Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (3): 267-284.DOI: 10.12211/2096-8280.2020-040
• Invited Review • Previous Articles Next Articles
Recent research progress in the design and construction of synthetic microbial consortia
QIAN Xiujuan1, CHEN Lin1, ZHANG Wenming1,2, ZHOU Jie1,2, DONG Weiliang1,2, XIN Fengxue1,2, JIANG Min1,2
- 1.State Key Laboratory of Materials-Oriented Chemical Engineering,College of Biotechnology and Pharmaceutical Engineering,Nanjing Tech University,Nanjing 211816,Jiangsu,China
2.Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM),Nanjing Tech University,Nanjing 211816,Jiangsu,China
-
Received:2020-04-05Revised:2020-08-03Online:2020-09-29Published:2020-06-30 -
Contact:XIN Fengxue, JIANG Min
人工多细胞体系设计与构建研究进展
钱秀娟1, 陈琳1, 章文明1,2, 周杰1,2, 董维亮1,2, 信丰学1,2, 姜岷1,2
- 1.南京工业大学材料化学工程国家重点实验室,江苏 南京 211816
2.南京工业大学江苏先进生物与化学制造协同创新中心(SICAM),江苏 南京 211816
-
通讯作者:信丰学,姜岷 -
作者简介:钱秀娟(1992—),女,博士,博士后,研究方向为代谢工程及合成生物学。E-mail:xiujuanqian@njtech.edu.cn
信丰学(1982—),男,博士,教授, 研究方向为生物化工与生物能源。E-mail:xinfengxue@njtech.edu.cn
姜岷(1972—),男,博士,教授,研究方向为生物转化与生物催化。E-mail:jiangmin@njtech.edu.cn -
基金资助:国家重点研发计划“合成生物学”重点专项(2018YFA0902200);国家自然科学基金项目(31961133017)
CLC Number:
Cite this article
QIAN Xiujuan, CHEN Lin, ZHANG Wenming, ZHOU Jie, DONG Weiliang, XIN Fengxue, JIANG Min. Recent research progress in the design and construction of synthetic microbial consortia[J]. Synthetic Biology Journal, 2020, 1(3): 267-284.
钱秀娟, 陈琳, 章文明, 周杰, 董维亮, 信丰学, 姜岷. 人工多细胞体系设计与构建研究进展[J]. 合成生物学, 2020, 1(3): 267-284.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-040
| 微生物组合 | 新产物 | 活性 | 是否可单独 培养获得 | 年份 | 参考文献 |
|---|---|---|---|---|---|
| Alternaria tenuissima &Nigrospora sphaerica | 间苯甲酚,异戊二烯醇 | 抗真菌 | 是 | 2013 | [ |
| Aspergillus fumigates &Streptomyces rapamycinicus | 富马环素A, B | 抗生素 | 否 | 2013 | [ |
| A. fumigatus & S. bullii | 麦角甾醇、二酮哌嗪生物碱 | 抗菌和抗原虫 | 否 | 2013 | [ |
| Fusarium tricinctum &Fusarium begoniae | 亚苯甲酸甲酯,亚苯甲酸乙酯 | 抗生素 | 否 | 2013 | [ |
| Trichophyton rubrum &Bionectria ochroleuca | 4″-羟基亚砜-2,2″-二甲基硫丙氨酸P | N/A | 否 | 2014 | [ |
| Streptomyces clavuligerus & Staphylococcus aureus | 全霉素 | 抗微生物 | 否 | 2012 | [ |
| Penicillium pinophilum &Trichoderma harzianum | 二氯戊菊酯C 青霉素、MC-141和施托美霉素 | N/A | 否 是 | 2011 | [ |
| A. fumigates & Streptomyces peucetius | 烟酰胺(1),N,N′-[(1Z,3Z)-1,4-双(4-甲氧基苯基)丁烷-1,3-二烯-2,3-二基]二甲酰胺(2) | (1)无活性 (2)细胞毒素 | 否 | 2011 | [ |
Tab. 1 Recent studies on mixed cultures identifying novel secondary metabolites
| 微生物组合 | 新产物 | 活性 | 是否可单独 培养获得 | 年份 | 参考文献 |
|---|---|---|---|---|---|
| Alternaria tenuissima &Nigrospora sphaerica | 间苯甲酚,异戊二烯醇 | 抗真菌 | 是 | 2013 | [ |
| Aspergillus fumigates &Streptomyces rapamycinicus | 富马环素A, B | 抗生素 | 否 | 2013 | [ |
| A. fumigatus & S. bullii | 麦角甾醇、二酮哌嗪生物碱 | 抗菌和抗原虫 | 否 | 2013 | [ |
| Fusarium tricinctum &Fusarium begoniae | 亚苯甲酸甲酯,亚苯甲酸乙酯 | 抗生素 | 否 | 2013 | [ |
| Trichophyton rubrum &Bionectria ochroleuca | 4″-羟基亚砜-2,2″-二甲基硫丙氨酸P | N/A | 否 | 2014 | [ |
| Streptomyces clavuligerus & Staphylococcus aureus | 全霉素 | 抗微生物 | 否 | 2012 | [ |
| Penicillium pinophilum &Trichoderma harzianum | 二氯戊菊酯C 青霉素、MC-141和施托美霉素 | N/A | 否 是 | 2011 | [ |
| A. fumigates & Streptomyces peucetius | 烟酰胺(1),N,N′-[(1Z,3Z)-1,4-双(4-甲氧基苯基)丁烷-1,3-二烯-2,3-二基]二甲酰胺(2) | (1)无活性 (2)细胞毒素 | 否 | 2011 | [ |
| 混菌体系 | 产物 | 相对单菌体系提升效果 | 发表年份 | 参考文献 |
|---|---|---|---|---|
| E. coli-E. coli | 黏康酸 | 产量提高19倍 | 2015 | [ |
| E. coli-E. coli | 4-羟基苯甲酸 | 产量提高8.6倍 | 2015 | [ |
| E. coli-E. coli | 乙酸苏氨酯 | 产量提高3.3倍,生产强度提高34倍 | 2015 | [ |
| E. coli-E. coli | 3-氨基苯甲酸 | 产量提高15倍 | 2016 | [ |
| E. coli-E. coli | 白藜芦醇 | 22.6 mg/L,以甘油为底物从头合成首次报道 | 2016 | [ |
| E. coli-E. coli | 类黄酮 | 产量提高970倍 | 2016 | [ |
| E. coli-E. coli | 水杨酸酯2-O-β-d-葡萄糖苷 | 2.5 g/L,首次混菌高产 | 2016 | [ |
| E. coli-E. coli | 柚皮素 | 产量提高1.5倍 | 2017 | [ |
| E. coli-E. coli | 咖啡醇 | 产量提高12倍 | 2017 | [ |
| E. coli-E. coli | 卡达维林 | 产量提高3倍 | 2018 | [ |
| E. coli-E. coli | 芹菜素 | 产量提高2.1倍 | 2018 | [ |
| E. coli-E. coli | 咖啡酰苹果酸 | 产量提高5倍 | 2018 | [ |
| E. coli-E. coli | 红景天苷 | 产量提高20多倍 | 2018 | [ |
| E. coli-E. coli | 白藜芦醇苷 | 产量提高2.9倍 | 2018 | [ |
| E. coli-E. coli | 双去甲氧基姜黄素 | 6.28 mg/L,以葡萄糖为底物从头合成首次报道 | 2018 | [ |
| E. coli-E. coli | 蒎烯 | 产量提高1.9倍 | 2018 | [ |
| E. coli-E. coli | 吡喃花青素 | 优于植物提取 | 2019 | [ |
| E. coli-E. coli | 3-羟基苯甲酸 | 产量提高5.3倍 | 2019 | [ |
| G. oxydans-K. vulgare | 2-酮-L-古洛糖酸 | 89.7% 的理论转化率,与双阶段发酵相当 | 2016 | [ |
| P. putida-P. putida | 2-羟基联苯 | 50%转化率 | 2016 | [ |
| E. coli-B. subtilis- | 电 | 0.28 g葡萄糖产生约550 mV电压供电15天 | 2017 | [ |
| S. oneidensis | ||||
| E. coli-E. coli-E. coli- E. coli | 花葵素 | 9.5 mg/L,微生物体系首次合成 | 2017 | [ |
| E. coli-E. coli-E. coli | 迷迭香酸 | 产量提高38倍 | 2019 | [ |
| P. pastoris-P. pastoris | 莫那可林J,洛伐他汀 | 莫那可林J产量提升55%,洛伐他汀产量提升71% | 2018 | [ |
| E. coli-S.cerevisiae | 氧化紫杉烷 | 33 mg/L,单细胞体系无法合成 | 2015 | [ |
| E. coli-S.cerevisiae | 柚皮素 | 产量提高8倍 | 2017 | [ |
Tab. 2 Summary of recent progress in valuable compound production applying synthetic microbial consortia
| 混菌体系 | 产物 | 相对单菌体系提升效果 | 发表年份 | 参考文献 |
|---|---|---|---|---|
| E. coli-E. coli | 黏康酸 | 产量提高19倍 | 2015 | [ |
| E. coli-E. coli | 4-羟基苯甲酸 | 产量提高8.6倍 | 2015 | [ |
| E. coli-E. coli | 乙酸苏氨酯 | 产量提高3.3倍,生产强度提高34倍 | 2015 | [ |
| E. coli-E. coli | 3-氨基苯甲酸 | 产量提高15倍 | 2016 | [ |
| E. coli-E. coli | 白藜芦醇 | 22.6 mg/L,以甘油为底物从头合成首次报道 | 2016 | [ |
| E. coli-E. coli | 类黄酮 | 产量提高970倍 | 2016 | [ |
| E. coli-E. coli | 水杨酸酯2-O-β-d-葡萄糖苷 | 2.5 g/L,首次混菌高产 | 2016 | [ |
| E. coli-E. coli | 柚皮素 | 产量提高1.5倍 | 2017 | [ |
| E. coli-E. coli | 咖啡醇 | 产量提高12倍 | 2017 | [ |
| E. coli-E. coli | 卡达维林 | 产量提高3倍 | 2018 | [ |
| E. coli-E. coli | 芹菜素 | 产量提高2.1倍 | 2018 | [ |
| E. coli-E. coli | 咖啡酰苹果酸 | 产量提高5倍 | 2018 | [ |
| E. coli-E. coli | 红景天苷 | 产量提高20多倍 | 2018 | [ |
| E. coli-E. coli | 白藜芦醇苷 | 产量提高2.9倍 | 2018 | [ |
| E. coli-E. coli | 双去甲氧基姜黄素 | 6.28 mg/L,以葡萄糖为底物从头合成首次报道 | 2018 | [ |
| E. coli-E. coli | 蒎烯 | 产量提高1.9倍 | 2018 | [ |
| E. coli-E. coli | 吡喃花青素 | 优于植物提取 | 2019 | [ |
| E. coli-E. coli | 3-羟基苯甲酸 | 产量提高5.3倍 | 2019 | [ |
| G. oxydans-K. vulgare | 2-酮-L-古洛糖酸 | 89.7% 的理论转化率,与双阶段发酵相当 | 2016 | [ |
| P. putida-P. putida | 2-羟基联苯 | 50%转化率 | 2016 | [ |
| E. coli-B. subtilis- | 电 | 0.28 g葡萄糖产生约550 mV电压供电15天 | 2017 | [ |
| S. oneidensis | ||||
| E. coli-E. coli-E. coli- E. coli | 花葵素 | 9.5 mg/L,微生物体系首次合成 | 2017 | [ |
| E. coli-E. coli-E. coli | 迷迭香酸 | 产量提高38倍 | 2019 | [ |
| P. pastoris-P. pastoris | 莫那可林J,洛伐他汀 | 莫那可林J产量提升55%,洛伐他汀产量提升71% | 2018 | [ |
| E. coli-S.cerevisiae | 氧化紫杉烷 | 33 mg/L,单细胞体系无法合成 | 2015 | [ |
| E. coli-S.cerevisiae | 柚皮素 | 产量提高8倍 | 2017 | [ |
| 12 | FLEMING A. On the antibacterial action of cultures of a penicillium with special reference to their use in isolation of B. influenzae [J]. Bull World Health Org, 2001,79:780-90 (reprinted from the Br J Exper Pathol, 1929,10:226-236). |
| 13 | PARSEK MR, Greenberg EP. Colloquium Paper: Acyl-homoserine lactone quorum sensing in Gram-negative bacteria: a signaling mechanism involved in associations with higher organisms [J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(16): 8789-8793. |
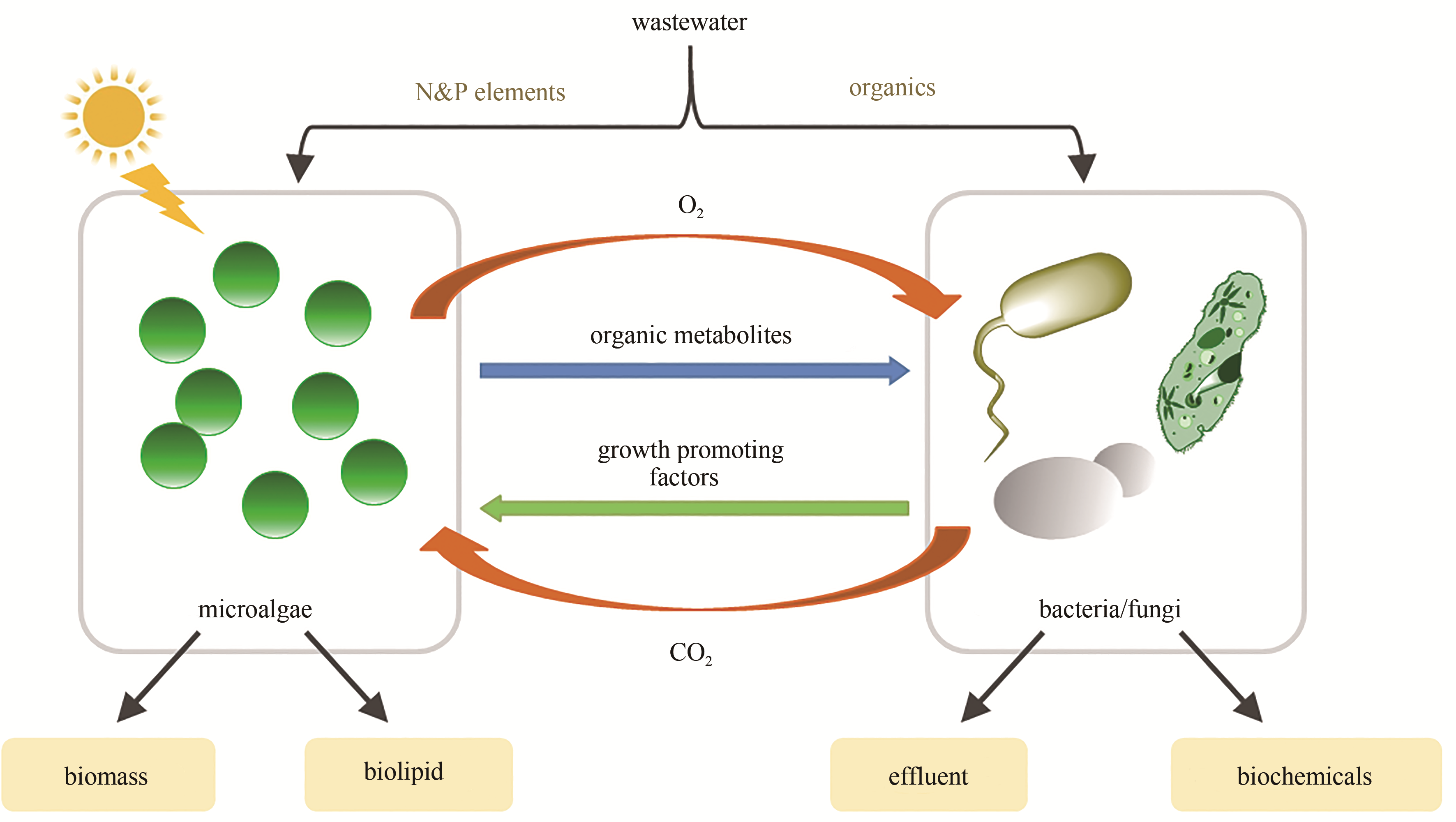
| 14 | SHOU W, RAM S, VILAR JMG. Synthetic cooperation in engineered yeast populations [J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104 (6) :1877-1882. |
| 15 | SCHROECKH V, SCHERLACH K, H-W NÜTZMANN, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009,106:14558-63. |
| 16 | MINTY JJ, SINGER ME, SCHOLZ SA, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110:14592-7. |
| 17 | PLOS ONE STAFF THE. Correction: applications and comparisons of four time series models in epidemiological surveillance data [J]. PLoS ONE, 2014, 9(2):e91629. |
| 18 | LLOYD-PRICE J, ABU-ALI G, HUTTENHOWER C. The healthy human microbiome [J]. Genome Medicine, 2016,8:51. |
| 19 | JONES JA, VERNACCHIO VR, COLLINS SM, et al. Complete biosynthesis of anthocyanins using E. coli polycultures [J]. MBio. 2017;8: e00621-17. |
| 20 | SHAHAB RL, LUTERBACHER JS, BRETHAUER S, et al. Consolidated bioprocessing of lignocellulosic biomass to lactic acid by a synthetic fungal-bacterial consortium [J]. Biotechnology and Bioengineering, 2018, 115. |
| 21 | JIAO F, XU B: Electrochemical ammonia synthesis and ammonia fuel cells [J]. Advanced materials, 2019, 31:1970221. |
| 22 | NETZKER T, FLAK M, KRESPACH M K, et al. Microbial interactions trigger the production of antibiotics [J]. Current Opinion in Microbiology, 2018, 45: 117-123. |
| 23 | TURNBAUGH P J, LEY R E, HAMADY M, et al. The human microbiome project [J]. Nature, 2007, 449: 804-810. |
| 24 | ZHOU K, QIAO K, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products [J]. Nature Biotechnology, 2015, 33: 377. |
| 25 | HAYS S G, PATRICK W G, ZIESACK M, et al. Better together: engineering and application of microbial symbioses [J]. Current Opinion in Biotechnology, 2015, 36: 40-49. |
| 26 | MCCARTY N S, LEDESMA-AMARO R. Synthetic biology tools to engineer microbial communities for biotechnology [J]. Trends in Biotechnology, 2019, 37: 181-197. |
| 27 | WANG J, LIN W, WRAY V, et al. Induced production of depsipeptides by co-culturing Fusarium tricinctum and Fusarium begoniae [J]. Tetrahedron Letters, 2013, 54: 2492-2496. |
| 28 | CHARUSANTI P, FONG N L, NAGARAJAN H, et al. Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction [J]. PLoS One, 2012, 7: e33727. |
| 29 | MARMANN A, ALY A, LIN Wenhan, et al. Co-cultivation-a powerful emerging tool for enhancing the chemical diversity of microorganisms [J]. Marine Drugs, 2014, 12: 1043-1065. |
| 30 | SPOHN M, KIRCHNER N, KULIK A, et al. Overproduction of ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17 [J]. Antimicrob Agents Chemother, 2014, 58: 6185-6196. |
| 31 | BERTRAND S, BOHNI N, SCHNEE S, et al. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery [J]. Biotechnology Advances, 2014, 32: 1180-1204. |
| 32 | CHAGAS F O, DIAS L G, PUPO M T. A mixed culture of endophytic fungi increases production of antifungal polyketides [J]. Journal of Chemical Ecology, 2013, 39: 1335-1342. |
| 33 | KÖNIG CC, SCHERLACH K, SCHROECKH V, et al. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus [J]. ChemBioChem, 2013, 14: 938-942. |
| 34 | RATEB M E, HALLYBURTON I, HOUSSEN W E, et al. Induction of diverse secondary metabolites in Aspergillus fumigatus by microbial co-culture [J]. RSC Advances, 2013, 3: 14444-14450. |
| 35 | NONAKA K, ABE T, IWATSUKI M, et al. Enhancement of metabolites productivity of Penicillium pinophilum FKI-5653, by co-culture with Trichoderma harzianum FKI-5655 [J]. The Journal of Antibiotics, 2011, 64: 769. |
| 36 | ZUCK KM, SHIPLEY S, NEWMAN DJ. Induced production of N-formyl alkaloids from Aspergillus fumigatus by co-culture with Streptomyces peucetius [J]. Journal of Natural Products, 2011, 74: 1653-1657. |
| 37 | TACCONELLI E, CARRARA E, SAVOLDI A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis [J]. The Lancet Infectious Diseases, 2018, 18(3): 318-327. |
| 38 | GILBERT J A, BLASER M J, CAPORASO J G, et al. Current understanding of the human microbiome [J]. Nature Medicine, 2013, 24: 392-400. |
| 39 | LLOYD-PRICE J, ABU-ALI G, HUTTENHOWER C. The healthy human microbiome [J]. Genome Medicine, 2016, 8: 51. |
| 40 | CHEN L, GARMAEVA S, ZHERANKOVA A, et al. A system biology perspective on environment-host-microbe interactions [J]. Human Molecular Genetics, 2018, 27(R2): R187-R194. |
| 41 | AJIKUMAR PK, XIAO W, KE TYO, et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli [J]. Science, 2010, 330: 70-74. |
| 42 | ZHANG H, PEREIRA B, LI Z, et al. Engineering Escherichia coli coculture systems for the production of biochemical products [J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112: 8266-8271. |
| 43 | SENGUPTA S, JONNALAGADDA S, GOONEWARDENA L, et al. Metabolic engineering of a novel muconic acid biosynthesis pathway via 4-hydroxybenzoic acid in Escherichia coli [J]. Applied & Environmental Microbiology, 2015, 81(23): 8037-8043. |
| 44 | WANG S, BILAL M, HU H, et al. 4-Hydroxybenzoic acid-a versatile platform intermediate for value-added compounds [J]. Applied Microbiology & Biotechnology, 2018, 102(8): 3561-3571. |
| 45 | WANG J, LU X, YING H, et al. A novel process for cadaverine bio-production using a consortium of two engineered Escherichia coli [J]. Frontiers in Microbiology, 2018, 9: 1312. |
| 46 | LIU X, LI X, JIANG J, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides [J]. Metabolic Engineering, 2018, 47: 243-253. |
| 47 | ZHANG H, LI Z, PEREIRA B, et al. Engineering E. coli-E. coli cocultures for production of muconic acid from glycerol [J]. Microbial Cell Factories, 2015, 14: 134. |
| 48 | AKDEMIR H, SILVA A, ZHA J, et al. Production of pyranoanthocyanins using Escherichia coli co-cultures [J]. Metabolic Engineering, 2019, 55: 290-298. |
| 49 | LI T, ZHOU W, BI H, et al. Production of caffeoylmalic acid from glucose in engineered Escherichia coli [J]. Biotechnology Letters, 2018, 40: 1057-1065. |
| 50 | NIU F, HE X, WU Y, et al. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering [J]. Frontiers in Microbiology, 2018, 9. |
| 1 | HALL G M, HOWE J. The impact of synthetic biology in chemical engineering-educational issues [J]. Education for Chemical Engineers, 2012, 7: e51-e55. |
| 2 | BHATIA S K, BHATIA R K, CHOI Yong-Keun, et al. Biotechnological potential of microbial consortia and future perspectives [J]. Critical Reviews in Biotechnology, 2018, 38: 1209-1229. |
| 3 | SHONG J, DIAZ M R J, COLLINS C H. Towards synthetic microbial consortia for bioprocessing [J]. Current Opinion in Biotechnology, 2012, 23: 798-802. |
| 4 | KAEBERLEIN T, LEWIS K, EPSTEIN S S. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment [J]. Science, 2002, 296: 1127-1129. |
| 5 | HANEMAAIJER M, RÖLING W F M, OLIVIER B G, ET AL. Systems modeling approaches for microbial community studies: from metagenomics to inference of the community structure [J]. Frontiers in Microbiology, 2013, 6: 213. |
| 6 | WANG Xin, SU Rui, CHEN Kequan, et al. Engineering a microbial consortium based whole-cell system for efficient production of glutarate from L-lysine [J]. Frontiers in Microbiology, 2019, 10: 341. |
| 7 | ZHANG H, WANG X. Modular co-culture engineering, a new approach for metabolic engineering [J]. Metabolic Engineering, 2016, 37: 114-121. |
| 8 | WANG E, LIU Y, MA Q, et al. Synthetic cell-cell communication in a three-species consortium for one-step vitamin C fermentation [J]. Biotechnology Letters, 2019, 41(8/9): 951-961. |
| 9 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor [J]. Microbial Cell Factories, 2019, 18: 35. |
| 10 | LI F, AN X, WU D, et al. Engineering microbial consortia for high-performance cellulosic hydrolyzates-fed microbial fuel cells [J]. Frontiers in Microbiology, 2019, 10: 409. |
| 11 | SONG H, DING M, JIA X, et al. Synthetic microbial consortia: from systematic analysis to construction and applications [J]. Chemical Society Reviews, 2014, 43: 6954-6981. |
| 51 | CAMACHO-ZARAGOZA J M, HERNÁNDEZ-CHÁVEZ G, MORENO-AVITIA F, et al. Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol [J]. Microbial Cell Factories, 2016, 15: 163. |
| 52 | WILLRODT C, HOSCHEK A, BÜHLER B, et al. Coupling limonene formation and oxyfunctionalization by mixed‐culture resting cell fermentation [J]. Biotechnology and Bioengineering, 2015, 112: 1738-1750. |
| 53 | ZHANG H, STEPHANOPOULOS G. Co‐culture engineering for microbial biosynthesis of 3‐amino‐benzoic acid in Escherichia coli [J]. Biotechnology Journal, 2016, 11: 981-987. |
| 54 | JONES J A, VERNACCHIO V R, SINKOE A L, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids [J]. Metabolic Engineering, 2016, 35: 55-63. |
| 55 | AHMADI M K, FANG Lei, MOSCATELLO N, et al. E. coli metabolic engineering for gram scale production of a plant-based anti-inflammatory agent [J]. Metabolic Engineering, 2016, 38: 382-388. |
| 56 | GANESAN V, LI Z, WANG X, et al. Heterologous biosynthesis of natural product naringenin by co-culture engineering [J]. Synthetic and Systems Biotechnology, 2017, 2: 236-242. |
| 57 | CHEN Z, SUN X, LI Ye, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols [J]. Metabolic Engineering, 2017, 39: 102-109. |
| 58 | THUAN N H, CHAUDHARY A K, CUONG D VAN, et al. Engineering co-culture system for production of apigetrin in Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45: 175-185. |
| 59 | THUAN N H, TRUNG N T, CUONG N X, et al. Escherichia coli modular coculture system for resveratrol glucosides production [J]. World Journal of Microbiology and Biotechnology, 2018, 34: 75. |
| 60 | FANG Z, JONES J A, ZHOU J, et al. Engineering Escherichia coli co‐cultures for production of curcuminoids from glucose [J]. Biotechnology Journal, 2018, 13: 1700576. |
| 61 | ZHOU YY, LI ZH, WANG XN, et al. Establishing microbial co‐cultures for 3‐hydroxybenzoic acid biosynthesis on glycerol [J]. Engineering in Life Sciences, 2019, 19: 389-395. |
| 62 | WANG EX, DING MZ, MA Q, et al. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation [J]. Microbial Cell Factories, 2016, 15: 21. |
| 63 | MARTÍNEZ I, E-S MOHAMED M, ROZAS D, et al. Engineering synthetic bacterial consortia for enhanced desulfurization and revalorization of oil sulfur compounds [J]. Metabolic Engineering, 2016, 35: 46-54. |
| 64 | LIU Y, DING MZ, LING W, et al. A three-species microbial consortium for power generation [J]. Energy & Environmental Science, 2017, 10: 1600-1609. |
| 65 | JONES JA, VERNACCHIO VR, COLLINS SM, et al. Complete biosynthesis of anthocyanins using E. coli polycultures [J]. mBio, 2017, 8(3): 28588129. |
| 66 | LI ZH, WANG XN, ZHANG HR. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering [J]. Energy & Environmental Science, 2019, 54: 1-11. |
| 67 | LIU YQ, TU XH, XU Q, et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol [J]. Metabolic Engineering, 2018, 45: 189-199. |
| 68 | ZHANG W, LIU H, LI X, et al. Production of naringenin from D‐xylose with co‐culture of E. coli and S. cerevisiae [J]. Engineering in Life Sciences, 2017, 17: 1021-1029. |
| 69 | CAMPBELL CD, VEDERAS JC. Biosynthesis of lovastatin and related metabolites formed by fungal iterative PKS enzymes [J]. Biopolymers, 2010, 93: 755-763. |
| 70 | RODRÍGUEZ-BUSTAMANTE E, MALDONADO-ROBLEDO G, ORTIZ M A, et al. Bioconversion of lutein using a microbial mixture-maximizing the production of tobacco aroma compounds by manipulation of culture medium [J]. Applied Microbiology and Biotechnology, 2005, 68: 174-182. |
| 71 | KLEIN-MARCUSCHAMER D, OLESKOWICZ-POPIEL P, SIMMONS B A, et al. The challenge of enzyme cost in the production of lignocellulosic biofuels [J]. Biotechnology and Bioengineering, 2012, 109: 1083-1087. |
| 72 | OLSON DG, MCBRIDE JE, SHAW AJ, et al. Recent progress in consolidated bioprocessing [J]. Current Opinion in Biotechnology, 2012, 23: 396-405. |
| 73 | XIN FX, CHEN TP, JIANG YJ, et al. Strategies for improved isopropanol-butanol production by a Clostridium strain from glucose and hemicellulose through consolidated bioprocessing [J]. Biotechnology for Biofuels, 2017, 10: 118. |
| 74 | Y-S JANG, LEE JY, LEE JM, et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum [J]. mBio, 2012, 3(5): e00314-12. |
| 75 | YANG XR, XU MM, YANG ST. Metabolic and process engineering of Clostridium cellulovorans for biofuel production from cellulose [J]. Metabolic Engineering, 2015, 32: 39-48. |
| 76 | ZHANG XZ, SATHITSUKSANOH N, ZHU ZG, et al. One-step production of lactate from cellulose as the sole carbon source without any other organic nutrient by recombinant cellulolytic Bacillus subtilis [J]. Metabolic Engineering, 2011, 13: 364-372. |
| 77 | EDWARDS MC, HENRIKSEN ED, YOMANO LP, et al. Addition of genes for cellobiase and pectinolytic activity in Escherichia coli for fuel ethanol production from pectin-rich lignocellulosic biomass [J]. Applied and Environmental Microbiology, 2011, 77(15): 5184-5191. |
| 78 | FAVARO L, VIKTOR MJ, ROSE SH, et al. Consolidated bioprocessing of starchy substrates into ethanol by industrial Saccharomyces cerevisiae strains secreting fungal amylases [J]. Biotechnology and Bioengineering, 2015, 112: 1751-1760. |
| 79 | HASUNUMA T, KONDO A. Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering [J]. Biotechnology Advances, 2012, 30: 1207-1218. |
| 80 | VECCHIO DD, QIAN YL, MURRAY RM, et al. Future systems and control research in synthetic biology [J]. Annual Reviews in Control, 2018, 45: 5-17. |
| 81 | VAN ZYL W H, HAAN R DEN, GRANGE DC LA. Developing cellulolytic organisms for consolidated bioprocessing of lignocellulosics [M]// GUPTA V K, TUOHY M G. Biofuel technologies. Berlin, Heidelberg: Springer, 2013: 189-220. |
| 82 | HAAN R DEN, RENSBURG E VAN, ROSE S H, et al. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing [J]. Current Opinion in Biotechnology, 2015, 33: 32-38. |
| 83 | GUO ZP, JULIEN R, SOPHIE D, et al. Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose [J]. Biotechnology for Biofuels, 2018, 11: 141. |
| 84 | SINGH N, MATHUR AS, GUPTA RP, et al. Enhanced cellulosic ethanol production via consolidated bioprocessing by Clostridium thermocellum ATCC 31924 [J]. Bioresource Technology, 2018, 250: 860-867. |
| 85 | SHAHAB R L, LUTERBACHER J S, BRETHAUER S, et al. Consolidated bioprocessing of lignocellulosic biomass to lactic acid by a synthetic fungal‐bacterial consortium [J]. Biotechnology and Bioengineering, 2018, 115: 1207-1215. |
| 86 | BUZZINI P. Batch and fed‐batch carotenoid production by Rhodotorula glutinis-Debaryomyces castellii co‐cultures in corn syrup [J]. Journal of Applied Microbiology, 2001, 90: 843-847. |
| 87 | BAYER TS, WIDMAIER DM, TEMME K, et al. Synthesis of methyl halides from biomass using engineered microbes [J]. Journal of the American Chemical Society, 2009, 131: 6508-6515. |
| 88 | SGOBBA E, STUMPF AK, VORTMANN M, et al. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for L-lysine production from starch and sucrose [J]. Bioresource Technology, 2018, 260: 302-310. |
| 89 | PATLE S, LAL B. Ethanol production from hydrolysed agricultural wastes using mixed culture of Zymomonas mobilis and Candida tropicalis [J]. Biotechnology Letters, 2007, 29: 1839-1843. |
| 90 | BRETHAUER S, STUDER MH. Consolidated bioprocessing of lignocellulose by a microbial consortium [J]. Energy & Environmental Science, 2014, 7: 1446-1453. |
| 91 | MINTY JJ, SINGER ME, SCHOLZ SA, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110: 14592-14597. |
| 92 | SURIYACHAI N, WEERASAIA K, LAOSIRIPOJANA N, et al. Optimized simultaneous saccharification and co-fermentation of rice straw for ethanol production by Saccharomyces cerevisiae and Scheffersomyces stipitis co-culture using design of experiments [J]. Bioresource Technology, 2013, 142: 171-178. |
| 93 | ZUROFF T R, XIQUES S B, CURTIS W R. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture [J]. Biotechnology for Biofuels, 2013, 6: 59. |
| 94 | VALDEZ-VAZQUEZ I, PÉREZ-RANGEL M, TAPIA A, et al. Hydrogen and butanol production from native wheat straw by synthetic microbial consortia integrated by species of Enterococcus and Clostridium [J]. Fuel, 2015, 159: 214-222. |
| 95 | PAPONE T, KOOKHUNTHOD S, PAUNGBUT M, et al. Producing of microbial oil by mixed culture of microalgae and oleaginous yeast using sugarcane molasses as carbon substrate [J]. Journal of Clean Energy Technologies, 2016, 4: 253-256. |
| 96 | IKE A, MURAKAWA T, KAWAGUCHI H, et al. Photoproduction of hydrogen from raw starch using a halophilic bacterial community [J]. Journal of Bioscience and Bioengineering, 1999, 88: 72-77. |
| 97 | PACHAPUR VL, SARMA SJ, BRAR SK, et al. Co‐culture strategies for increased biohydrogen production [J]. International Journal of Energy Research, 2015, 39: 1479-1504. |
| 98 | MASSET J, CALUSINSKA M, HAMILTON C, et al. Fermentative hydrogen production from glucose and starch using pure strains and artificial co-cultures of Clostridium spp . [J]. Biotechnology for Biofuels, 2012, 5: 35. |
| 99 | KUMAR R, SINGH S, SINGH O V. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives [J]. Journal of Industrial Microbiology & Biotechnology, 2008, 35: 377-391. |
| 100 | GUPTE A, MADAMWAR D. Solid state fermentation of lignocellulosic waste for cellulase and β‐Glucosidase production by cocultivation of Aspergillus ellipticus and Aspergillus fumigatus [J]. Biotechnology Progress, 1997, 13: 166-169. |
| 101 | VERMA P, MADAMWAR D. Production of ligninolytic enzymes for dye decolorization by cocultivation of white-rot fungi Pleurotus ostreatus and Phanerochaete chrysosporium under solid-state fermentation [J]. Applied Biochemistry and Biotechnology, 2002, 102: 109-118. |
| 102 | HU HL, BRINK J VAN DEN, GRUBEN B S, et al. Improved enzyme production by co-cultivation of Aspergillus niger and Aspergillus oryzae and with other fungi [J]. International Biodeterioration & Biodegradation, 2011, 65: 248-252. |
| 103 | AHMED N, THOMPSON S, GLASER M. Global aquaculture productivity, environmental sustainability, and climate change adaptability [J]. Environmental Management, 2019, 63: 159-172. |
| 104 | CAMPBELL-LENDRUM D, PRÜSS-USTÜN A. Climate change, air pollution and noncommunicable diseases [J]. Bulletin of the World Health Organization, 2019, 97: 160. |
| 105 | WALKER D, BAUMGARTNER D, GERBA C, et al. Surface water pollution [M]// BRUSSEAU M L, PEPPER I L, GERBA C P. Environmental and pollution science. 3rd ed. Cambridge, Massachusetts:Elsevier, 2019: 261-292. |
| 106 | AZUBUIKE C C, CHIKERE C B, OKPOKWASILI G C. Bioremediation techniques-classification based on site of application: principles, advantages, limitations and prospects [J]. World Journal of Microbiology and Biotechnology, 2016, 32: 180. |
| 107 | VILLEGAS L B, MARTÍNEZ M A, RODRÍGUEZ A, et al. consortia Microbial, a viable alternative for cleanup of contaminated soils [M]// ALVAREZ A, POLTI M A. Bioremediation in Latin America. Berlin, Germany:Springer, 2014: 135-148. |
| 108 | MUJTABA G, LEE K. Advanced treatment of wastewater using symbiotic co-culture of microalgae and bacteria [J]. Applied Chemistry for Engineering, 2016, 27(1): 1-9. |
| 109 | GONÇALVES A L, PIRES J C, SIMÕES M. A review on the use of microalgal consortia for wastewater treatment [J]. Algal Research, 2017, 24(B): 403-415. |
| 110 | K-J CHOI, HAN T H, YOO G, et al. Co-culture consortium of Scenedesmus dimorphus and nitrifiers enhances the removal of nitrogen and phosphorus from artificial wastewater [J]. KSCE Journal of Civil Engineering, 2018, 22: 3215-3221. |
| 111 | MUJTABA G, RIZWAN M, LEE K. Removal of nutrients and COD from wastewater using symbiotic co-culture of bacterium Pseudomonas putida and immobilized microalga Chlorella vulgaris [J]. Journal of Industrial and Engineering Chemistry, 2017, 49: 145-151. |
| 112 | REN HY, LIU BF, KONG FY, et al. Hydrogen and lipid production from starch wastewater by co-culture of anaerobic sludge and oleaginous microalgae with simultaneous COD, nitrogen and phosphorus removal [J]. Water Research, 2015, 85: 404-412. |
| 113 | ABINANDAN S, SUBASHCHANDRABOSE S R, VENKATESWARLU K, et al. Nutrient removal and biomass production: advances in microalgal biotechnology for wastewater treatment [J]. Critical Reviews in Biotechnology, 2018, 38: 1244-1260. |
| 114 | BORDEL S, GUIEYSSE B, MUNOZ R. Mechanistic model for the reclamation of industrial wastewaters using algal-bacterial photobioreactors [J]. Environmental Science & Technology, 2009, 43: 3200-3207. |
| 115 | BHATIA S K, BHATIA R K, YANG Y-H. An overview of microdiesel-a sustainable future source of renewable energy [J]. Renewable and Sustainable Energy Reviews, 2017, 79: 1078-1090. |
| 116 | WREDE D, TAHA M, MIRANDA A F, et al. Co-cultivation of fungal and microalgal cells as an efficient system for harvesting microalgal cells, lipid production and wastewater treatment [J]. PLoS One, 2014, 9: e113497. |
| 117 | STILES W A, STYLES D, CHAPMAN S P, et al. Using microalgae in the circular economy to valorise anaerobic digestate: challenges and opportunities [J]. Bioresource Technology, 2018, 267: 732-742. |
| 118 | OSUNDEKO O, ANSOLIA P, GUPTA S K, et al. Promises and challenges of growing microalgae in wastewater [M]// SINGH R P, KOLOK A S, BARTELT-HUNT S L. Water Conservation, Recycling and Reuse: Issues and Challenges. Singapore: Springer, 2019: 29-53. |
| 119 | SATHIYANARAYANAN G, BHATIA S K, KIM H J, et al. Metal removal and reduction potential of an exopolysaccharide produced by Arctic psychrotrophic bacterium Pseudomonas sp. PAMC 28620 [J]. RSC Advances, 2016, 6: 96870-96881. |
| 120 | SHAFIQUE M, JAWAID A, REHMAN Y. As(V) reduction, As (III) oxidation, and Cr (VI) reduction by multi-metal-resistant Bacillus subtilis, Bacillus safensis, and Bacillus cereus species isolated from wastewater treatment plant [J]. Geomicrobiology Journal, 2017, 34: 687-694. |
| 121 | KIPIGROCH K, JANOSZ-RAJCZYK M, WYKROTA L. Biosorption of heavy metals with the use of mixed algal population [J]. Archives of Environmental Protection, 2012, 38: 3-10. |
| 122 | ILAMATHI R, NIRMALA G, MURUGANANDAM L. Heavy metals biosorption in liquid solid fluidized bed by immobilized consortia in alginate beads [J]. International Journal of Chem Tech Research, 2014, 6: 652-662. |
| 123 | SENAN R C, ABRAHAM T E. Bioremediation of textile azo dyes by aerobic bacterial consortium aerobic degradation of selected azo dyes by bacterial consortium [J]. Biodegradation, 2004, 15: 275-280. |
| 124 | KHAN R, BHAWANA P, FULEKAR M. Microbial decolorization and degradation of synthetic dyes: a review [J]. Reviews in Environmental Science and Bio/Technology, 2013, 12: 75-97. |
| 125 | SOLÍS M, SOLÍS A, PÉREZ H I, al et, Microbial decolouration of azo dyes : a review [J]. Process Biochemistry, 2012, 47: 1723-1748. |
| 126 | SHANMUGAM B K, EASWARAN S N, LAKRA R, et al. Metabolic pathway and role of individual species in the bacterial consortium for biodegradation of azo dye: a biocalorimetric investigation [J]. Chemosphere, 2017, 188: 81-89. |
| 127 | CHAN G F, RASHID N A, KOAY L L, et al. Identification and optimization of novel NAR-1 bacterial consortium for the biodegradation of Orange II [J]. Insight Biotechnol, 2011, 1: 7-16. |
| 128 | SARATALE R, SARATALE G, KALYANI D, et al. Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR [J]. Bioresource Technology, 2009, 100: 2493-2500. |
| 129 | SARATALE R, SARATALE G, CHANG J, et al. Decolorization and biodegradation of reactive dyes and dye wastewater by a developed bacterial consortium [J]. Biodegradation, 2010, 21: 999-1015. |
| 130 | SENTHILVELAN T, KANAGARAJ J, PANDA R C, et al. Biodegradation of phenol by mixed microbial culture: an eco-friendly approach for the pollution reduction [J]. Clean Technologies and Environmental Policy, 2014, 16: 113-126. |
| 131 | XU GM, LI YY, ZHENG W, et al. Mineralization of chlorpyrifos by co-culture of Serratia and Trichosporon spp [J]. Biotechnology Letters, 2007, 29: 1469-1473. |
| 132 | JIN D F, HU H, LIU D F, et al. Optimization of a bacterial consortium for nitrobenzene degradation [J]. Water Science and Technology, 2012, 65: 795-801. |
| 133 | GONZÁLEZ N, SIMARRO R, MOLINA M, et al. Effect of surfactants on PAH biodegradation by a bacterial consortium and on the dynamics of the bacterial community during the process [J]. Bioresource Technology, 2011, 102: 9438-9446. |
| 134 | JOINT I, TAIT K, CALLOW ME, et al. Cell-to-cell communication across the prokaryote-eukaryote boundary [J]. Science, 2002, 298: 1207-1207. |
| 135 | W-L NG, BASSLER B L. Bacterial quorum-sensing network architectures [J]. Annual Review of Genetics, 2009, 43: 197-222. |
| 136 | BALAGADDÉ F K, SONG Hao, OZAKI J, et al. A synthetic Escherichia coli predator-prey ecosystem [J]. Molecular Systems Biology, 2008, 4. |
| 137 | BASU S, MEHREJA R, THIBERGE S, et al. Spatiotemporal control of gene expression with pulse-generating networks [J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101: 6355-6360. |
| 138 | YOU Lingchong, COX R S, WEISS R, et al. Programmed population control by cell-cell communication and regulated killing [J]. Nature, 2004, 428: 868-871. |
| 139 | THOENDEL M, HORSWILL A R. Biosynthesis of peptide signals in gram-positive bacteria [J]. Advances in Applied Microbiology, 2010, 71: 91-112. |
| 140 | MICHIE K L, CORNFORTH D M, WHITELEY M. Bacterial Tweets and Podcasts #signaling#eavesdropping#microbialfightclub [J]. Molecular & Biochemical Parasitology, 2016, 208(1): 41-48. |
| 141 | FISCHER J, MUELLER S Y, NETZKER T, et al. Fungal chromatin mapping identifies BasR, as the regulatory node of bacteria-induced fungal secondary metabolism [J/OL]. BioRxiv, 2018, 211979. |
| 142 | SCHROECKH V, SCHERLACH K, H-W NÜTZMANN, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106: 14558-14563. |
| 143 | TOYOFUKU M. Bacterial communication through membrane vesicles [J]. Bioscience, Biotechnology, and Biochemistry, 2019, 83: 1-7. |
| 144 | NÜTZMANN H W, REYES-DOMINGUEZ Y, SCHERLACH K, et al. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation [J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108: 14282-14287. |
| 145 | KUROSAWA K, GHIVIRIGA I, SAMBANDAN T, et al. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians [J]. Journal of the American Chemical Society, 2008, 130: 1126-1127. |
| 146 | SHAHAB R L, LUTERBACHER J S, BRETHAUER, et al. Labor division in engineered cross-kingdom consortia: consolidated bioprocessing of lignocellulosic biomass to carboxylic acids [D]. Lausanne, Switzerland:École Polytechnique Fédérale de Lausanne(EPFL), 2019. |
| 147 | WANG C, LI YZ, TAN H, et al. A novel microbe consortium, nano-visible light photocatalyst and microcapsule system to degrade PAHs [J]. Chemical Engineering Journal, 2019, 359: 1065-1074. |
| 148 | LINDEMANN S R, BERNSTEIN H C, SONG Hyun-Seob, et al l. Engineering microbial consortia for controllable outputs [J]. The ISME Journal, 2016, 10: 2077. |
| 149 | SHAHAB R L, BRETHAUER S, LUTERBACHER J S, et al. Engineering of ecological niches to create stable artificial consortia for complex biotransformations [J]. Current Opinion in Biotechnology, 2020, 62: 129-136. |
| 150 | AGAPAKIS C M, BOYLE P M, SILVER P A. Natural strategies for the spatial optimization of metabolism in synthetic biology [J]. Nature Chemical Biology, 2012, 8: 527. |
| 151 | JOHNS N I, BLAZEJEWSKI T, GOMES A L, et al. Principles for designing synthetic microbial communities [J]. Current Opinion in Microbiology, 2016, 31: 146-153. |
| 152 | MACLEOD F, GUIOT S, COSTERTON J. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor [J]. Applied & Environmental Microbiology, 1990, 56: 1598-1607. |
| 153 | DAS A A, BOVILL J, AYESH M, et al. Fabrication of living soft matter by symbiotic growth of unicellular microorganisms [J]. Journal of Materials Chemistry B, 2016, 4: 3685-3694. |
| 154 | AZEREDO J, AZEVEDO N F, BRIANDET R, et al. Critical review on biofilm methods [J]. Critical reviews in Microbiology, 2017, 43: 313-351. |
| 155 | H-C FLEMMING, WINGENDER J. The biofilm matrix [J]. Nature Reviews Microbiology, 2010, 8: 623. |
| 156 | WONDRACZEK L, POHNERT G, SCHACHER F H, et al. Artificial microbial arenas: materials for observing and manipulating microbial consortia [J]. Advanced Materials, 2019, 31(24): 1900284. |
| 157 | KIM H J, BOEDICKER J Q, CHOI Jang Wook, et al. Defined spatial structure stabilizes a synthetic multispecies bacterial community [J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105: 18188-18193. |
| 158 | CONNELL J L, RITSCHDORFF E T, WHITELEY M, et al. 3D printing of microscopic bacterial communities [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110: 18380-18385. |
| [1] | ZHU Huawei, LI Yin. Biophotovoltaics: an environmentally friendly technology for solar energy utilization [J]. Synthetic Biology Journal, 2023, 4(6): 1259-1280. |
| [2] | Mingzhu DING, Bingzhi LI, Ying WANG, Zexiong XIE, Duo LIU, Yingjin YUAN. Significant research progress in synthetic biology [J]. Synthetic Biology Journal, 2020, 1(1): 7-28. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||