合成生物学 ›› 2020, Vol. 1 ›› Issue (5): 556-569.DOI: 10.12211/2096-8280.2020-044
酿酒酵母基因组进化的研究进展
夏思杨1,2( ), 江丽红1,2, 蔡谨1, 黄磊1, 徐志南1, 连佳长1,2
), 江丽红1,2, 蔡谨1, 黄磊1, 徐志南1, 连佳长1,2
- 1.浙江大学化学工程与生物工程学院,生物质化工教育部重点实验室,浙江 杭州 310027
2.浙江大学化学工程与生物工程学院,合成生物学研究中心,浙江 杭州 310027
-
收稿日期:2020-04-08修回日期:2020-09-28出版日期:2020-10-31发布日期:2020-12-03 -
通讯作者:蔡谨,连佳长 -
作者简介:作者简介:夏思杨(1996—),女,硕士研究生。研究方向为基因组进化研究。E-mail:21828174@zju.edu.cn
蔡谨(1960—),男,博士,副教授。研究方向为工业微生物学。E-mail:caij@zju.edu.cn
连佳长(1984—),男,博士,研究员。研究方向为合成生物学。E-mail:jzlian@zju.edu.cn -
基金资助:国家重点研发计划(2018YFA0901800);国家自然科学基金(21808199);浙江省自然科学基金(R20B060006)
Advances in genome evolution of Saccharomyces cerevisiae
XIA Siyang1,2( ), JIANG Lihong1,2, CAI Jin1, HUANG Lei1, XU Zhinan1, LIAN Jiazhang1,2
), JIANG Lihong1,2, CAI Jin1, HUANG Lei1, XU Zhinan1, LIAN Jiazhang1,2
- 1.Key Laboratory of Biomass Chemical Engineering of Ministry of Education,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310027,Zhejiang,China
2.Center for Synthetic Biology,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310027,Zhejiang,China
-
Received:2020-04-08Revised:2020-09-28Online:2020-10-31Published:2020-12-03 -
Contact:CAI Jin, LIAN Jiazhang
摘要:
由于细胞代谢和调控网络的复杂性,尤其是对于多基因调控的复杂性状和遗传工具有限的生物系统而言,基因组进化在微生物细胞工厂的构建中起着至关重要的作用。基因组进化通过人为创造多样化性状以及功能筛选的迭代循环,在实验室中模拟且加速自然进化的过程,从而快速获得满足目标需求的进化突变体。酿酒酵母是代谢工程中重要的底盘细胞,全基因组进化是对其进行系统性改造的最有效合成生物学手段之一。本文总结了基因组进化在构建高效的酿酒酵母细胞工厂中的技术进展和应用,包括基因组改组、转座子插入诱变和全局转录机制工程(gTME)等基于随机突变的非理性基因组进化以及诸如酵母寡核苷酸介导的基因组工程(YOGE),真核基因组多重位点自动改造技术(eMAGE)、RNAi辅助的基因组进化方法(RAGE)以及基于CRISPR体系的基因组规模改造技术(CHAnGE、MAGIC和MAGESTIC)等可示踪的半理性基因组进化,并简要介绍了基因组进化面临的挑战和高通量筛选方法的发展前景。
中图分类号:
引用本文
夏思杨, 江丽红, 蔡谨, 黄磊, 徐志南, 连佳长. 酿酒酵母基因组进化的研究进展[J]. 合成生物学, 2020, 1(5): 556-569.
XIA Siyang, JIANG Lihong, CAI Jin, HUANG Lei, XU Zhinan, LIAN Jiazhang. Advances in genome evolution of Saccharomyces cerevisiae[J]. Synthetic Biology Journal, 2020, 1(5): 556-569.
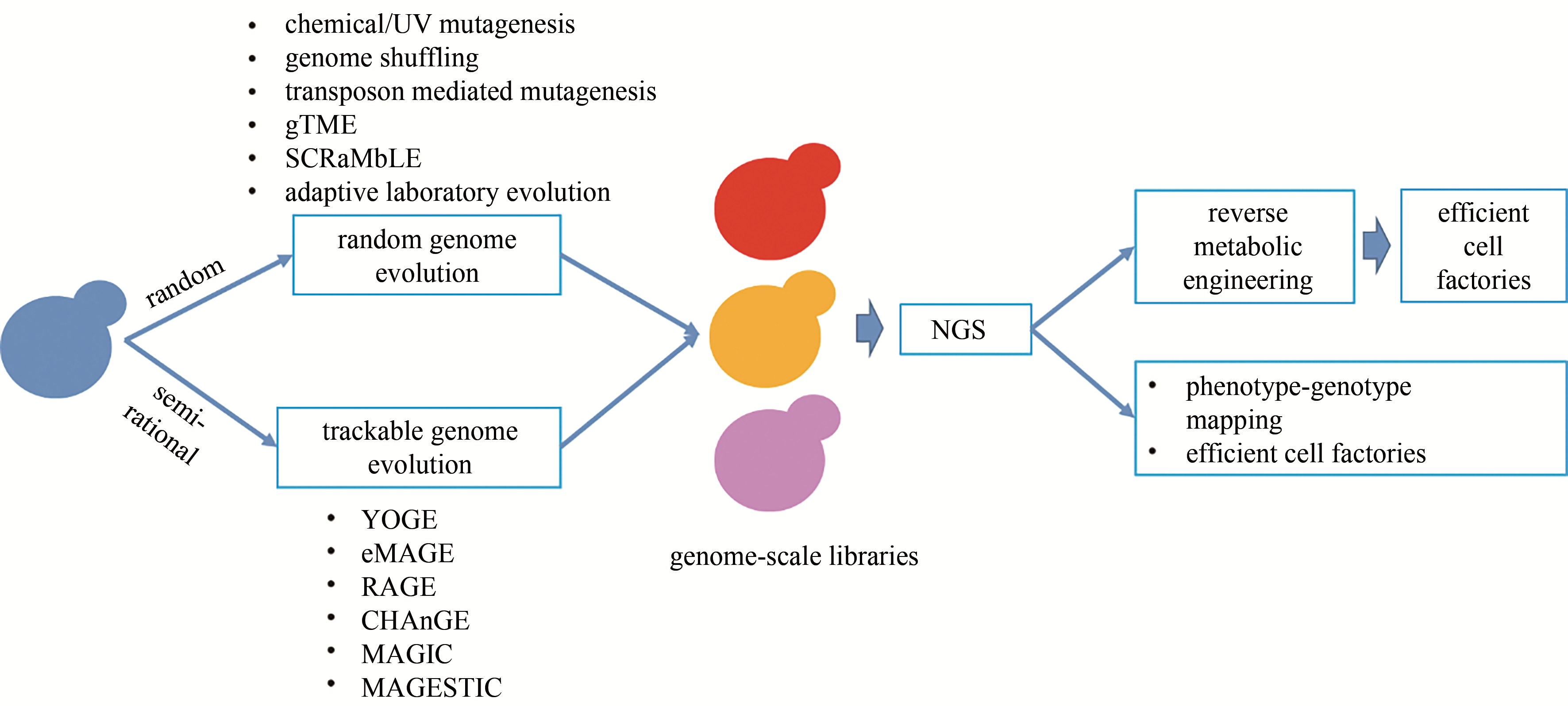
图1 酿酒酵母基因组进化的主要技术策略(基因组进化主要包括基于随机突变的基因组进化技术和基于高效基因组编辑技术的可示踪基因组进化技术)gTME—全局转录机制工程;SCRaMbLE—LoxP介导的合成染色体重组和修饰进化系统;YOGE—酵母寡核苷酸介导的基因组工程;eMAGE—真核多重自动化基因组工程;RAGE—RNAi辅助基因组进化;CHAnGE—CRISPR/Cas9和同源定向修复辅助的全基因组进化;MAGIC—多功能全基因组CRISPR系统; MAGESTIC—基于短、可示踪、整合的细胞条形码的多重精准基因编辑技术; NGS—二代测序
Fig. 1 Major technologies for genome evolution in Saccharomyces cerevisiae, including random genome evolution and trackable genome evolutiongTME—global transcription machinery engineering; SCRaMbLE—synthetic chromosome recombination and modification by LoxP-mediated evolution; YOGE—yeast oligo-mediated genome engineering; eMAGE—eukaryotic multiplex automated genome engineering; RAGE—RNAi-assisted genome evolution; CHAnGE—CRISPR/Cas9- and homology-directed-repair-assisted genome-scale engineering; MAGIC—multi-functional genome-wide CRISPR system; MAGESTIC—multiplexed accurate genome editing with short, trackable, integrated cellular barcodes; NGS—next-generation sequencing
| 技术名称 | 进化策略 | 技术特征 | 参考文献 |
|---|---|---|---|
| gTME | 对转录复合体中的关 键转录元件定向进化 | 靶向反式作用因子,无需修饰靶基因座,在转录水平上产生全基因组规模的多样性;不易阐述其分子机制 | [ |
| 实验室适 应性进化 | 环境压力 | 不需要考虑错综复杂的代谢网络,只需根据目标设计相应的选择压力,适用广泛;多重突变的存在使得分析进化表型的分子机制难度较大 | [ |
| 逆向代谢工程 | 基于全基因组突变分 析构建酵母细胞工厂 | 全面、系统地确定优良突变菌株的基因型-表型关系,可去除不良突变来最大程度地减少进化弊端;瓶颈在于如何在大量随机突变中确定有益突变 | [ |
| YOGE | 合成单链DNA库 | 酿酒酵母中首次由单链寡核苷酸介导重组的基因组工程;编辑效率低 | [ |
| eMAGE | 合成单链DNA库 | 比YOGE更加精确和高效的单链寡核苷酸整合技术;靶序列需要紧密接近复制起点以及URA3标记的共同选择 | [ |
| RAGE | 合成全基因组基因的 反向全长cDNA库 | 全长cDNA的表达可实现基因过表达,全长反义RNA的转录(RNAi)可实现基因下调;cDNA文库突变率有待进一步提高 | [ |
| CHAnGE | 基于全基因组构建 gRNA和同源模板库 | 基于CRISPR/Cas9的同源修复实现全基因组进化,可利用独特的条形码实现可追踪的编辑;只有基因敲除单一功能调控 | [ |
| MAGIC | 基于全基因组构建抑制、 激活和敲除的gRNA库 | 基于CRISPR/Cas9构建了最全面最多样化的酵母基因组文库,gRNA作为独特的基因条形码,可通过二代测序进行追踪;基因激活效率有待进一步提升 | [ |
| MAGESTIC | 基于目标序列构建 gRNA和同源模板库 | 通过基因组条形码整合进行追踪,可对上百万个细胞进行高通量基因编辑;其在基因组进化的应用有待进一步验证 | [ |
表1 酿酒酵母基因组进化策略
Tab. 1 Genome evolution strategies for S. cerevisiae
| 技术名称 | 进化策略 | 技术特征 | 参考文献 |
|---|---|---|---|
| gTME | 对转录复合体中的关 键转录元件定向进化 | 靶向反式作用因子,无需修饰靶基因座,在转录水平上产生全基因组规模的多样性;不易阐述其分子机制 | [ |
| 实验室适 应性进化 | 环境压力 | 不需要考虑错综复杂的代谢网络,只需根据目标设计相应的选择压力,适用广泛;多重突变的存在使得分析进化表型的分子机制难度较大 | [ |
| 逆向代谢工程 | 基于全基因组突变分 析构建酵母细胞工厂 | 全面、系统地确定优良突变菌株的基因型-表型关系,可去除不良突变来最大程度地减少进化弊端;瓶颈在于如何在大量随机突变中确定有益突变 | [ |
| YOGE | 合成单链DNA库 | 酿酒酵母中首次由单链寡核苷酸介导重组的基因组工程;编辑效率低 | [ |
| eMAGE | 合成单链DNA库 | 比YOGE更加精确和高效的单链寡核苷酸整合技术;靶序列需要紧密接近复制起点以及URA3标记的共同选择 | [ |
| RAGE | 合成全基因组基因的 反向全长cDNA库 | 全长cDNA的表达可实现基因过表达,全长反义RNA的转录(RNAi)可实现基因下调;cDNA文库突变率有待进一步提高 | [ |
| CHAnGE | 基于全基因组构建 gRNA和同源模板库 | 基于CRISPR/Cas9的同源修复实现全基因组进化,可利用独特的条形码实现可追踪的编辑;只有基因敲除单一功能调控 | [ |
| MAGIC | 基于全基因组构建抑制、 激活和敲除的gRNA库 | 基于CRISPR/Cas9构建了最全面最多样化的酵母基因组文库,gRNA作为独特的基因条形码,可通过二代测序进行追踪;基因激活效率有待进一步提升 | [ |
| MAGESTIC | 基于目标序列构建 gRNA和同源模板库 | 通过基因组条形码整合进行追踪,可对上百万个细胞进行高通量基因编辑;其在基因组进化的应用有待进一步验证 | [ |

图2 基于RNAi的酿酒酵母自动化多重基因组进化技术[31-32](a)酵母RNAi工作原理,核酸内切酶Dicer切割双链RNA(dsRNA)生成短干扰RNAs (siRNA),siRNA与效应蛋白Argonaute形成RNA诱导的沉默复合体RISC,siRNA反义链与靶标mRNA结合,指导Dicer干扰靶标基因的转录;(b)基于RNAi机制构建全基因组规模调控文库,在组成型启动子下定向克隆全长的cDNA文库,有义和反义构型分别引起目标基因的过表达或敲降;(c)基于RNAi的多重基因组突变方法,编码各种遗传修饰的基因调节部分的侧翼是同源δ序列,用于迭代和多重整合到重复的基因组序列。为了使CRISPR/Cas能进行高效且无需选择的δ整合,将Cas9表达盒整合到含有RNAi机制的酵母菌株中
Fig. 2 Scheme of automated RNAi-assisted genome evolution in yeast(a) RNAi mechanism in yeast. The double-stranded RNA (dsRNA) is digested by the endonuclease Dicer into short interference RNA (siRNA), which binds to the effector protein Argonaute to form the RNA-induced silencing complex (RISC). The non-sense strand of siRNA binds to the target mRNA, leading to the degradation and interference of the transcription of the target gene. (b) Construction of a genome-wide modulation part library in the yeast strain with the reconstituted RNAi machinery. Full-length cDNA library was directionally cloned under the control of a constitutive promoter. The sense and anti-sense configurations resulted in genetic overexpression and knockdown, respectively. (c) RNAi-assisted multiplex genomic mutations in yeast. Gene modulation parts were flanked by homologous arms for iterative and multiplex δ integration into the repetitive genomic sequences. To enable efficient and selection-free δ integration, a Cas9 expression cassette was integrated into the RNAi harboring yeast strain
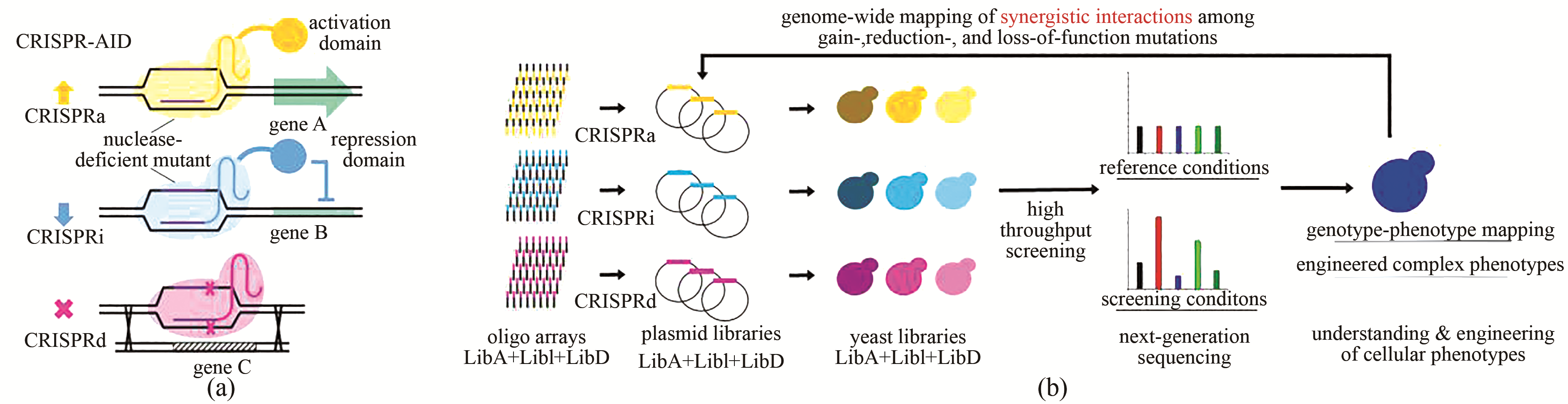
图3 MAGIC用于全基因组基因型-表型关系研究[34, 42](a)使用三种正交的CRISPR蛋白开发CRISPR-AID体系,其中CRISPRa由核酸酶缺陷的CRISPR蛋白与激活域融合(dLbCpf1-VP)实现转录激活,CRISPRi由核酸酶缺陷的突变体与抑制域融合(dSpCas9-RD1152)实现转录抑制,CRISPRd由具有催化活性的CRISPR蛋白(SaCas9)实现基因敲除;(b)通过DNA芯片寡核苷酸阵列的形式合成用于基因组规模激活(橙色)、干扰(浅蓝色)和缺失(品红色)的gRNA文库,并克隆到相应的gRNA表达质粒中。通过将质粒文库转入CRISPR-AID整合的酵母菌株中构建MAGIC文库,通过高通量筛选或者生长富集,并结合二代测序分析富集的gRNA序列。MAGIC可用于更好地理解、设计和改造复杂表型
Fig. 3 MAGIC for genome-wide mapping genotype-phenotype relationships[34,42](a) Development of CRISPR-AID using three orthogonal CRISPR proteins, a nuclease-deficient CRISPR protein fused with an activation domain (dLbCpf1-VP) for CRISPRa, a nuclease-deficient mutant fused with a repression domain (dSpCas9-RD1152) for CRISPRi, and a catalytically active CRISPR protein (SaCas9) for CRISPRd. (b) Guide sequences for genome-wide activation (orange), interference (light blue), and deletion (magenta) were synthesized as arrayed oligos on DNA chip and cloned into the corresponding gRNA expression plasmids. The transformation of the pooled plasmid libraries into the CRISPR-AID integrated yeast strain resulted in the construction of the MAGIC library. The MAGIC library was subject to growth enrichment or high throughput screening, and the corresponding enrichment or depletion of guide sequences were profiled using next-generation sequencing. MAGIC can be employed to better understand and engineer complex phenotypes
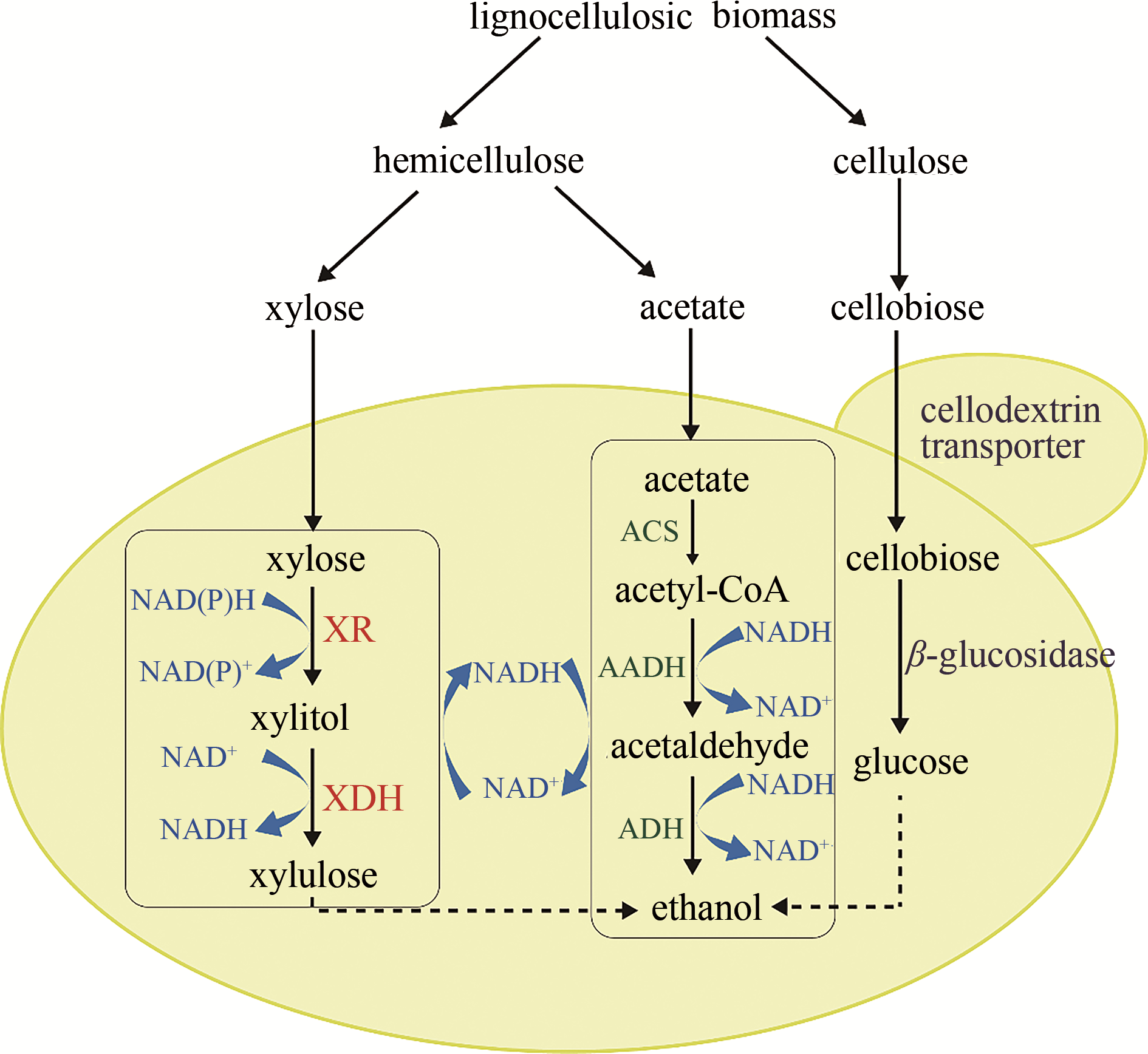
图4 同时利用木质纤维素碳源,如纤维二糖、木糖和乙酸的酵母菌株的构建XR—木糖还原酶;XDH—木糖醇脱氢酶;ACS—乙酰辅酶A合成酶;AADH—乙酰化乙醛脱氢酶;ADH—醇脱氢酶
Fig. 4 Construction of a recombinant yeast strain for simultaneous utilization of lignocellulosic carbons, such as cellobiose, xylose, and acetateXR—xylose reductase; XDH—xylitol dehydrogenase; ACS—acetyl-CoA synthetase; AADH—acetylating acetaldehyde dehydrogenase; ADH—alcohol dehydrogenase
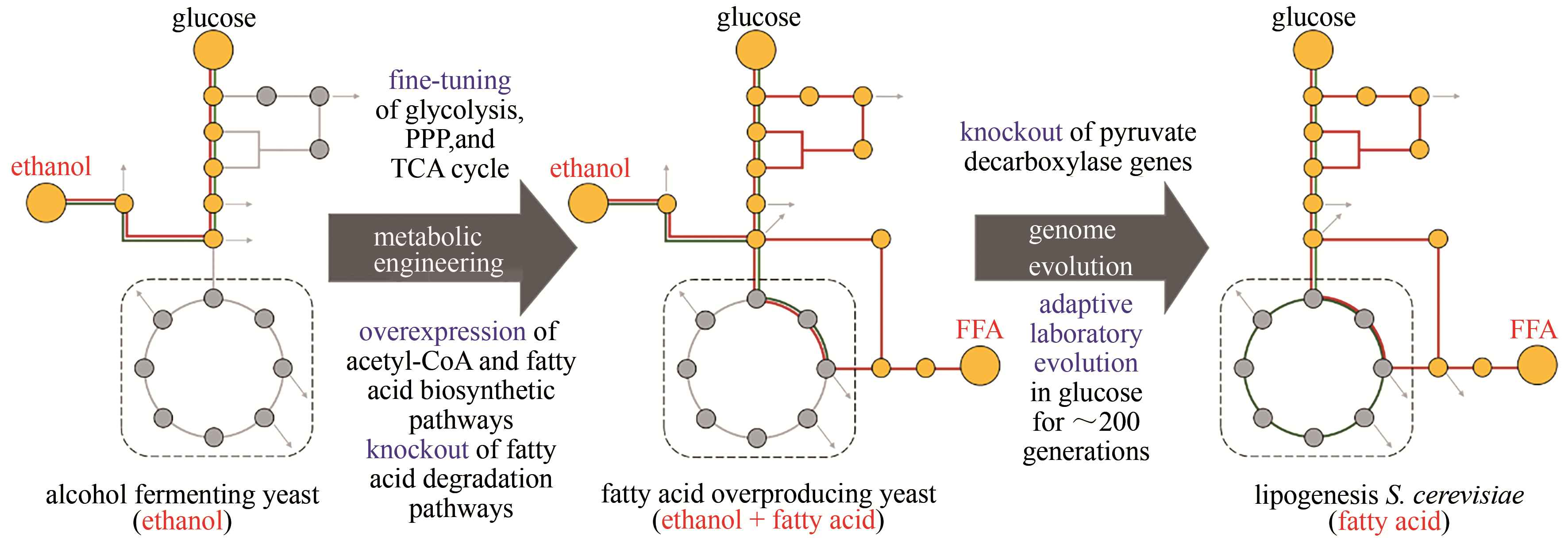
图5 高效生产FFA酵母菌株的构建[48](野生型酿酒酵母中,中心碳代谢是“葡萄糖转化为乙醇”;高产脂肪酸酿酒酵母中,为了实现“葡萄糖转化为油”的代谢模型,建立了FFA生产的有效途径;产油酿酒酵母中,通过适应性进化,酒精发酵成功地重新编程为纯脂肪酸合成)PPP—磷酸戊糖途径;TCA—三羧酸循环;FFA—游离脂肪酸
Fig. 5 Construction of yeast cell factories for an efficient production of FFA[48](In wild-type yeast, the major carbon metabolism is to drive ethanol fermentation from glucose. In the fatty acid-overproducing yeast, metabolic engineering strategies were employed to establish efficient biosynthetic pathway from glucose to fatty acid. In the lipogenesis yeast, genome evolution was performed to completely reprogram the cellular metabolism from alcohol fermentation to fatty acid biosynthesis)PPP—pentose phosphate pathway; TCA—tricarboxylic acid; FFA—free fatty acid
| 1 | KERKHOVEN E J, LAHTVEE P J, NIELSEN J. Applications of computational modeling in metabolic engineering of yeast [J]. FEMS Yeast Research, 2014, 15(1): 12199. |
| 2 | GOFFEAU A, BARRELL B G, BUSSEY H, et al. Life with 6000 genes [J]. Science, 1996, 274(5287): 546, 563-567. |
| 3 | HONG Kuk-Ki, NIELSEN J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries [J]. Cellular and Molecular Life Sciences, 2012, 69(16): 2671-2690. |
| 4 | WANG Yajie, YU Xiaowei, ZHAO Huimin. Biosystems design by directed evolution [J]. AIChE Journal, 2020, 66(3): e16716. |
| 5 | PATNAIK R. Engineering complex phenotypes in industrial strains [J]. Biotechnology Progress, 2008, 24(1): 38-47. |
| 6 | HASHIMOTO S, OGURA M, ARITOMI K, et al. Isolation of auxotrophic mutants of diploid industrial yeast strains after UV mutagenesis [J]. Applied and Environmental Microbiology, 2005, 71(1): 312-319. |
| 7 | ROUS C V, SNOW R, KUNKEE R E. Reduction of higher alcohols by fermentation with a leucine-auxotrophic mutant of wine yeast [J]. Journal of the Institute of Brewing, 1983, 89(4): 274-278. |
| 8 | BIOT PELLETIER D, MARTIN V J. Evolutionary engineering by genome shuffling [J]. Applied Microbiology and Biotechnology, 2014, 98(9): 3877-3887. |
| 9 | ZHANG Yingxin, PERRY K, VINCI V A, et al. Genome shuffling leads to rapid phenotypic improvement in bacteria [J]. Nature, 2002, 415(6872): 644-646. |
| 10 | SHI Dongjian, WANG Changlu, WANG Kuiming. Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae [J]. Journal of Industrial Microbiology & Biotechnology, 2009, 36(1): 139-147. |
| 11 | ZHENG D Q, WU X C, WANG P M, et al. Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae [J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(3): 415-422. |
| 12 | KUMAR A, SERINGHAUS M, BIERY M C, et al. Large-scale mutagenesis of the yeast genome using a Tn7-derived multipurpose transposon [J]. Genome Research, 2004, 14(10a): 1975-1986. |
| 13 | NI Haiying, LAPLAZA J M, JEFFRIES T W. Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on D-xylose [J]. Applied and Environmental Microbiology, 2007, 73(7): 2061-2066. |
| 14 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production [J]. Science, 2006, 314(5805): 1565-1568. |
| 15 | NAGY A. Cre recombinase: the universal reagent for genome tailoring [J]. Genesis, 2000, 26(2): 99-109. |
| 16 | TURAN S, BODE J. Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications [J]. FASEB Journal, 2011, 25(12): 4088-4107. |
| 17 | DYMOND J S, RICHARDSON S M, COOMBES C E, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design [J]. Nature, 2011, 477(7365): 471-476. |
| 18 | JIA Bin, WU Yi, LI Bingzhi, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast [J]. Nature Communications, 2018, 9(1): 1933. |
| 19 | Eun Joong OH, SKERKER J M, KIM Soo Rin, et al. Gene amplification on demand accelerates cellobiose utilization in engineered Saccharomyces cerevisiae [J]. Applied and Environmental Microbiology, 2016, 82(12): 3631-3639. |
| 20 | KIM Soo Rin, SKERKER J M, KANG Wei, et al. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae [J]. PLoS One, 2013, 8(2): e57048. |
| 21 | ENQUIST NEWMAN M, FAUST A M, BRAVO D D, et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform [J]. Nature, 2014, 505(7482): 239-243. |
| 22 | VOORDECKERS K, KOMINEK J, DAS A, et al. Adaptation to high ethanol reveals complex evolutionary pathways [J]. PLoS Genetics, 2015, 11(11): e1005635. |
| 23 | GONZÁLEZ-RAMOS D, DE VRIES A R G, GRIJSEELS S S, et al. A new laboratory evolution approach to select for constitutive acetic acid tolerance in Saccharomyces cerevisiae and identification of causal mutations [J]. Biotechnology for Biofuels, 2016, 9: 173. |
| 24 | FLETCHER E, FEIZI A, BISSCHOPS M M, et al. Evolutionary engineering reveals divergent paths when yeast is adapted to different acidic environments [J]. Metabolic Engineering, 2017, 39: 19-28. |
| 25 | HACISALIHOĞLU B, HOLYAVKIN C, TOPALOĞLU A, et al. Genomic and transcriptomic analysis of a coniferyl aldehyde-resistant Saccharomyces cerevisiae strain obtained by evolutionary engineering [J]. FEMS Yeast Research, 2019, 19(3): foz021. |
| 26 | CASPETA L, CHEN Yun, GHIACI P, et al. Altered sterol composition renders yeast thermotolerant [J]. Science, 2014, 346(6205): 75-78. |
| 27 | OUD B, MARIS A J VAN, DARAN J M, et al. Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast [J]. FEMS Yeast Research, 2012, 12(2): 183-196. |
| 28 | HONG Min Eui, Ki Sung LEE, YU Byung Jo, et al. Identification of gene targets eliciting improved alcohol tolerance in Saccharomyces cerevisiae through inverse metabolic engineering [J]. Journal of Biotechnology, 2010, 149(1/2): 52-59. |
| 29 | DICARLO J E, CONLEY A J, PENTTILA M, et al. Yeast oligo-mediated genome engineering (YOGE) [J]. ACS Synthetic Biology, 2013, 2(12): 741-749. |
| 30 | BARBIERI E M, MUIR P, AKHUETIE-ONI B O, et al. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes [J]. Cell, 2017, 171(6): 1453-1467 |
| 31 | SI Tong, LUO Yunzi, BAO Zehua, et al. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering [J]. ACS Synthetic Biology, 2015, 4(3): 283-291. |
| 32 | SI Tong, CHAO Ran, MIN Yuhao, et al. Automated multiplex genome-scale engineering in yeast [J]. Nature Communications, 2017, 8(1): 15187. |
| 33 | BAO Zehua, HAMEDIRAD M, XUE Pu, et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision [J]. Nature Biotechnology, 2018, 36(6): 505-508. |
| 34 | LIAN Jiazhang, SCHULTZ C, CAO Mingfeng, et al. Multi-functional genome-wide CRISPR system for high throughput genotype-phenotype mapping [J]. Nature Communications, 2019, 10(1): 5794. |
| 35 | ROY K R, SMITH J D, VONESCH S C, et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast [J]. Nature Biotechnology, 2018, 36(6): 512-520. |
| 36 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution [J]. Nature, 2009, 460(7257): 894-898. |
| 37 | PIJKEREN J P VAN, BRITTON R A. High efficiency recombineering in lactic acid bacteria [J]. Nucleic Acids Research, 2012, 40(10): e76. |
| 38 | DRINNENBERG I A, WEINBERG D E, XIE K T, et al. RNAi in budding yeast [J]. Science, 2009, 326(5952): 544-550. |
| 39 | XIAO Han, ZHAO Huimin. Genome-wide RNAi screen reveals the E3 SUMO-protein ligase gene SIZ1 as a novel determinant of furfural tolerance in Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2014, 7(1): 78. |
| 40 | BAO Zehua, XIAO Han, LIANG Jing, et al. Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2015, 4(5): 585-594. |
| 41 | RUSSA M F LA, QI L S. The new state of the art: Cas9 for gene activation and repression [J]. Molecular and Cellular Biology, 2015, 35(22): 3800-3809. |
| 42 | LIAN Jiazhang, HAMEDIRAD M, HU Sumeng, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system [J]. Nature Communications, 2017, 8(1): 1688. |
| 43 | Suk Jin HA, GALAZKA J M, KIM Soo Rin, et al. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation [J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(2): 504-509. |
| 44 | WEI Na, QUARTERMAN J, KIM Soo Rin, et al. Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast [J]. Nature Communications, 2013, 4(1): 2580. |
| 45 | WEI Na, Eun Joong OH, MILLION G, et al. Simultaneous utilization of cellobiose, xylose, and acetic acid from lignocellulosic biomass for biofuel production by an engineered yeast platform [J]. ACS Synthetic Biology, 2015, 4(6): 707-713. |
| 46 | REYES L H, GOMEZ J M, KAO K C. Improving carotenoids production in yeast via adaptive laboratory evolution [J]. Metabolic Engineering, 2014, 21: 26-33. |
| 47 | PATZSCHKE A, STEIGER M G, HOLZ C, et al. Enhanced glutathione production by evolutionary engineering of Saccharomyces cerevisiae strains [J]. Biotechnology Journal, 2015, 10(11): 1719-1726. |
| 48 | YU Tao, ZHOU Yongjin J, HUANG Mingtao, et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis [J]. Cell, 2018, 174(6): 1549-1558 |
| 49 | ZHU Zhiwei, HU Yating, TEIXEIRA P G, et al. Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids [J]. Nature Catalysis, 2020, 3(1): 64-74. |
| 50 | WANG Yanfeng, ZHANG Shuxian, LIU Huaqing, et al. Changes and roles of membrane compositions in the adaptation of Saccharomyces cerevisiae to ethanol [J]. Journal of Basic Microbiology, 2015, 55(12): 1417-1426. |
| 51 | SNOEK T, PICCA NICOLINO M, BREMT S VAN DEN, et al. Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance [J]. Biotechnology for Biofuels, 2015, 8(1): 32. |
| 52 | LING Hua, JUWONO N K P, Wei Suong TEO, et al. Engineering transcription factors to improve tolerance against alkane biofuels in Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2015, 8(1): 231. |
| 53 | BRENNAN T C, WILLIAMS T C, SCHULZ B L, et al. Evolutionary engineering improves tolerance for replacement jet fuels in Saccharomyces cerevisiae [J]. Applied and Environmental Microbiology, 2015, 81(10): 3316-3325. |
| 54 | BRACHER J M, DE HULSTER E, KOSTER C C, et al. Laboratory evolution of a biotin-requiring Saccharomyces cerevisiae strain for full biotin prototrophy and identification of causal mutations [J]. Applied and Environmental Microbiology, 2017, 83(16): e00892-00817. |
| 55 | LI Sijin, SI Tong, WANG Meng, et al. Development of a synthetic malonyl-CoA Sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening [J]. ACS Synthetic Biology, 2015, 4(12): 1308-1315. |
| 56 | WANG Meng, LI Sijin, ZHAO Huimin. Design and engineering of intracellular-metabolite-sensing/regulation gene circuits in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2016, 113(1): 206-215. |
| 57 | MUKHERJEE K, BHATTACHARYYA S, PERALTA-YAHYA P. GPCR-based chemical biosensors for medium-chain fatty acids [J]. ACS Synthetic Biology, 2015, 4(12): 1261-1269. |
| 58 | KIM Hee-Jung, Sura HA, Hee Yoon LEE, et al. ROSics: chemistry and proteomics of cysteine modifications in redox biology [J]. Mass Spectrometry Reviews, 2015, 34(2): 184-208. |
| 59 | LEAVITT J M, WAGNER J M, TU C C, et al. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae [J]. Biotechnology Journal, 2017, 12(10): 1600687. |
| 60 | MAIR P, GIELEN F, HOLLFELDER F. Exploring sequence space in search of functional enzymes using microfluidic droplets [J]. Current Opinion in Chemical Biology, 2017, 37: 137-144. |
| 61 | CHEN B, Sungwon LIM, KANNAN A, et al. High-throughput analysis and protein engineering using microcapillary arrays [J]. Nature Chemical Biology, 2016, 12(2): 76-81. |
| 62 | LARSEN A C, DUNN M R, HATCH A, et al. A general strategy for expanding polymerase function by droplet microfluidics [J]. Nature Communications, 2016, 7(1): 11235. |
| 63 | DORR M, FIBINGER M P, LAST D, et al. Fully automatized high-throughput enzyme library screening using a robotic platform [J]. Biotechnology and Bioengineering, 2016, 113(7): 1421-1432. |
| [1] | 晏雄鹰, 王振, 娄吉芸, 张皓瑜, 黄星宇, 王霞, 杨世辉. 生物燃料高效生产微生物细胞工厂构建研究进展[J]. 合成生物学, 2023, 4(6): 1082-1121. |
| [2] | 吴玉洁, 刘欣欣, 刘健慧, 杨开广, 随志刚, 张丽华, 张玉奎. 基于高通量液相色谱质谱技术的菌株筛选与关键分子定量分析研究进展[J]. 合成生物学, 2023, 4(5): 1000-1019. |
| [3] | 孙梦楚, 陆亮宇, 申晓林, 孙新晓, 王佳, 袁其朋. 基于荧光检测的高通量筛选技术和装备助力细胞工厂构建[J]. 合成生物学, 2023, 4(5): 947-965. |
| [4] | 高纤云, 牛灵雪, 见妮, 管宁子. 微生物合成生物学在疾病诊疗上的应用进展[J]. 合成生物学, 2023, 4(2): 263-282. |
| [5] | 潘颖佳, 夏思杨, 董昌, 蔡谨, 连佳长. 基因增变器驱动的酿酒酵母基因组连续进化[J]. 合成生物学, 2023, 4(1): 225-240. |
| [6] | 任师超, 孙秋艳, 冯旭东, 李春. 微生物细胞工厂合成五环三萜皂苷类化合物[J]. 合成生物学, 2022, 3(1): 168-183. |
| [7] | 陈久洲, 王钰, 蒲伟, 郑平, 孙际宾. 5-氨基乙酰丙酸生物合成技术的发展及展望[J]. 合成生物学, 2021, 2(6): 1000-1016. |
| [8] | 熊亮斌, 宋璐, 赵云秋, 刘坤, 刘勇军, 王风清, 魏东芝. 甾体化合物绿色生物制造:从生物转化到微生物从头合成[J]. 合成生物学, 2021, 2(6): 942-963. |
| [9] | 郭亮, 高聪, 柳亚迪, 陈修来, 刘立明. 大肠杆菌生产饲用氨基酸的研究进展[J]. 合成生物学, 2021, 2(6): 964-981. |
| [10] | 孙文涛, 张昕哲, 万盛通, 王茹雯, 李春. Ⅱ型细胞色素P450酶氧化β-香树脂醇的选择性调控研究[J]. 合成生物学, 2021, 2(5): 804-814. |
| [11] | 李晓东, 杨成帅, 王平平, 严兴, 周志华. 构建酿酒酵母细胞工厂从头合成倍半萜类化合物α-新丁香三环烯和β-石竹烯[J]. 合成生物学, 2021, 2(5): 792-803. |
| [12] | 李祎, 林振泉, 刘子鹤. 酿酒酵母适应性实验室进化工具的最新进展[J]. 合成生物学, 2021, 2(2): 287-301. |
| [13] | 盛月, 张根林. 酵母终止子工程:从机理探索到人工设计[J]. 合成生物学, 2020, 1(6): 709-721. |
| [14] | 袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673. |
| [15] | 许可, 王靖楠, 李春. 智能抗逆微生物细胞工厂与绿色生物制造[J]. 合成生物学, 2020, 1(4): 427-439. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
