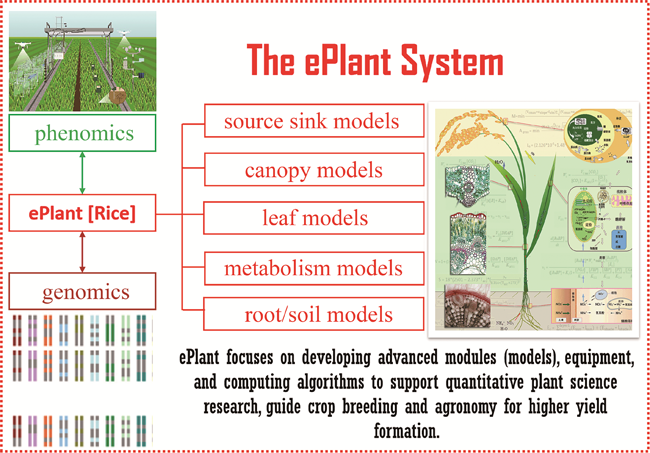
Plant systems and synthetic biology has gained more and more attention in recent years as a result of the rapid advances in genomics, proteomics, metabolomics, epigenomics and genome editing technologies. Plant systems and synthetic biology for quantitative studies of plant systems differs from traditional plant science that mainly focuses on descriptive and qualitative studies of plant systems. In addition, plant systems and synthetic biology also emphasizes the design and creation of new biochemical pathways, regulatory circuits, signaling pathways, and even biological structures that are not exist in native plants hosts for desired properties. A number of mega projects in plant synthetic biology have been initiated in recent years, all with a common goal of boosting crop yield and developing technologies for Green Revolution of agriculture. With these projects, it was evident that to maximize the benefit of plant synthetic biology, we ought to develop a robust system model for plant growth and development: ePlant, which includes the simulation not only for molecular, biochemical, physiological, and physical processes in different organs of a plant, but also for interaction between plant and environment as highlighted by the system of soil, root, plant and air. The ePlant model needs to be developed using a divide-and-conquer approach followed by the modular construction, and once developed, it can be used as a major tool for basic research in plant science, studies for interaction among genes, environment and management practices, identification of new options to engineer crops for desired properties, and finally design of ideotypes desired for crop breeding. To expedite the development of ePlant model, we propose a number of priorities and strategies, which include developing basic models for plant growth and development, collecting systems data related to partitioning of similarities among different organs, creating new methods to integrate modules at different temporal and spatial scales, building up public online platforms to support the development of ePlant, developing algorithms to support effective integration of ePlant models with phenomics data, and finally forming policies to nurture the development of communities on plant systems modeling. We emphasize a strong demand for an online platform or portal to support the development of ePlant models and their applications. This platform needs to include not only the basic models, data supporting model development and application and high-performance super-computing resources to enable the models' widely applications by the plant science research community. We also advocate for using rice as the model species to construct such an ePlant, considering the large quantities of studies on rice genomics, genetics, physiology, breeding and agronomics, which will expedite the development, validation and application of ePlant models. After ePlant model developed for rice (eRice), we can use it to guide dissection of mechanism underlying high yield formation in current rice lines, to design ideotypes for super-high-yield rice breeding, and to format optimal agronomic practices.