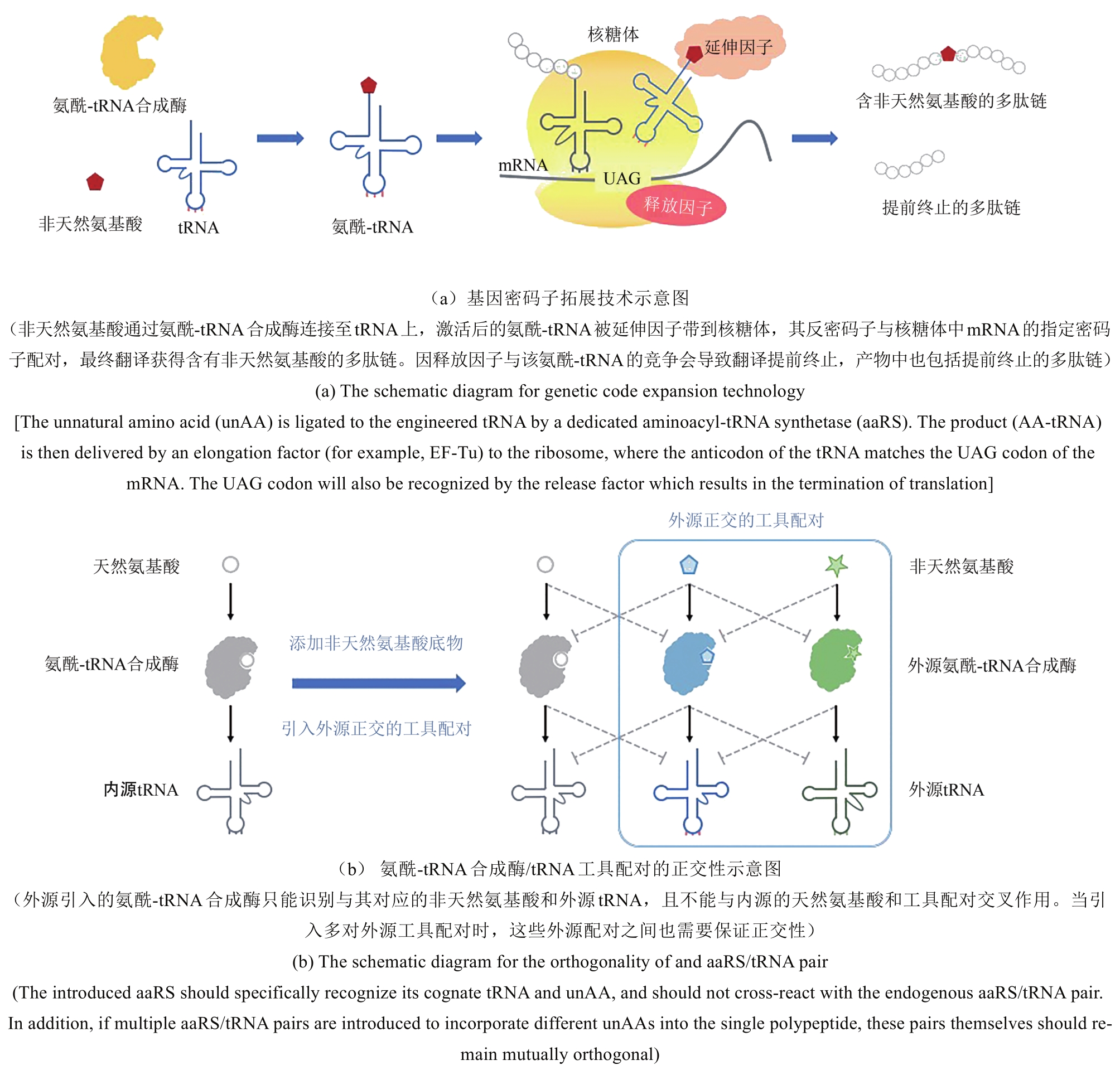
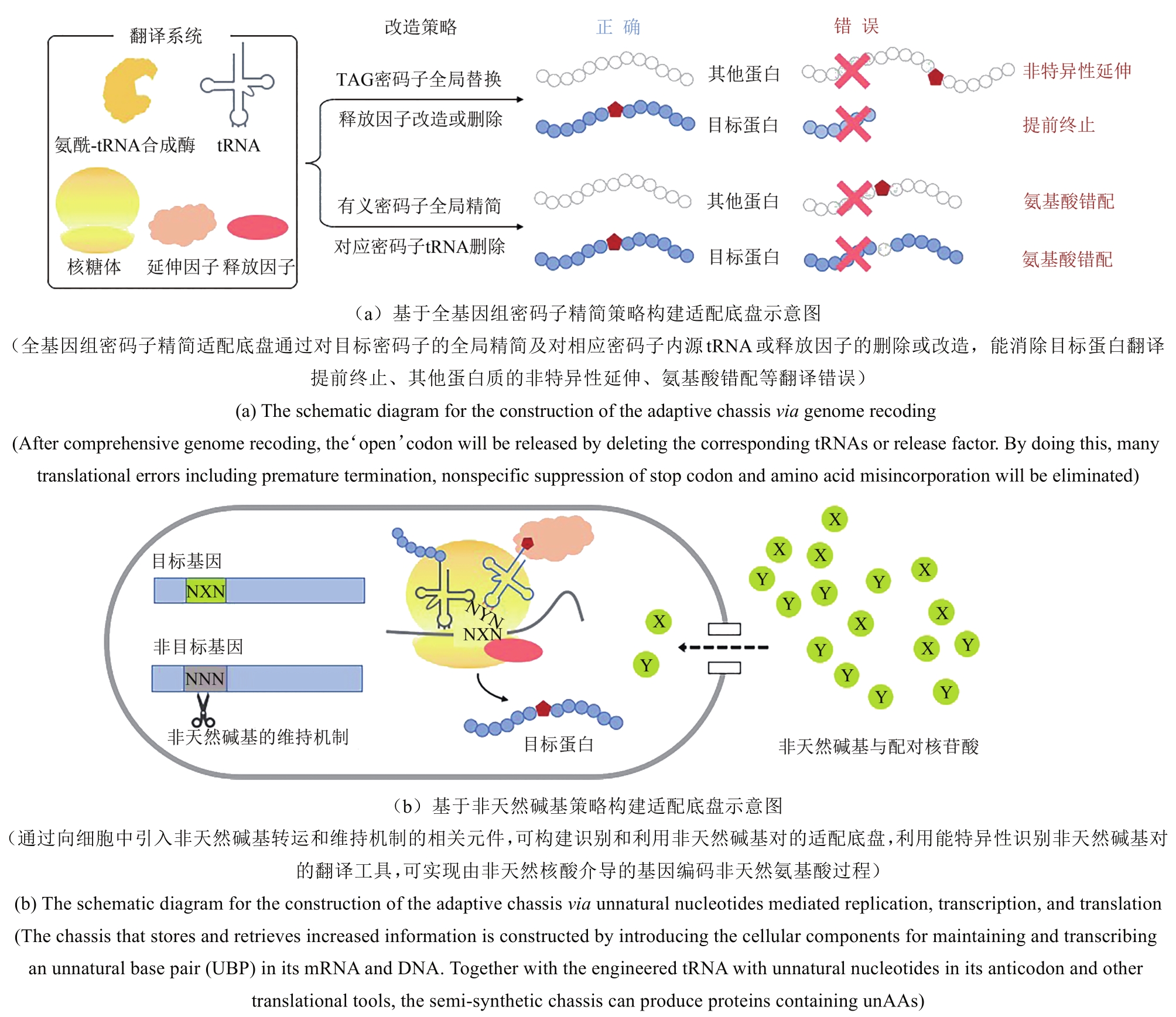
Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (1): 103-119.DOI: 10.12211/2096-8280.2020-007
• Invited Review • Previous Articles
Progress in the study of genetic code expansion related methods, principles and applications
FU Xian1,2, LIN Tao1, ZHANG Fan3, ZHANG Huiming1, ZHANG Wenwei1, YANG Huanming1,4, ZHU Shida1,5, XU Xun1,2, SHEN Yue1,2,5,6
- 1.BGI-Shenzhen, Shenzhen 518083, Guangdong, China
2.Guangdong Provincial Key Laboratory of Genome Read and Write, BGI-Shenzhen, Shenzhen 518120, Guangdong, China
3.BGI Education Center, University of Chinese Academy of Sciences, Shenzhen 518083, Guangdong, China
4.Guangdong Provincial Academician Workstation of BGI Synthetic Genomics, BGI-Shenzhen, Shenzhen 518120, Guangdong, China
5.Shenzhen Engineering Laboratory for Innovative Molecular Diagnostics, BGI-Shenzhen, Shenzhen 518120, Guangdong, China
6.China National GeneBank, BGI-Shenzhen, Shenzhen 518120, Guangdong, China
-
Received:2020-02-28Revised:2020-03-25Online:2020-07-07Published:2020-02-29 -
Contact:SHEN Yue
基因密码子拓展技术的方法原理和前沿应用研究进展
付宪1,2, 林涛1, 张帆3, 张惠铭1, 章文蔚1, 杨焕明1,4, 朱师达1,5, 徐讯1,2, 沈玥1,2,5,6
- 1.深圳华大生命科学研究院, 广东 深圳 518083
2.广东省高通量基因组测序与合成编辑应用重点实验室,深圳华大生命科学研究院,广东 深圳 518120
3.中国科学院大学华大教育中心,广东 深圳 518083
4.(广东省 )华大基因合成基因组学院士工作站, 深圳华大基因科技有限公司,广东 深圳 518120
5.深圳创新分子诊断技术工程实验室,深圳华大生命科学研究院,广东 深圳 518120
6.深圳国家基因库,广东 深圳 518120
-
通讯作者:沈玥 -
作者简介:付宪(1989-),男,博士,副研究员,研究方向为合成生物学、合成基因组学、蛋白质定向进化。E-mail:fuxian1@genomics.cn
沈玥(1986-),女,博士,研究员,研究方向为合成生物学、合成基因组学、DNA合成技术与工具开发。E-mail:shenyue@genomics.cn -
基金资助:国家重点研发计划(2018YFA0900100)国家自然科学基金?(31901029、31800078)
CLC Number:
Cite this article
FU Xian, LIN Tao, ZHANG Fan, ZHANG Huiming, ZHANG Wenwei, YANG Huanming, ZHU Shida, XU Xun, SHEN Yue. Progress in the study of genetic code expansion related methods, principles and applications[J]. Synthetic Biology Journal, 2020, 1(1): 103-119.
付宪, 林涛, 张帆, 张惠铭, 章文蔚, 杨焕明, 朱师达, 徐讯, 沈玥. 基因密码子拓展技术的方法原理和前沿应用研究进展[J]. 合成生物学, 2020, 1(1): 103-119.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-007
| 1 | MACINO G, CORUZZI G, NOBREGA F G, et al. Use of the UGA terminator as a tryptophan codon in yeast mitochondria[J]. PNAS, 1979, 76(8): 3784-3785. |
| 2 | BOCK A and STADTMAN T C. Selenocysteine, a highly specific component of certain enzymes, is incorporated by a UGA-directed co-translational mechanism[J]. Biofactors, 1988, 1(3): 245-250. |
| 3 | NOREN C, ANTHONY-CAHILL S, GRIFFITH M, et al. A general method for site-specific incorporation of unnatural amino acids into proteins[J]. Science, 1989, 244(4901): 182-188. |
| 4 | BUDISA N, MINKS C, ALEFELDER S, et al. Toward the experimental codon reassignment in vivo: protein building with an expanded amino acid repertoire[J]. FASEB J., 1999, 13(1): 41-51. |
| 5 | CORNISH V W, MENDEL D, SCHULTZ P G. Probing protein structure and function with an expanded genetic code[J]. Cheminform, 1995, 34(6): 621-633. |
| 6 | CHIN J W. Expanding and reprogramming the genetic code of cells and animals[J]. Annu. Rev. Biochem., 2014, 83: 379-408. |
| 7 | CHIN J W. Expanding and reprogramming the genetic code[J]. Nature, 2017, 550(7674): 53-60. |
| 8 | ARRANZ-GIBERT P, PATEL J R, ISAACS F J. The role of orthogonality in genetic code expansion[J]. Life, 2019, 9(3): 58. |
| 9 | GIEGÉ R, SISSLER M, FLORENTZ C. Universal rules and idiosyncratic features in tRNA identity[J]. Nucleic Acids Res., 1998, 26(22): 5017-5035. |
| 10 | LIU D R, MAGLIERY T J, PASTRNAK M, et al. Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo [J]. PNAS, 1997, 94(19): 10092-10097. |
| 11 | WANG L, BROCK A, HERBERICH B, et al. Expanding the genetic code of Escherichia coli [J]. Science, 2001, 292(5516): 498-500. |
| 12 | STEER B A, SCHIMMEL P. Major anticodon-binding region missing from an archaebacterial tRNA synthetase[J]. Journal of Biological Chemistry, 1999, 274(50): 35601-35606. |
| 13 | JAKUBOWSKI H, GOLDMAN E. Editing of errors in selection of amino acids for protein synthesis[J]. Microbiology and Molecular Biology Reviews, 1992, 56(3): 412-429. |
| 14 | XIE J, SCHULTZ P G. An expanding genetic code[J]. Methods, 2005, 36(3): 227-238. |
| 15 | CHIN J W, CROPP T A, ANDERSON J C, et al. An expanded eukaryotic genetic code[J]. Science, 2003, 301(5635): 964-967. |
| 16 | CHIN J W, CROPP T A, CHU S, et al. Progress toward an expanded eukaryotic genetic code[J]. Chemistry & Biology, 2003, 10(6): 511-519. |
| 17 | SAKAMOTO K, HAYASHI A, SAKAMOTO A, et al. Site‐specific incorporation of an unnatural amino acid into proteins in mammalian cells[J]. Nucleic Acids Research, 2002, 30(21): 4692-4699. |
| 18 | LIU W R, SCHULTZ P G. Genetic incorporation of unnatural amino acids into proteins in mammalian cells[J]. Nature Methods, 2007, 4(3): 239-244. |
| 19 | WU N, DEITERS A, CROPP T A, et al. A genetically encoded photocaged amino acid[J]. Journal of the American Chemical Society, 2004, 126(44): 14306-14307. |
| 20 | AMBROGELLY A, GUNDLLAPALLI S, HERRING S, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons[J]. PNAS, 2007, 104(9): 3141-3146. |
| 21 | SUZUKI T, MILLER C, GUO L T, et al. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase[J]. Nature Chemical Biology, 2017, 13(12): 1261-1266. |
| 22 | ITALIA J S, ADDY P S, ERICKSON S B, et al. Mutually orthogonal nonsense-suppression systems and conjugation chemistries for precise protein labeling at up to three distinct sites[J]. Journal of the American Chemical Society, 2019, 141(15): 6204-6212. |
| 23 | NEUMANN H, PEAK-CHEW S Y, CHIN J W. Genetically encoding N ε -acetyllysine in recombinant proteins[J]. Nature Chemical Biology, 2008, 4(4): 232-234. |
| 24 | WAN W, HUANG Y, WANG Z, et al. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli [J]. Angewandte Chemie International Edition, 2010, 49(18): 3211-3214. |
| 25 | CHATTERJEE A, XIAO H, SCHULTZ P G. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli [J]. PNAS, 2012, 109(37): 14841-14846. |
| 26 | WILLIS J C W, CHIN J W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs[J]. Nat. Chem., 2018, 10(8): 831-837. |
| 27 | MEINEKE B, HEIMGÄRTNER J, LAFRANCHI L, et al. Methanomethylophilus alvus Mx1201 provides basis for mutual orthogonal pyrrolysyl tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells[J]. ACS Chemical Biology, 2018, 13(11): 3087-3096. |
| 28 | VARGAS-RODRIGUEZ O, SEVOSTYANOVA A, SÖLL D, et al. Upgrading aminoacyl-tRNA synthetases for genetic code expansion[J]. Current Opinion in Chemical Biology, 2018, 46: 115-122. |
| 29 | GUO L, WANG Y, NAKAMURA A, et al. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution[J]. PNAS, 2014, 111(47): 16724-16729. |
| 30 | WANG L, SCHULTZ P G. Expanding the genetic code[J]. Angewandte Chemie International Edition, 2005, 44(1): 34-66. |
| 31 | WAN W, THARP J M, LIU W R. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool[J]. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2014, 1844(6): 1059-1070. |
| 32 | DUMAS A, LERCHER L, SPICER C D, et al. Designing logical codon reassignment-expanding the chemistry in biology[J]. Chemical Science, 2015, 6(1): 50-69. |
| 33 | LIU C C, SCHULTZ P G. Adding new chemistries to the genetic code[J]. Annual Review of Biochemistry, 2010, 79: 413-444. |
| 34 | DULIC M, CVETESIC N, ZIVKOVIC I, et al. Kinetic origin of substrate specificity in post-transfer editing by leucyl-tRNA synthetase[J]. Journal of Molecular Biology, 2018, 430(1): 1-16. |
| 35 | O'DONOGHUE P, LING J, WANG Y S, et al. Upgrading protein synthesis for synthetic biology[J]. Nature Chemical Biology, 2013, 9(10): 594-598. |
| 36 | AMIRAM M, HAIMOVICH A D, FAN C, et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids[J]. Nature Biotechnology, 2015, 33(12): 1272. |
| 37 | BRYSON D I, FAN C, GUO L T, et al. Continuous directed evolution of aminoacyl-tRNA synthetases[J]. Nature Chemical Biology, 2017, 13(12): 1253. |
| 38 | PACKER M S, LIU D R. Methods for the directed evolution of proteins[J]. Nature Reviews Genetics, 2015, 16(7): 379-394. |
| 39 | BADRAN A H, LIU D R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra[J]. Nature Communications, 2015, 6: 8425. |
| 40 | ALDAG C, BRÖCKER M J, HOHN M J, et al. Rewiring translation for elongation factor Tu‐dependent selenocysteine incorporation[J]. Angewandte Chemie International Edition, 2013, 52(5): 1441-1445. |
| 41 | MILLER C, BRÖCKER M J, PRAT L, et al. A synthetic tRNA for EF‐Tu mediated selenocysteine incorporation in vivo and in vitro [J]. FEBS letters, 2015, 589(17): 2194-2199. |
| 42 | GUO J, MELANÇON III C E, LEE H S, et al. Evolution of amber suppressor tRNAs for efficient bacterial production of proteins containing nonnatural amino acids[J]. Angewandte Chemie International Edition, 2009, 48(48): 9148-9151. |
| 43 | THARP J M, EHNBOM A, LIU W R. tRNAPyl: structure, function, and applications[J]. RNA Biology, 2018, 15(4/5): 441-452. |
| 44 | FAN C, XIONG H, REYNOLDS N M, et al. Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids[J]. Nucleic Acids Research, 2015, 43(22): e156. |
| 45 | PETRY S, BRODERSEN D E, MURPHY IV F V, et al. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon[J]. Cell, 2005, 123(7): 1255-1266. |
| 46 | WANG K, NEUMANN H, PEAK-CHEW S Y, et al. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion[J]. Nature Biotechnology, 2007, 25(7): 770-777. |
| 47 | ORELLE C, CARLSON E D, SZAL T, et al. Protein synthesis by ribosomes with tethered subunits[J]. Nature, 2015, 524(7563): 119-124. |
| 48 | FRIED S D, SCHMIED W H, UTTAMAPINANT C, et al. Ribosome subunit stapling for orthogonal translation in E. coli [J]. Angewandte Chemie International Edition, 2015, 54(43): 12791-12794. |
| 49 | PARK H S, HOHN M J, UMEHARA T, et al. Expanding the genetic code of Escherichia coli with phosphoserine[J]. Science, 2011, 333(6046): 1151-1154. |
| 50 | MUKAI T, HAYASHI A, IRAHA F, et al. Codon reassignment in the Escherichia coli genetic code[J]. Nucleic Acids Res., 2010, 38(22): 8188-8195. |
| 51 | SCHMIED W H, ELSÄSSER S J, UTTAMAPINANT C, et al. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1[J]. Journal of the American Chemical Society, 2014, 136(44): 15577-15583. |
| 52 | ARRANZ-GIBERT P, VANDERSCHUREN K, ISAACS F J. Next-generation genetic code expansion[J]. Current Opinion in Chemical Biology, 2018, 46: 203-211. |
| 53 | SHIMIZU Y, INOUE A, TOMARI Y, et al. Cell-free translation reconstituted with purified components[J]. Nature Biotechnology, 2001, 19(8): 751-755. |
| 54 | 高伟,卜宁,卢元. 无细胞体系非天然蛋白质合成研究进展[J]. 生物工程学报, 2018, 34(9):1371-1385. |
| GAO W, BU N, LU Y. Recent advances in cell-free unnatural protein synthesis[J]. Chinese Journal of Biotechnology, 2018, 34(9): 1371-1385. | |
| 55 | NEUMANN H, WANG K H, DAVIS L, et al. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome[J]. Nature, 2010, 464(7287): 441-444. |
| 56 | RACKHAM O, CHIN J W. A network of orthogonal ribosome·mRNA pairs[J]. Nature Chemical Biology, 2005, 1(3): 159-166. |
| 57 | SCHMIED W H, TNIMOV Z, UTTAMAPINANT C, et al. Controlling orthogonal ribosome subunit interactions enables evolution of new function[J]. Nature, 2018, 564(7736): 444-448. |
| 58 | REINKEMEIER C D, GIRONA G E, LEMKE E A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes[J]. Science, 2019, 363(6434): eaaw2644. |
| 59 | JOHNSON D B, XU J, SHEN Z, et al. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites[J]. Nature Chemical Biology, 2011, 7(11): 779-786. |
| 60 | RUAN B, PALIOURA S, SABINA J, et al. Quality control despite mistranslation caused by an ambiguous genetic code[J]. PNAS, 2008, 105(43): 16502-16507. |
| 61 | LAJOIE M J, ROVNER A J, GOODMAN D B, et al. Genomically recoded organisms expand biological functions[J]. Science, 2013, 342(6156): 357-360. |
| 62 | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G, et al. Design of a synthetic yeast genome[J]. Science, 2017, 355(6329): 1040-1044. |
| 63 | KOMAR A A. The Yin and Yang of codon usage[J]. Human Molecular Genetics, 2016, 25(R2): R77. |
| 64 | CHANEY J L, CLARK P L. Roles for synonymous codon usage in protein biogenesis[J]. Annual Review of Biophysics, 2015, 44: 143-166. |
| 65 | QUAX T E, CLAASSENS N J, SOLL D, et al. Codon bias as a means to fine-tune gene expression[J]. Molecular Cell, 2015, 59(2): 149-161. |
| 66 | CHO B K, ZENGLER K, QIU Y, et al. The transcription unit architecture of the Escherichia coli genome[J]. Nat. Biotechnol., 2009, 27(11): 1043-1049. |
| 67 | LI G W, OH E, WEISSMAN J S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria[J]. Nature, 2012, 484(7395): 538-541. |
| 68 | ZHOU M, GUO J, CHA J, et al. Non-optimal codon usage affects expression, structure and function of clock protein FRQ[J]. Nature, 2013, 495(7439): 111-115. |
| 69 | PRESNYAK V, ALHUSAINI N, CHEN Y H, et al. Codon optimality is a major determinant of mRNA stability[J]. Cell, 2015, 160(6): 1111-1124. |
| 70 | BOËL G, LETSO R, NEELY H, et al. Codon influence on protein expression in E. coli correlates with mRNA levels[J]. Nature, 2016, 529(7586): 358-363. |
| 71 | MISHIMA Y, TOMARI Y. Codon usage and 3' UTR length determine maternal mRNA stability in Zebrafish[J]. Molecular Cell, 2016, 61(6): 874-885. |
| 72 | NAPOLITANO M G, LANDON M, GREGG C J, et al. Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli [J]. PNAS, 2016, 113(38): E5588-E5597. |
| 73 | KUDLA G, MURRAY A W, TOLLERVEY D, et al. Coding-sequence determinants of gene expression in Escherichia coli [J]. Science, 2009, 324(5924): 255-258. |
| 74 | ZHOU Z, DANG Y, ZHOU M, et al. Codon usage is an important determinant of gene expression levels largely through its effects on transcription[J]. PNAS, 2016, 113(41): E6117-E6125. |
| 75 | CHEN S, LI K, CAO W, et al. Codon-resolution analysis reveals a direct and context-dependent impact of individual synonymous mutations on mRNA level[J]. Molecular Biology and Evolution, 2017, 34(11): 2944-2958. |
| 76 | SORENSEN M A, KURLAND C G, PEDERSEN S. Codon usage determines translation rate in Escherichia coli [J]. Journal of Molecular Biology, 1989, 207(2): 365-377. |
| 77 | SORENSEN M A, PEDERSEN S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate[J]. Journal of Molecular Biology, 1991, 222(2): 265-280. |
| 78 | TSAI C J, SAUNA Z E, KIMCHI-SARFATY C, et al. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima[J]. Journal of Molecular Biology, 2008, 383(2): 281-291. |
| 79 | BUHR F, JHA S, THOMMEN M, et al. Synonymous codons direct cotranslational folding toward different protein conformations[J]. Molecular Cell, 2016, 61(3): 341-351. |
| 80 | ZALUCKI Y M, BEACHAM I R, JENNINGS M P. Biased codon usage in signal peptides: a role in protein export[J]. Trends Microbiol, 2009, 17(4): 146-150. |
| 81 | ZALUCKI Y M, BEACHAM I R, JENNINGS M P. Coupling between codon usage, translation and protein export in Escherichia coli [J]. Biotechnol J., 2011, 6(6): 660-667. |
| 82 | PECHMANN S, CHARTRON J W, FRYDMAN J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo [J]. Nature Structure & Molecular Biology, 2014, 21(12): 1100-1105. |
| 83 | WANNIER T M, KUNJAPUR A M, RICE D P, et al. Adaptive evolution of genomically recoded Escherichia coli [J]. PNAS, 2018, 115(12): 3090-3095. |
| 84 | WANG K, FREDENS J, BRUNNER S F, et al. Defining synonymous codon compression schemes by genome recoding[J]. Nature, 2016, 539(7627): 59-64. |
| 85 | GOODMAN D B, CHURCH G M, KOSURI S. Causes and effects of N-terminal codon bias in bacterial genes[J]. Science, 2013, 342(6157): 475-479. |
| 86 | OSTROV N, LANDON M, GUELL M, et al. Design, synthesis, and testing toward a 57-codon genome[J]. Science, 2016, 353(6301): 819-822. |
| 87 | FREDENS J, WANG K, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| 88 | VENETZ J E, MEDICO L DEL, WÖLFLE A, et al. Chemical synthesis rewriting of a bacterial genome to achieve design flexibility and biological functionality[J]. PNAS, 2019, 116(16): 8070-8079. |
| 89 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363(6429): 884-887. |
| 90 | MALYSHEV D A, DHAMI K, LAVERGNE T, et al. A semi-synthetic organism with an expanded genetic alphabet[J]. Nature, 2014, 509: 385-388. |
| 91 | ZHANG Y, PTACIN J L, FISCHER E C, et al. A semi-synthetic organism that stores and retrieves increased genetic information[J]. Nature, 2017, 551: 644. |
| 92 | DIEN V T, HOLCOMB M, FELDMAN A W, et al. Progress toward a semi-synthetic organism with an unrestricted expanded genetic alphabet[J]. Journal of the American Chemical Society, 2018, 140(47): 16115-16123. |
| 93 | ZHOU A X Z, SHENG K, FELDMAN A W, et al. Progress toward eukaryotic semisynthetic organisms: translation of unnatural codons[J]. Journal of the American Chemical Society, 2019, 141(51): 20166-20170. |
| 94 | ZHANG Y, LAMB B M, FELDMAN A W, et al. A semisynthetic organism engineered for the stable expansion of the genetic alphabet[J]. PNAS, 2017, 114(6): 1317-1322. |
| 95 | ROMESBERG F E. Synthetic biology: the chemist's approach[J]. Israel Journal of Chemistry, 2019, 59(1/2): 91-94. |
| 96 | BROWNING D F, GODFREY R E, RICHARDS K L, et al. Exploitation of the Escherichia coli lac operon promoter for controlled recombinant protein production[J]. Biochemical Society Transaction, 2019, 47(2): 755-763. |
| 97 | SUZUKI T, ASAMI M, PATEL S G, et al. Switchable genome editing via genetic code expansion[J]. Scientific Report, 2018, 8(1): 10051. |
| 98 | HEMPHILL J, BORCHARDT E K, BROWN K, et al. Optical control of CRISPR/Cas9 gene editing[J]. Journal of the American Chemical Society, 2015, 137(17): 5642-5645. |
| 99 | EDWARDS W F, YOUNG D D, DEITERS A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells[J]. ACS Chemistry Biology, 2009, 4(6): 441-445. |
| 100 | BROWN W, LIU J, TSANG M, et al. Cell-lineage tracing in zebrafish embryos with an expanded genetic code[J]. ChemBioChem, 2018, 19(12): 1244-1249. |
| 101 | WANG J, LIU Y, LIU Y, et al. Time-resolved protein activation by proximal decaging in living systems[J]. Nature, 2019, 569(7757): 509-513. |
| 102 | MOK J, KIM P M, LAM H Y, et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs[J]. Sci. Signal., 2010, 3(109): ra12. |
| 103 | PARK H S, HOHN M J, UMEHARA T, et al. Expanding the genetic code of Escherichia coli with phosphoserine[J]. Science, 2011, 333(6046): 1151-1154. |
| 104 | PIRMAN N L, BARBER K W, AERNI H R, et al. A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation[J]. Nature Communications, 2015, 6: 8130. |
| 105 | ROGERSON D T, SACHDEVA A, WANG K, et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog[J]. Nature Chemical Biology, 2015, 11(7): 496-503. |
| 106 | YANG A, HA S, AHN J, et al. A chemical biology route to site-specific authentic protein modifications[J]. Science, 2016, 354(6312): 623-626. |
| 107 | ZHANG M S, BRUNNER S F, HUGUENIN-DEZOT N, et al. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing[J]. Nature Methods, 2017, 14(7): 729-736. |
| 108 | LUO X, FU G, WANG R E, et al. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria[J]. Nature Chemical Biology, 2017, 13(8): 845-849. |
| 109 | WANG Z A, KURRA Y, WANG X, et al. A versatile approach for site-specific lysine acylation in proteins[J]. Angewandte Chemie International Edition, 2017, 56(6): 1643-1647. |
| 110 | TSIEN R Y. The green fluorescent protein[J]. Annu. Rev. Biochem., 1998, 67: 509-544. |
| 111 | CAMPBELL R E, TOUR O, PALMER A E, et al. A monomeric red fluorescent protein[J]. PNAS, 2002, 99(12): 7877-7882. |
| 112 | LANG K, DAVIS L, TORRES-KOLBUS J, et al. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction[J]. Nat. Chem., 2012, 4(4): 298-304. |
| 113 | LUKINAVICIUS G, UMEZAWA K, OLIVIER N, et al. A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins[J]. Nat. Chem., 2013, 5(2): 132-139. |
| 114 | SIEVERS E L, SENTER P D. Antibody-drug conjugates in cancer therapy[J]. Annu. Rev. Med., 2013, 64: 15-29. |
| 115 | LAMBERT J M. Drug-conjugated antibodies for the treatment of cancer[J]. Br. J. Clin. Pharmacol., 2013, 76(2): 248-262. |
| 116 | WANG L, ZHANG Z, BROCK A, et al. Addition of the keto functional group to the genetic code of Escherichia coli [J]. PNAS, 2003, 100(1): 56-61. |
| 117 | KOLB H C, FINN M G, SHARPLESS K B. Click chemistry: diverse chemical function from a few good reactions[J]. Angewandte Chemie International Edition, 2001, 40(11): 2004-2021. |
| 118 | AGARWAL P, KUDIRKA R, ALBERS A E, et al. Hydrazino-Pictet-Spengler ligation as a biocompatible method for the generation of stable protein conjugates[J]. Bioconjug. Chem., 2013, 24(6): 846-851. |
| 119 | SCHMIDT M J, WEBER A, POTT M, et al. Structural basis of furan-amino acid recognition by a polyspecific aminoacyl-tRNA-synthetase and its genetic encoding in human cells[J]. ChemBioChem, 2014, 15(12): 1755-1760. |
| 120 | TEY S K. Adoptive T-cell therapy: adverse events and safety switches[J]. Clin. Transl. Immunology, 2014, 3(6): e17. |
| 121 | MA J S, KIM J Y, KAZANE S A, et al. Versatile strategy for controlling the specificity and activity of engineered T cells[J]. PNAS, 2016, 113(4): E450-E458. |
| 122 | JANG Y H, SEONG B L. Principles underlying rational design of live attenuated influenza vaccines[J]. Clin. Exp. Vaccine Res., 2012, 1(1): 35-49. |
| 123 | SI L, XU H, ZHOU X, et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines[J]. Science, 2016, 354(6316): 1170-1173. |
| 124 | LI Q, WU Y J. A fluorescent, genetically engineered microorganism that degrades organophosphates and commits suicide when required[J]. Appl. Microbiol. Biotechnol., 2009, 82(4): 749-756. |
| 125 | RONCHEL M C, RAMOS J L. Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation[J]. Appl. Environ. Microbiol., 2001, 67(6): 2649-2656. |
| 126 | WRIGHT O, STAN G B, ELLIS T. Building-in biosafety for synthetic biology[J]. Microbiology, 2013, 159(Pt 7): 1221-1235. |
| 127 | DIWO C, BUDISA N. Alternative biochemistries for alien life: basic concepts and requirements for the design of a robust biocontainment system in genetic isolation[J]. Genes (Basel), 2019, 10(1):17. |
| 128 | GAN F, LIU R, WANG F, et al. Functional replacement of histidine in proteins to generate noncanonical amino acid dependent organisms[J]. Journal of American Chemical Society, 2018, 140(11): 3829-3832. |
| 129 | MANDELL D J, LAJOIE M J, MEE M T, et al. Biocontainment of genetically modified organisms by synthetic protein design[J]. Nature, 2015, 518(7537): 55-60. |
| 130 | KOH M, YAO A, GLEASON P R, et al. A general strategy for engineering noncanonical amino acid dependent bacterial growth[J]. Journal of American Chemical Society, 2019, 141(41): 16213-16216. |
| 131 | ROVNER A J, HAIMOVICH A D, KATZ S R, et al. Recoded organisms engineered to depend on synthetic amino acids[J]. Nature, 2015, 518(7537): 89-93. |
| 132 | KATO Y. An engineered bacterium auxotrophic for an unnatural amino acid: a novel biological containment system[J]. PeerJ., 2015, 3: e1247. |
| [1] | YUAN Feiyan, YU Yang, LI Chun. Artificial enzyme designs and its application based on non-natural structural elements [J]. Synthetic Biology Journal, 2020, 1(6): 685-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||