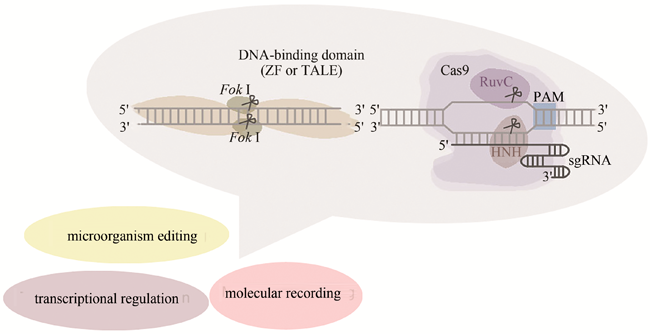
A vast amount of genetic information has been garnered with the rapid development of sequencing technology over the past decades. To decipher this information, genome editing tools that could functionally perturb specific genomic sequences have been developed. By recognizing DNA sequence, ZFN and TALEN emerged early as primitive genome editing approaches. Recently, the CRISPR/Cas9 system has become the most widely used genome editing tool due to its convenience of assembly and high efficiency. As a novel interdisciplinary field, synthetic biology has been developed through integrating engineering principles and biological fundamentals with the help from biotechnology tools. With the aim of improving our ability to decode and reprogram biological systems, synthetic biology has a tremendous demand for DNA synthesis, assembly and editing. With genome editing tools, synthetic biology promises innovations in the fields of biology, medicine, chemistry, agriculture, energy, environment, etc. In this review, we briefly describe the early genome editing tools: ZFN and TALEN. Furthermore, we comprehensively introduce the discovery, principle, development, optimization, derivative tools and applications of CRISPR/Cas9 system. Especially, we review the applications of these genome editing tools in synthetic biology from three aspects. The first is the application of genome editing tools in transcriptional regulation, such as precise gene expression regulation in dynamic biological process; the second is the application in engineering microbial strains, such as boosting the yield of antibiotic drugs and discovering potential resource for active natural products by manipulating specific gene or synthetic pathway; the last is their application in molecular recording, such as various approaches to record occurrence of detected signal or dynamic transcriptional information and to complete lineage tracing in live cells. Finally, we discuss current challenages and potential improvements of genome editing tools and envisage the future development of genome editing technology in synthetic biology.