵 | 含20 mmol/L乙胺的基础培养基 | L-丁醇 | 25.5×10-3 | 2016 | [ |
| 扭脱甲基杆菌AM1 | 补料分批发酵 | 含有3 g/L甲醇的矿物盐培养基 | 2-羟基异丁酸 | 2.1 | 2016 | [ |
| 扭脱甲基杆菌AM1 | 摇瓶发酵 | 含125 mmol/L甲醇的基础培养基 | 3-羟基丙酸 | 69.8×10-3 | 2017 | [ |
| α-变形杆菌甲基杆菌AM1 | 补料分批发酵 | 含5%(体积分数)甲醇的基础培养基 | 甲羟戊酸 | 2.3 | 2018 | [ |
| 毕赤酵母 | 试管发酵 | 酵母/蛋白胨/葡萄糖培养基 | D-乳酸 | 3.5 | 2019 | [ |
表1 由甲基营养性菌发酵生产特定有机醇与有机酸实例
Tab. 1 Summary of methylotrophic production data
| 菌株 | 方法 | 培养基 | 产物 | 产量/(g/L) | 时间 | 文献 |
|---|---|---|---|---|---|---|
| 甲醇芽孢杆菌 | 补料分批发酵 | 含200 mmol/L甲醇的MVcM培养基 | γ-氨基丁酸 | 9.0 | 2016 | [ |
| α-变形杆菌甲基杆菌AM1 | 摇瓶发酵 | 含20 mmol/L乙胺的基础培养基 | L-丁醇 | 25.5×10-3 | 2016 | [ |
| 扭脱甲基杆菌AM1 | 补料分批发酵 | 含有3 g/L甲醇的矿物盐培养基 | 2-羟基异丁酸 | 2.1 | 2016 | [ |
| 扭脱甲基杆菌AM1 | 摇瓶发酵 | 含125 mmol/L甲醇的基础培养基 | 3-羟基丙酸 | 69.8×10-3 | 2017 | [ |
| α-变形杆菌甲基杆菌AM1 | 补料分批发酵 | 含5%(体积分数)甲醇的基础培养基 | 甲羟戊酸 | 2.3 | 2018 | [ |
| 毕赤酵母 | 试管发酵 | 酵母/蛋白胨/葡萄糖培养基 | D-乳酸 | 3.5 | 2019 | [ |
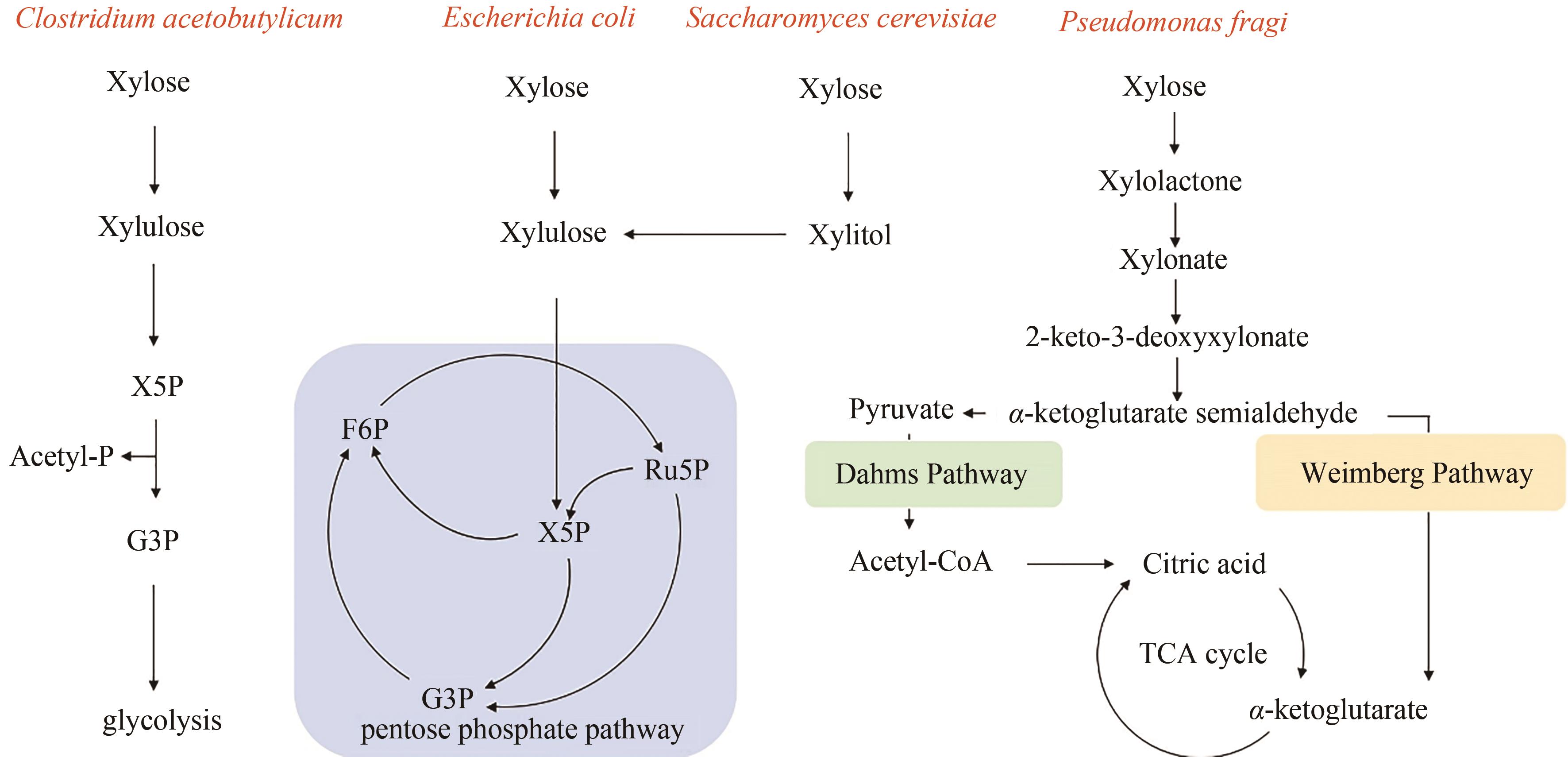
图2 不同微生物的木糖代谢途径X5P—Xylulose 5-phosphate; G3P—glyceraldehyde-3-phosphate; F6P—fructose-6-phosphate; Ru5P—ribulose-5-phosphate; Acetyl-P—acetylphosphate
Fig. 2 Metabolic pathways of xylose in different microorganisms
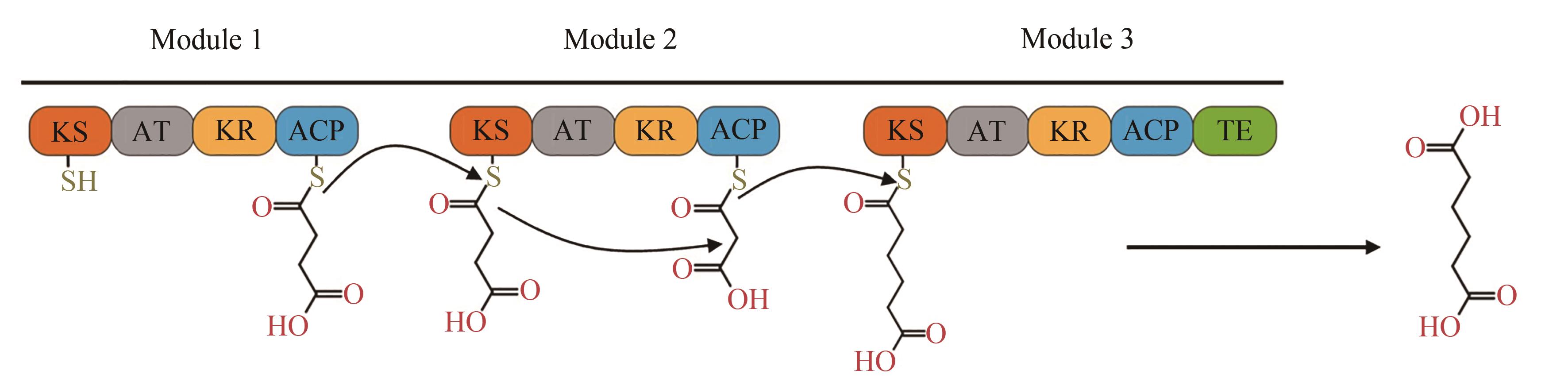
图6 以合成己二酸为例的PKS途径[其中一共有3个模块,底物通过硫酯交换反应从前一个模块转移到下一个模块,最后通过硫酯酶(TEp)结构域释放]
Fig. 6 PKS pathway for production of adipic acid[There are 3 respective modules. Substrate is transferred from previous module to next one via thioester exchange reaction, and finally released by thioesterase (TEp) domain]
| 1 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 2 | PACALA S, SOCOLOW R. Stabilization wedges: Solving the climate problem for the next 50 years with current technologies[J]. Science, 2004, 305(5686): 968-972. |
| 3 | O'NEILL B C, OPPENHEIMER M. Climate change. Dangerous climate impacts and the Kyoto protocol[J]. Science, 2002, 296(5575): 1971-1972. |
| 4 | 任光. 我国煤制甲醇的工业现状及发展趋势分析[J]. 化肥设计, 2016, 54(5): 5-7. |
| REN G. Analysis on present situation and development trend of coal methanol industry in China[J]. Chemical Fertilizer Design, 2016, 54(5): 5-7. | |
| 5 | 高姣丽, 潘子鹤, 成怀刚, 等. CO2催化制甲酸反应设备的现状与研究进展[J]. 化工机械, 2020, 47(6): 737-741. |
| GAO J L, PAN Z H, CHENG H G, et al. Status and research progress of reaction equipment for preparation of formic acid catalyzed by CO2 [J]. Chemical Engineering & Machinery, 2020, 47(6): 737-741. | |
| 6 | CHEN X, LIU Y, WU J W. Sustainable production of formic acid from biomass and carbon dioxide[J]. Molecular Catalysis, 2020, 483: 110716. |
| 7 | LI H, OPGENORTH P H, WERNICK D G, et al. Integrated electromicrobial conversion of CO2 to higher alcohols[J]. Science, 2012, 335(6076): 1596. |
| 8 | SAWATDEENARUNAT C, NGUYEN D, SURENDRA K C, et al. Anaerobic biorefinery: current status, challenges, and opportunities[J]. Bioresource Technology, 2016, 215: 304-313. |
| 9 | CLOMBURG J M, CRUMBLEY A M, GONZALEZ R. Industrial biomanufacturing: the future of chemical production [J]. Science, 2017, 355(6320): aag0804. |
| 10 | ZENG A P. New bioproduction systems for chemicals and fuels: Needs and new development[J]. Biotechnology Advances, 2019, 37(4): 508-518. |
| 11 | CHEN F Y H, JUNG H W, TSUEI C Y, et al. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol[J]. Cell, 2020, 182(4): 933-946. |
| 12 | BODEN R, CUNLIFFE M, SCANLAN J, et al. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09[J]. Journal of Bacteriology, 2011, 193(24): 7001-7002. |
| 13 | PEEL D, QUAYLE J R. Microbial growth on C1 compounds (I): Isolation and characterization of Pseudomonas AM 1[J]. Biochemical Journal, 1961, 81(3): 465-469. |
| 14 | STROM T, FERENCI T, QUAYLE J R. The carbon assimilation pathways of Methylococcus capsulatus, Pseudomonas methanica and Methylosinus trichosporium (OB3B) during growth on methane[J]. The Biochemical Journal, 1974, 144(3): 465-476. |
| 15 | WHITAKER W B, JONES J A, BENNETT R K, et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli [J]. Metabolic Engineering, 2017, 39: 49-59. |
| 16 | KALYUZHNAYA M G, YANG S, ROZOVA O N, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium[J]. Nature Communications, 2013, 4(1): 2785. |
| 17 | MEYER F, KELLER P, HARTL J, et al. Methanol-essential growth of Escherichia coli [J]. Nature Communications, 2018, 9(1): 1508. |
| 18 | WOOLSTON B M, KING J R, REITER M, et al. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli [J]. Nature Communications, 2018, 9(1): 2387-2398. |
| 19 | YU H, LIAO J C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9(1): 3992. |
| 20 | 郭姝媛, 吴良焕, 刘香健, 等. 微生物中一碳代谢网络构建的进展与挑战[J/OL]. 合成生物学: 1-24, doi: 10.12211/2096-8280.2021-079 . . |
| GUO Shuyuan, WU Lianghuan, LIU Xiangjian, et al. The biotransformation outlook of C1-based metabolic networks in methylotrophy[J/OL]. Synthetic Biology Journal: 1-24, doi: 10.12211/2096-8280.2021-079 . . | |
| 21 | GOYAL N, ZHOU Z, KARIMI I A. Metabolic processes of Methanococcus maripaludis and potential applications[J]. Microbial Cell Factories, 2016, 15(1): 107. |
| 22 | SOHN Y J, SON J, JO S Y, et al. Chemoautotroph Cupriavidus necator as a potential game-changer for global warming and plastic waste problem: a review[J]. Bioresource Technology, 2021, 340: 125693. |
| 23 | KOZLOWSKI J T, DAVIS R J. Heterogeneous catalysts for the guerbet coupling of alcohols[J]. ACS Catalysis, 2013, 3(7): 1588-1600. |
| 24 | LIAO J C, MI L, PONTRELLI S, et al. Fuelling the future: microbial engineering for the production of sustainable biofuels [J]. Nature Reviews Microbiology, 2016, 14(5): 288-304. |
| 25 | IRLA M, NÆRDAL I, BRAUTASET T, et al. Methanol-based γ-aminobutyric acid (GABA) production by genetically engineered Bacillus methanolicus strains[J]. Industrial Crops and Products, 2017, 106: 12-20. |
| 26 | HU B, YANG Y M, BECK D A C, et al. Comprehensive molecular characterization of Methylobacterium extorquens AM1 adapted for 1-butanol tolerance[J]. Biotechnology for Biofuels, 2016, 9(1): 84-97. |
| 27 | ROHDE M T, TISCHER S, HARMS H, et al. Production of 2-hydroxyisobutyric acid from methanol by Methylobacterium extorquens AM1 expressing (R)-3-hydroxybutyryl-CoA isomerizing enzymes [J]. Applied & Environmental Microbiology, 2017, 83(3): AEM.02622-16. |
| 28 | YANG Y M, CHEN W J, YANG J, et al. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route[J]. Microbial Cell Factories, 2017, 16(1): 179. |
| 29 | CUI L Y, WANG S S, GUAN C G, et al. Breeding of methanol-tolerant Methylobacterium extorquens AM1 by atmospheric and room temperature plasma mutagenesis combined with adaptive laboratory evolution[J]. Biotechnology Journal, 2018, 13(6): e1700679. |
| 30 | YAMADA R, OGURA K, KIMOTO Y, et al. Toward the construction of a technology platform for chemicals production from methanol: D-lactic acid production from methanol by an engineered yeast Pichia pastoris [J]. World Journal of Microbiology and Biotechnology, 2019, 35(2): 37-45. |
| 31 | GODDARD A W. Cortical and subcortical gamma amino acid butyric acid deficits in anxiety and stress disorders: clinical implications[J]. World Journal of Psychiatry, 2016, 6(1): 43-53. |
| 32 | BOGORAD I W, CHEN C T, THEISEN M K, et al. Building carbon-carbon bonds using a biocatalytic methanol condensation cycle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(45): 15928-15933. |
| 33 | TILMAN D, SOCOLOW R, FOLEY J A, et al. Beneficial biofuels—the food, energy, and environment trilemma[J]. Science, 2009, 325(5938): 270-271. |
| 34 | GRAHAM-ROWE D. Agriculture: beyond food versus fuel[J]. Nature, 2011, 474(7352): S6-S8. |
| 35 | PERLACK R D, WRIGHT L L, TURHOLLOW A F, et al. Biomass as feedstock for a bioenergy and bioproducts industry: The technical feasibility of a billion-ton annual supply[R]. Office of Scientific and Technical Information (OSTI), 2005. |
| 36 | SOMERVILLE C. The billion-ton biofuels vision[J]. Science, 2006, 312(5778): 1277. |
| 37 | RAGAUSKAS A J, WILLIAMS C K, DAVISON B H, et al. The path forward for biofuels and biomaterials[J]. Science, 2006, 311(5760): 484-489. |
| 38 | GALL D L, RALPH J, DONOHUE T J, et al. Biochemical transformation of lignin for deriving valued commodities from lignocellulose[J]. Current Opinion in Biotechnology, 2017, 45: 120-126. |
| 39 | QIN L, LI W C, LIU L, et al. Inhibition of lignin-derived phenolic compounds to cellulase[J]. Biotechnology for Biofuels, 2016, 9: 70. |
| 40 | LI W C, ZHU J Q, ZHAO X, et al. Improving co-fermentation of glucose and xylose by adaptive evolution of engineering xylose-fermenting Saccharomyces cerevisiae and different fermentation strategies[J]. Renewable Energy, 2019, 139: 1176-1183. |
| 41 | LIU H, ZHU J Q, LI X, et al. Hybridization improves inhibitor tolerance of xylose-fermenting Saccharomyces cerevisiae [J]. BioResources, 2017, 12(3): 4737-4753. |
| 42 | JEFFRIES T W. Engineering yeasts for xylose metabolism[J]. Current Opinion in Biotechnology, 2006, 17(3): 320-326. |
| 43 | MATSUSHIKA A, INOUE H, KODAKI T, et al. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives [J]. Applied Microbiology and Biotechnology, 2009, 84(1): 37-53. |
| 44 | LIU L X, ZHANG L, TANG W, et al. Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis[J]. Journal of Bacteriology, 2012, 194(19): 5413-5422. |
| 45 | TANAKA K, KOMIYAMA A, SONOMOTO K, et al. Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1[J]. Applied Microbiology and Biotechnology, 2002, 60(1/2): 160-167. |
| 46 | SCALCINATI G, OTERO J M, VLEET J R H VAN, et al. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption[J]. FEMS Yeast Research, 2012, 12(5): 582-597. |
| 47 | WEIMBERG R. Pentose oxidation by Pseudomonas fragi [J]. Journal of Biological Chemistry, 1961, 236(3): 629-635. |
| 48 | DAHMS A S. 3-Deoxy-D-pentulosonic acid aldolase and its role in a new pathway of D-xylose degradation [J]. Biochemical and Biophysical Research Communications, 1974, 60(4): 1433-9. |
| 49 | KUHAD R C, GUPTA R, KHASA Y P, et al. Bioethanol production from pentose sugars: current status and future prospects[J]. Renewable and Sustainable Energy Reviews, 2011, 15(9): 4950-4962. |
| 50 | KIM S R, PARK Y C, JIN Y S, et al. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism[J]. Biotechnology Advances, 2013, 31(6): 851-861. |
| 51 | QI X, ZHA J, LIU G-G, et al. Heterologous xylose isomerase pathway and evolutionary engineering improve xylose utilization in Saccharomyces cerevisiae [J]. Frontiers in Microbiology, 2015, 6: 1165. |
| 52 | GOPINARAYANAN V E, NAIR N U. Pentose metabolism in Saccharomyces cerevisiae: the need to engineer global regulatory systems[J]. Biotechnology Journal, 2019, 14(1): e1800364. |
| 53 | KWAK S, JIN Y S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective [J]. Microbial Cell Factories, 2017, 16(1): 82-96. |
| 54 | ZHA J, SHEN M H, HU M L, et al. Enhanced expression of genes involved in initial xylose metabolism and the oxidative pentose phosphate pathway in the improved xylose-utilizing Saccharomyces cerevisiae through evolutionary engineering[J]. Journal of Industrial Microbiology & Biotechnology, 2014, 41(1): 27-39. |
| 55 | HOU J, SHEN Y, JIAO C L, et al. Characterization and evolution of xylose isomerase screened from the bovine rumen metagenome in Saccharomyces cerevisiae [J]. Journal of Bioscience and Bioengineering, 2016, 121(2): 160-165. |
| 56 | KATAHIRA S, MURAMOTO N, MORIYA S, et al. Screening and evolution of a novel protist xylose isomerase from the termite Reticulitermes speratus for efficient xylose fermentation in Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2017, 10(1): 203-220. |
| 57 | SONDEREGGER M, SAUER U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose[J]. Applied and Environmental Microbiology, 2003, 69(4): 1990-1998. |
| 58 | MADHAVAN A, TAMALAMPUDI S, USHIDA K, et al. Xylose isomerase from polycentric fungus Orpinomyces: gene sequencing, cloning, and expression in Saccharomyces cerevisiae for bioconversion of xylose to ethanol[J]. Applied Microbiology and Biotechnology, 2009, 82(6): 1067-1078. |
| 59 | KUYPER M, TOIRKENS M J, DIDERICH J A, et al. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain[J]. FEMS Yeast Research, 2005, 5(10): 925-934. |
| 60 | HOU J, QIU C X, SHEN Y, et al. Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose[J]. FEMS Yeast Research, 2017, 17(4): fox034. |
| 61 | WANG X, JIN M J, BALAN V, et al. Comparative metabolic profiling revealed limitations in xylose-fermenting yeast during co-fermentation of glucose and xylose in the presence of inhibitors[J]. Biotechnology and Bioengineering, 2014, 111(1): 152-164. |
| 62 | LÜ Y J, WANG X, MA Q, et al. Proteomic analysis reveals complex metabolic regulation in Saccharomyces cerevisiae cells against multiple inhibitors stress[J]. Applied Microbiology and Biotechnology, 2014, 98(5): 2207-2221. |
| 63 | ZHA J, LI B Z, SHEN M H, et al. Optimization of CDT-1 and XYL1 expression for balanced co-production of ethanol and xylitol from cellobiose and xylose by engineered Saccharomyces cerevisiae [J]. PLoS One, 2013, 8(7): e68317. |
| 64 | DUNN K L, RAO C V. High-throughput sequencing reveals adaptation-induced mutations in pentose-fermenting strains of Zymomonas mobilis [J]. Biotechnology and Bioengineering, 2015, 112(11): 2228-2240. |
| 65 | DUNN K L, RAO C V. Expression of a xylose-specific transporter improves ethanol production by metabolically engineered Zymomonas mobilis [J]. Applied Microbiology and Biotechnology, 2014, 98(15): 6897-6905. |
| 66 | YOUNG E M, TONG A, BUI H, et al. Rewiring yeast sugar transporter preference through modifying a conserved protein motif[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(1): 131-136. |
| 67 | SARKAR P, MUKHERJEE M, GOSWAMI G, et al. Adaptive laboratory evolution induced novel mutations in Zymomonas mobilis ATCC ZW658: a potential platform for co-utilization of glucose and xylose[J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(3): 329-341. |
| 68 | SARKAR P, GOSWAMI G, MUKHERJEE M, et al. Heterologous expression of xylose specific transporter improves xylose utilization by recombinant Zymomonas mobilis strain in presence of glucose[J]. Process Biochemistry, 2021, 102: 190-198. |
| 69 | TAI Y S, XIONG M, JAMBUNATHAN P, et al. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products[J]. Nature Chemical Biology, 2016, 12(4): 247-253. |
| 70 | LIU H W, LU T. Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli [J]. Metabolic Engineering, 2015, 29: 135-141. |
| 71 | WANG J, JAIN R, SHEN X L, et al. Rational engineering of diol dehydratase enables 1,4-butanediol biosynthesis from xylose[J]. Metabolic Engineering, 2017, 40: 148-156. |
| 72 | WANG J, SHEN X L, JAIN R, et al. Establishing a novel biosynthetic pathway for the production of 3,4-dihydroxybutyric acid from xylose in Escherichia coli [J]. Metabolic Engineering, 2017, 41: 39-45. |
| 73 | WANG J, SHEN X L, LIN Y H, et al. Investigation of the synergetic effect of xylose metabolic pathways on the production of glutaric acid[J]. ACS Synthetic Biology, 2018, 7(1): 24-29. |
| 74 | BAI W Q, TAI Y S, WANG J Y, et al. Engineering nonphosphorylative metabolism to synthesize mesaconate from lignocellulosic sugars in Escherichia coli [J]. Metabolic Engineering, 2016, 38: 285-292. |
| 75 | WEI N, QUARTERMAN J, KIM S R, et al. Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast[J]. Nature Communications, 2013, 4: 2580. |
| 76 | MEILGAARD M C. Flavor chemistry of beer (II): Flavor and threshold of 239 aroma volatiles [J]. Techquartmasterbrewassocam, 1975, 12: 22-8. |
| 77 | EHRLICH F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweißaufbau der Hefe [J]. Berichte der Deutschen Chemischen Gesellschaft, 1907, 40(1): 1027-1047. |
| 78 | HOHMANN S. Characterisation of PDC2, a gene necessary for high level expression of pyruvate decarboxylase structural genes in Saccharomyces cerevlsiae [J]. Molecular and General Genetics MGG, 1993, 241(5/6): 657-666. |
| 79 | HOHMANN S. Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae [J]. Journal of Bacteriology, 1991, 173(24): 7963-7969. |
| 80 | HOHMANN S, CEDERBERG H. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5 [J]. European Journal of Biochemistry, 1990, 188(3): 615-621. |
| 81 | VURALHAN Z, MORAIS M A, TAI S L, et al. Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae [J]. Applied and Environmental Microbiology, 2003, 69(8): 4534-4541. |
| 82 | VURALHAN Z, LUTTIK M A, TAI S L, et al. Physiological characterization of the ARO10-dependent, broad-substrate-specificity 2-oxo acid decarboxylase activity of Saccharomyces cerevisiae [J]. Applied and Environmental Microbiology, 2005, 71(6): 3276-3284. |
| 83 | IRAQUI I, VISSERS S, ANDRÉ B, et al. Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae [J]. Molecular and Cellular Biology, 1999, 19(5): 3360-3371. |
| 84 | MOJZITA D, HOHMANN S. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae [J]. Molecular Genetics and Genomics, 2006, 276(2): 147-161. |
| 85 | NOSAKA K, ONOZUKA M, KONNO H, et al. Genetic regulation mediated by thiamin pyrophosphate-binding motif in Saccharomyces cerevisiae [J]. Molecular Microbiology, 2005, 58(2): 467-479. |
| 86 | SMIT G, SMIT B A, ENGELS W J M. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products[J]. FEMS Microbiology Reviews, 2005, 29(3): 591-610. |
| 87 | ATSUMI S, HANAI T, LIAO J C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels[J]. Nature, 2008, 451(7174): 86-89. |
| 88 | KÖNIG S. Subunit structure, function and organisation of pyruvate decarboxylases from various organisms[J]. Biochimica et Biophysica Acta, 1998, 1385(2): 271-286. |
| 89 | CONNOR M R, CANN A F, LIAO J C. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation[J]. Applied Microbiology and Biotechnology, 2010, 86(4): 1155-1164. |
| 90 | ATSUMI S, WU T Y, ECKL E M, et al. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes[J]. Applied Microbiology and Biotechnology, 2010, 85(3): 651-657. |
| 91 | CANN A F, LIAO J C. Production of 2-methyl-1-butanol in engineered Escherichia coli [J]. Applied Microbiology and Biotechnology, 2008, 81(1): 89-98. |
| 92 | ZHANG K C, SAWAYA M R, EISENBERG D S, et al. Expanding metabolism for biosynthesis of nonnatural alcohols[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(52): 20653-20658. |
| 93 | XIONG M, DENG J, WOODRUFF A P, et al. A bio-catalytic approach to aliphatic ketones[J]. Scientific Reports, 2012, 2: 311. |
| 94 | WENDISCH V F. Metabolic engineering advances and prospects for amino acid production[J]. Metabolic Engineering, 2020, 58: 17-34. |
| 95 | LIN P P, MI L, MORIOKA A H, et al. Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum [J]. Metabolic Engineering, 2015, 31: 44-52. |
| 96 | LI S S, WEN J P, JIA X Q. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression[J]. Applied Microbiology and Biotechnology, 2011, 91(3): 577-589. |
| 97 | MATSUDA F, ISHII J, KONDO T, et al. Increased isobutanol production in Saccharomyces cerevisiae by eliminating competing pathways and resolving cofactor imbalance [J]. Microbial Cell Factories, 2013, 12: 119-129. |
| 98 | WANG J, LI C Y, ZOU Y S, et al. Bacterial synthesis of C3-C5 diols via extending amino acid catabolism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(32): 19159-19167. |
| 99 | CHAE T U, AHN J H, KO Y S, et al. Metabolic engineering for the production of dicarboxylic acids and diamines[J]. Metabolic Engineering, 2020, 58: 2-16. |
| 100 | CHAE T U, KO Y S, HWANG K S, et al. Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams[J]. Metabolic Engineering, 2017, 41: 82-91. |
| 101 | YU J L, XIA X X, ZHONG J J, et al. A novel synthetic pathway for glutarate production in recombinant Escherichia coli [J]. Process Biochemistry, 2017, 59: 167-171. |
| 102 | YU J L, XIA X X, ZHONG J J, et al. Enhanced production of C5 dicarboxylic acids by aerobic-anaerobic shift in fermentation of engineered Escherichia coli [J]. Process Biochemistry, 2017, 62: 53-58. |
| 103 | SCHULZ H. Beta oxidation of fatty acids[J]. Biochimica et Biophysica Acta, 1991, 1081(2): 109-120. |
| 104 | KATIYAR S S, PORTER J W. Mechanism of fatty acid synthesis[J]. Life Sciences, 1977, 20(5): 737-759. |
| 105 | CLOMBURG J M, VICK J E, BLANKSCHIEN M D, et al. A synthetic biology approach to engineer a functional reversal of the β-oxidation cycle[J]. ACS Synthetic Biology, 2012, 1(11): 541-554. |
| 106 | GULEVICH A Y, SKOROKHODOVA A Y, SUKHOZHENKO A V, et al. Metabolic engineering of Escherichia coli for 1-butanol biosynthesis through the inverted aerobic fatty acid β-oxidation pathway[J]. Biotechnology Letters, 2012, 34(3): 463-469. |
| 107 | LIAN J Z, ZHAO H M. Reversal of the β-oxidation cycle in Saccharomyces cerevisiae for production of fuels and chemicals[J]. ACS Synthetic Biology, 2015, 4(3): 332-341. |
| 108 | ZHUANG Q Q, WANG Q, LIANG Q F, et al. Synthesis of polyhydroxyalkanoates from glucose that contain medium-chain-length monomers via the reversed fatty acid β-oxidation cycle in Escherichia coli [J]. Metabolic Engineering, 2014, 24: 78-86. |
| 109 | CINTOLESI A, CLOMBURG J M, GONZALEZ R. In silico assessment of the metabolic capabilities of an engineered functional reversal of the β-oxidation cycle for the synthesis of longer-chain (C≥4) products[J]. Metabolic Engineering, 2014, 23: 100-115. |
| 110 | DELLOMONACO C, CLOMBURG J M, MILLER E N, et al. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals[J]. Nature, 2011, 476(7360): 355-359. |
| 111 | CLOMBURG J M, BLANKSCHIEN M D, VICK J E, et al. Integrated engineering of β-oxidation reversal and ω-oxidation pathways for the synthesis of medium chain ω-functionalized carboxylic acids[J]. Metabolic Engineering, 2015, 28: 202-212. |
| 112 | CHEONG S, CLOMBURG J M, GONZALEZ R. Energy-and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions[J]. Nature Biotechnology, 2016, 34(5): 556-561. |
| 113 | STEEN E J, KANG Y, BOKINSKY G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass[J]. Nature, 2010, 463(7280): 559-562. |
| 114 | LENNEN R M, BRADEN D J, WEST R A, et al. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes[J]. Biotechnology and Bioengineering, 2010, 106(2): 193-202. |
| 115 | CAI W L, ZHANG W J. Engineering modular polyketide synthases for production of biofuels and industrial chemicals[J]. Current Opinion in Biotechnology, 2018, 50: 32-38. |
| 116 | ZHANG Z, PAN H X, TANG G L. New insights into bacterial type II polyketide biosynthesis[J]. F1000Research, 2017, 6: 172. |
| 117 | SHIMIZU Y, OGATA H, GOTO S. Type III polyketide synthases: functional classification and phylogenomics[J]. ChemBioChem, 2017, 18(1): 50-65. |
| 118 | KHOSLA C, HERSCHLAG D, CANE D E, et al. Assembly line polyketide synthases: Mechanistic insights and unsolved problems[J]. Biochemistry, 2014, 53(18): 2875-2883. |
| 119 | ZHANG W J, LIU J. Recent advances in understanding and engineering polyketide synthesis[J]. F1000Research, 2016, 5: 208. |
| 120 | WEISSMAN K J. Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology [J]. Natural Product Reports, 2016, 33(2): 203-230. |
| 121 | WONG F T, KHOSLA C. Combinatorial biosynthesis of polyketides-a perspective[J]. Current Opinion in Chemical Biology, 2012, 16(1/2): 117-123. |
| 122 | HERTWECK C. Decoding and reprogramming complex polyketide assembly lines: prospects for synthetic biology[J]. Trends in Biochemical Sciences, 2015, 40(4): 189-199. |
| 123 | KALKREUTER E, CROWETIPTON J M, LOWELL A N, et al. Engineering the substrate specificity of a modular polyketide synthase for installation of consecutive non-natural extender units[J]. Journal of the American Chemical Society, 2019, 141(5): 1961-1969. |
| 124 | ENG C H, YUZAWA S, WANG G, et al. Alteration of polyketide stereochemistry from anti to syn by a ketoreductase domain exchange in a type I modular polyketide synthase subunit[J]. Biochemistry, 2016, 55(12): 1677-1680. |
| 125 | ZARGAR A, LAL R, VALENCIA L, et al. Chemoinformatic-guided engineering of polyketide synthases[J]. Journal of the American Chemical Society, 2020, 142(22): 9896-9901. |
| 126 | LÜNNE F, NIEHAUS E M, LIPINSKI S, et al. Identification of the polyketide synthase PKS7 responsible for the production of lecanoric acid and ethyl lecanorate in Claviceps purpurea [J]. Fungal Genetics and Biology, 2020, 145: 103481. |
| 127 | HAGEN A, POUST S, ROND T D, et al. Engineering a polyketide synthase for in vitro production of adipic acid[J]. ACS Synthetic Biology, 2016, 5(1): 21-27. |
| 128 | FARRELL A E, PLEVIN R J, TURNER B T, et al. Ethanol can contribute to energy and environmental goals [J]. Science, 2006, 311(5760): 506-508. |
| [1] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [2] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [3] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [4] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [5] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [6] | 禹伟, 高教琪, 周雍进. 一碳生物转化合成有机酸的研究进展[J]. 合成生物学, 2024, 5(5): 1169-1188. |
| [7] | 陈锡玮, 张华然, 邹懿. 真菌源非核糖体肽类药物生物合成及代谢工程[J]. 合成生物学, 2024, 5(3): 571-592. |
| [8] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [9] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| [10] | 赵静宇, 张健, 祁庆生, 王倩. 基于细菌双组分系统的生物传感器的研究进展[J]. 合成生物学, 2024, 5(1): 38-52. |
| [11] | 孙绘梨, 崔金玉, 栾国栋, 吕雪峰. 面向高效光驱固碳产醇的蓝细菌合成生物技术研究进展[J]. 合成生物学, 2023, 4(6): 1161-1177. |
| [12] | 晏雄鹰, 王振, 娄吉芸, 张皓瑜, 黄星宇, 王霞, 杨世辉. 生物燃料高效生产微生物细胞工厂构建研究进展[J]. 合成生物学, 2023, 4(6): 1082-1121. |
| [13] | 程真真, 张健, 高聪, 刘立明, 陈修来. 代谢工程改造微生物利用甲酸研究进展[J]. 合成生物学, 2023, 4(4): 756-778. |
| [14] | 刘家宇, 杨智晗, 杨蕾, 朱丽英, 朱政明, 江凌. 合成生物技术驱动酪丁酸梭菌细胞工厂开发的研究进展[J]. 合成生物学, 2022, 3(6): 1174-1200. |
| [15] | 郭姝媛, 吴良焕, 刘香健, 王博, 于涛. 微生物中一碳代谢网络构建的进展与挑战[J]. 合成生物学, 2022, 3(1): 116-137. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||