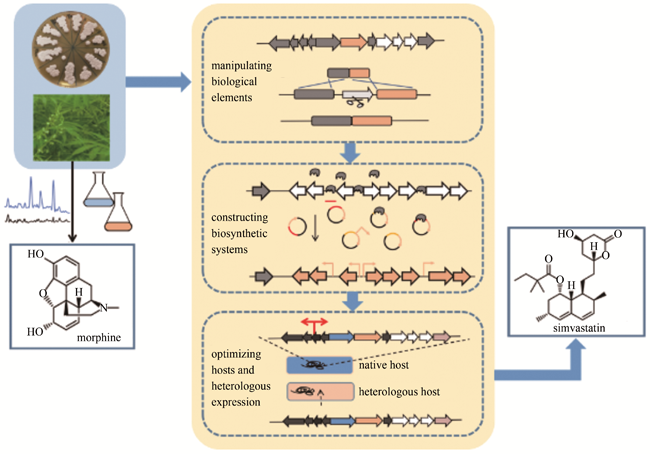
As an important source of clinical medicine and drug candidates, natural products originated from microorganisms and plants have a variety of biological activities, such as anti-infection, anticancer, immunosuppression and others. However, the physiochemical properties of natural products are usually not favorable for drug discovery and development, which has seriously limited the development of natural products for clinical applications. These hurdles include low aqueous solubility, lower potency, complexed structural analogs, and limited availability. Because all drugs should possess certain degree of aqueous solubility, the inherited low aqueous solubility of natural products markedly limits their druggability. Meanwhile, the efficacy of natural products is generally low, which requires significant improvements for therapeutic usefulness. Furthermore, the accumulation of various structural analogs of natural products leads to the difficulty of quality control for desired natural products. Moreover, in many cases, natural products are easily obtained or accessed for preclinical and clinical evaluations and subsequent clinical supply. Nevertheless, traditional strategy for natural product isolation has resulted in highly repeated rediscovery and the waste of time and resources, failing to deliver valuable bioactive leads and drug candidates. Recently, synthetic biology has become an emerging and valuable tool to address these limitations. Through the combination of genetic engineering, metabolic engineering, bioinformatics, systems biology, synthetic organic chemistry and computational biology, synthetic biology has been explored to improve various properties of natural products.In this review, we focus on the major factors that hinder the druggability of natural products and briefly summarize the progress made by approaches of synthetic biology in recent years. Based on structure-activity analysis, the structures of natural products can be modified or optimized by enzymes with different functions to produce favorable derivatives. Meanwhile, the manipulation of the synthetic and regulatory elements, the construction of a series of modules and the optimization of metabolic fluxes can significantly promote the production of natural product derived molecules. Moreover, de novo design of biosynthetic pathways under artificial regulation of the transcription and metabolism in coupling with suitable hosts and heterologous expression can further expand the biosynthetic potential towards natural products for their druggability. Given the diversity of structure and activity, natural products will continue to be an important source of bioactive compounds and new drugs in the future. With the rapid and prosperous development of synthetic biology technologies, together with the assistance of pharmaceutical sciences and computational technologies, a new era of natural product discovery and engineering can be foreseen.