Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (5): 583-592.DOI: 10.12211/2096-8280.2020-019
• Invited Review • Previous Articles Next Articles
Synthetic biology approaches to improve druggability of natural products
- Laboratory of Chemical Biology,School of Life Science and Technology,China Pharmaceutical University,Nanjing 211198,Jiangsu,China
-
Received:2020-03-08Revised:2020-09-23Online:2020-12-03Published:2020-10-31 -
Contact:CHEN Yijun
天然产物成药性的合成生物学改良
- 中国药科大学生命科学与技术学院化学生物学研究室,江苏 南京 211198
-
通讯作者:陈依军 -
作者简介:作者简介:王清(1994—),女,硕士研究生。研究方向:化学生物学。E-mail:499836857@qq.com
陈依军(1962—),男,教授,主要从事药物合成生物学研究。E-mail:yjchen@cpu.edu.cn -
基金资助:“重大新药创制”国家科技重大专项(2019ZX09721001-004-001);国家自然科学基金(21778076)
CLC Number:
Cite this article
WANG Qing, CHEN Yijun. Synthetic biology approaches to improve druggability of natural products[J]. Synthetic Biology Journal, 2020, 1(5): 583-592.
王清, 陈依军. 天然产物成药性的合成生物学改良[J]. 合成生物学, 2020, 1(5): 583-592.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-019
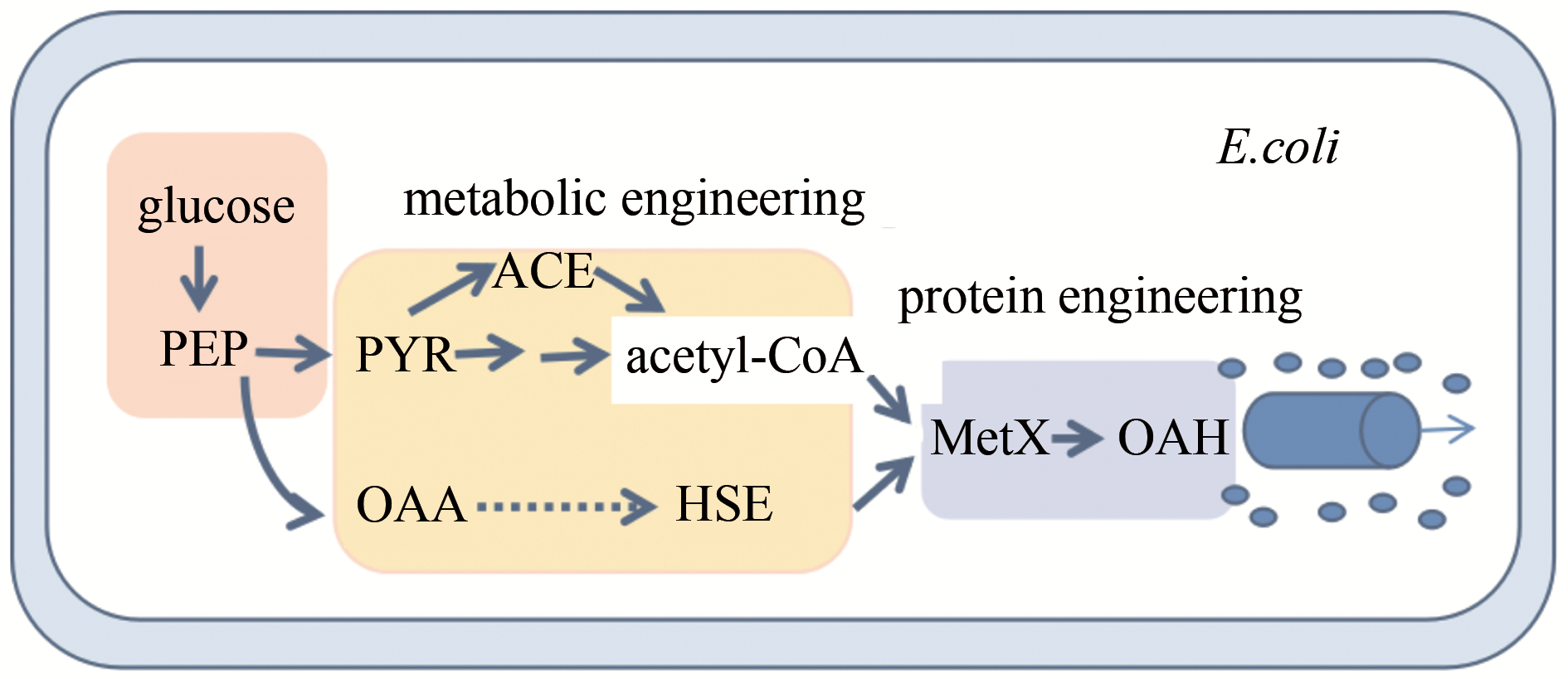
Fig.4 Synthetic biology strategy for high level production of O-acetylhomoserine in Escherichia coli PEP—phosphoenolpyruvate; PYR—pyruvate; OAA—oxaloacetate; ACE—acetate; HSE—L-homoserine; MetX—acetyltransferase
| 1 | DU Lin, ROBLES A J, KING J B, et al. Crowdsourcing natural products discovery to access uncharted dimensions of fungal metabolite diversity [J]. Angewandte Chemie International Edition, 2014, 53(3): 804-809. |
| 2 | BAUER A, BRÖNSTRUP M. Industrial natural product chemistry for drug discovery and development [J]. Natural Product Reports, 2014, 31(1): 35-60. |
| 3 | BROWN D G, LISTER T, MAY-DRACKA T L. New natural products as new leads for antibacterial drug discovery [J]. Bioorganic & Medicinal Chemistry Letters, 2014, 24(2): 413-418. |
| 4 | SCHEEPSTRA M, NIETO L, HIRSCH A K, et al. A natural-product switch for a dynamic protein interface [J]. Angewandte Chemie International Edition, 2014, 53(25): 6443-6448. |
| 5 | HARVEY A L, EDRADA-EBEL R A, QUINN R J. The re-emergence of natural products for drug discovery in the genomics era [J]. Nature Reviews Drug Discovery, 2015, 14(2): 111-129. |
| 6 | ZIMMERMANN T J, ROY S, MARTINEZ N E, et al. Biology-oriented synthesis of a tetrahydroisoquinoline-based compound collection targeting microtubule polymerization [J]. ChemBioChem, 2013, 14(3): 295-300. |
| 7 | LOWE D B. Drug discovery: combichem all over again [J]. Nature Chemistry, 2014, 6(10): 851-852. |
| 8 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the 30 years from 1981 to 2010 [J]. Journal of Natural Products, 2012, 75(3): 311-335. |
| 9 | RODRIGUES T, REKER D, SCHNEIDER P, et al. Counting on natural products for drug design [J]. Nature Chemistry, 2016, 8(6): 531-534. |
| 10 | DEITERS A, CROPP T A, SUMMERER D, et al. Site-specific PEGylation of proteins containing unnatural amino acids [J]. Bioorganic & Medicinal Chemistry Letters, 2004, 14(23): 5743-5745. |
| 11 | NEUMANN H, WANG Kaihang, DAVIS L, et al. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome [J]. Nature, 2010, 464(7287): 441-444. |
| 12 | BALTZ R H. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways [J]. ACS Synthetic Biology, 2014, 3(10): 748-758. |
| 13 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age [J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 14 | BREITLING R, TAKANO E. Synthetic biology advances for pharmaceutical production [J]. Current Opinion in Biotechnology, 2015, 35: 46-51. |
| 15 | TANG Xiaolong, DAI Hong, ZHU Yongxiang, et al. Maytansine-loaded star-shaped folate-core PLA-TPGS nanoparticles enhancing anticancer activity [J]. American Journal of Translational Research, 2014, 6(5): 528-537. |
| 16 | JARAPRAKASH NG, SUROLIA A. Role of glycosylation in nucleating protein folding and stability [J]. Biochemical Journal, 2017, 474(14): 2333-2347. |
| 17 | JI Shuai, LIANG Wenfei, LI Ziwei, et al. Effcient and selective glucosylation of prenylated phenolic compounds by Mucor hiemalis [J]. RSC Advances, 2016, 6(25): 20791-20799. |
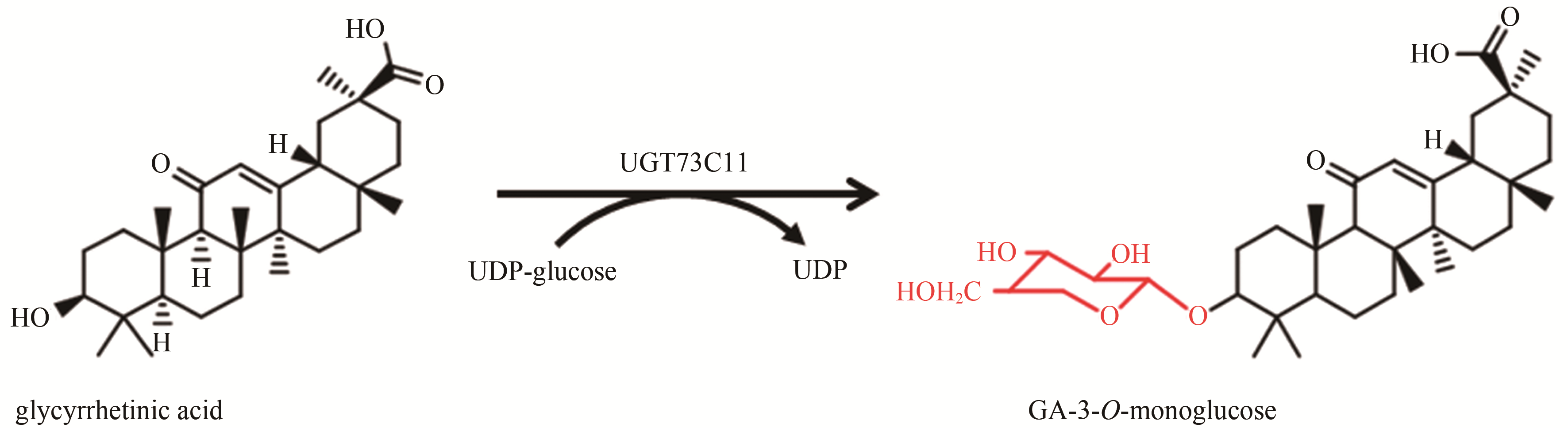
| 18 | LIU Xiaochen, ZHANG Liang. Biosynthesis of glycyrrhetinic acid-3-O-monoglucose using glycosyltransferase UGT73C11 from Barbarea vulgaris [J]. Industrial & Engineering Chemistry Research, 2017, 56(52): 14949-14958. |
| 19 | LIANG Wenfei, LI Ziwei, JI Shuai, et al. Microbial glycosylation of tanshinone IIA by Cunninghamella elegans AS 3.2028 [J]. RSC Advances, 2015, 5(78): 63753-63756. |
| 20 | HARMS J M, WILSON D N, SCHLUENZEN F, et al. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin [J]. Molecular Cell, 2008, 30(1): 26-38. |
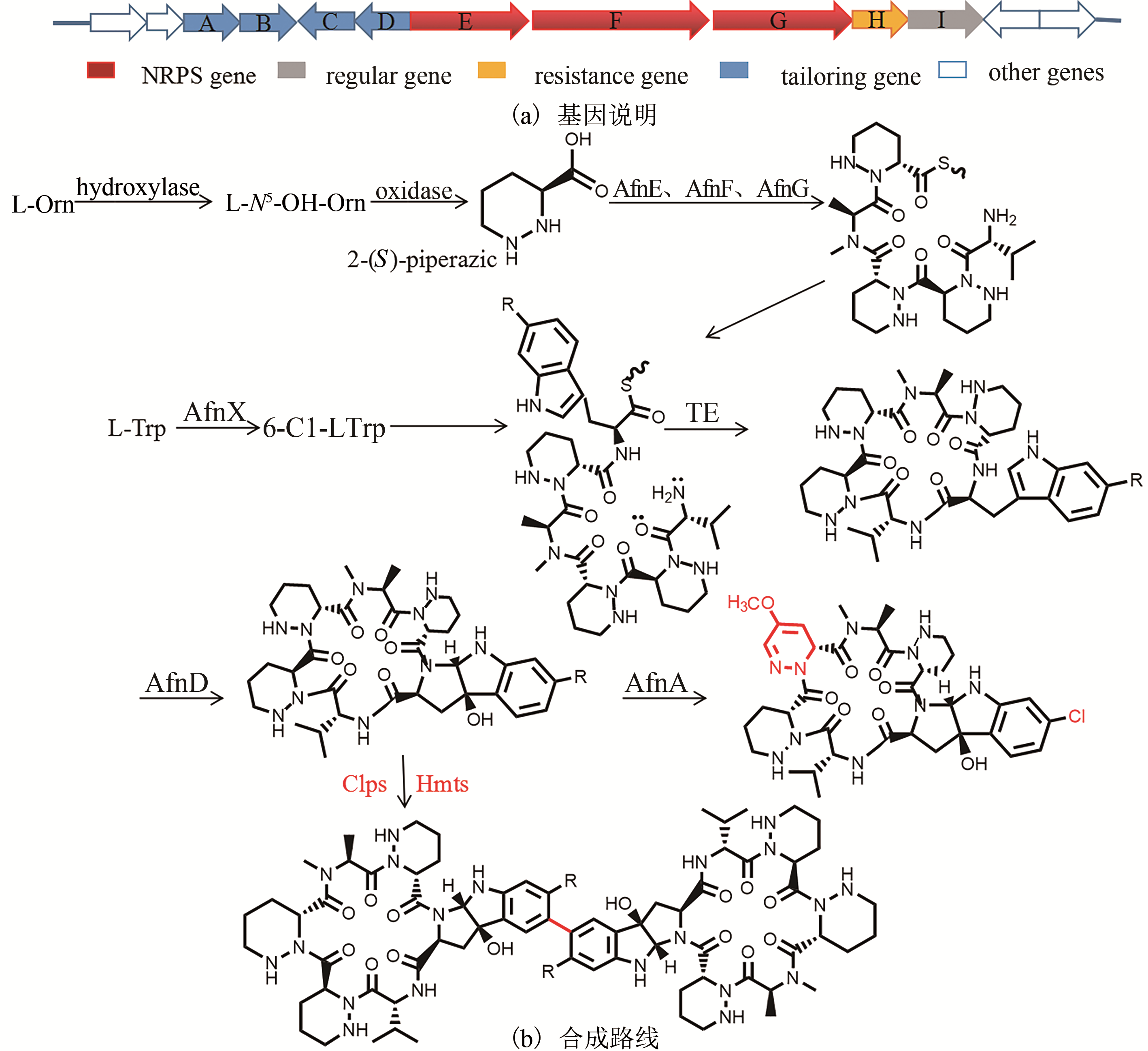
| 21 | ZHENG Qingfei, WANG Shoufeng, LIAO Rijing, et al. Precursor-directed mutational biosynthesis facilitates the functional assignment of two cytochromes P450 in thiostrepton biosynthesis [J]. ACS Chemical Biology, 2016, 11: 2673–2678. |
| 22 | WANG Shoufeng, ZHENG Qingfei, WANG Jianfeng, et al. Target-oriented design and biosynthesis of thiostrepton-derived thiopeptide antibiotics with improved pharmaceutical properties [J]. Organic Chemistry Frontiers, 2015, 2: 106-109. |
| 23 | EVANS B, CHEN Yunqiu, METCALF W, et al. Directed evolution of the nonribosomal peptide synthetase AdmK generates new Andrimid derivatives in vivo [J]. Chemistry & Biology, 2011, 18(5): 601-607. |
| 24 | MATSUDA Y, GOTFREDSEN C H, LARSEN T O. Genetic characterization of neosartorin biosynthesis provides insight into heterodimeric natural product generation [J]. Organic Letters, 2018, 20(22): 7197-7200. |
| 25 | JI Zhiqin, WEI Shaopeng, FAN Lixia, et al. Three novel cyclic hexapeptides from Streptomyces alboflavus 313 and their antibacterial activity [J]. European Journal of Medicinal Chemistry, 2012, 50: 296-303. |
| 26 | FAN Lixia, JI Zhiqin, GUO Zhengyan, et al. NW-G12, a novel nonchlorinated cyclohexapeptide from Streptomyces alboflavus313 [J]. Chemistry of Natural Compounds, 2013, 49(5): 910-913. |
| 27 | GUO Zhengyan, LI Pengwei, CHEN Guozhu, et al. Design and biosynthesis of dimeric alboflavusins with biaryl linkages via regiospecific C—C bond coupling [J]. Journal of the American Chemical Society, 2018, 140(51): 18009-18015. |
| 28 | HINDRA, YANG Dong, TENG Qihui, et al. Genome mining of Streptomyces mobaraensis DSM40847 as a bleomycin producer providing a biotechnology platform to engineer designer bleomycin analogues [J]. Organic Letters, 2017, 19(6): 1386-1389. |
| 29 | BUTLER M S, ROBERTSON A A B, COOPER M A. Natural product and natural product derived drugs in clinical trials [J]. Natural Product Reports, 2014, 31(11): 1612-1661. |
| 30 | CHEN Yun, DENG Wei, WU Jiequn, et al. Genetic modulation of the overexpression of tailoring genes eryK and eryG leading to the improvement of erythromycin A purity and production in Saccharopolyspora erythraea fermentation [J]. Applied and Environmental Microbiology, 2008, 74(6): 1820-1828. |
| 31 | 张万祥, 汪焰胜, 吴杭, 等. 前体代谢工程提高红霉素产量的研究进展[J]. 生物技术通讯, 2019, 30(1): 140-145. |
| ZHANG Wanxiang, WANG Yansheng, WU Hang, et al. Advances in metabolic engineering of precursors for improving Erythromycin production [J]. Letters in Biotechnology, 2019, 30(1): 140-145. | |
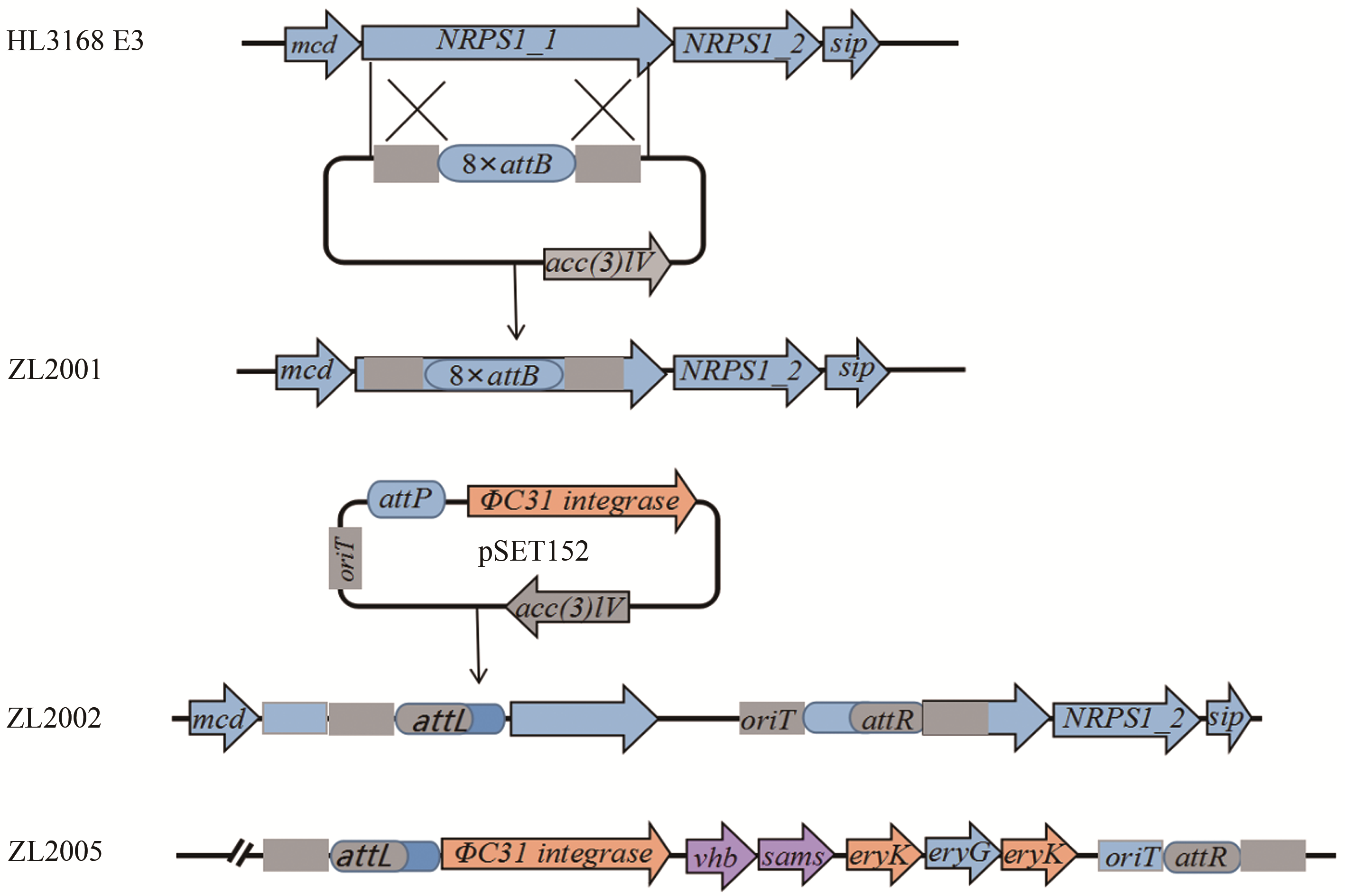
| 32 | WU Jiequn, ZHANG Qinglin, DENG Wei, et al. Toward improvement of erythromycin A production in an industrial Saccharopolyspora erythraea strain via facilitation of genetic manipulation with an artificial attB site for specific recombination [J]. Applied and Environmental Microbiology, 2011, 77(21): 7508-7516. |
| 33 | LIAN Jiazhang, SI Tong, NAIR N U, et al. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains [J]. Metabolic Engineering, 2014, 24: 139-149. |
| 34 | KRIVORUCHKO A, ZHANG Yiming, SIEWERS V, et al. Microbial acetyl-CoA metabolism and metabolic engineering [J]. Metabolic Engineering, 2015, 28: 28-42. |
| 35 | LIU Yiqi, BAI Chenxiao, LIU Qi, et al. Engineered ethanol-driven biosynthetic system for improving production of acetyl-CoA derived drugs in Crabtree-negative yeast [J]. Metabolic Engineering, 2019, 54: 275-284. |
| 36 | 朱灵英, 郭娟, 张爱丽, 等. 参与植物三萜生物合成的细胞色素P450酶研究进展[J]. 中草药, 2019, 50(22): 5597-5610. |
| ZHU Lingying, GUO Juan, ZHANG Aili, et al. Research progress on CYP450 involved in medicinal plant triterpenoid biosynthesis [J]. Chinese Traditional and Herbal Drugs, 2019, 50(22): 5597-5610. | |
| 37 | 漆丽华, 张媚, 潘海学, 等. 基于生物合成途径改造的一个三欣卡辛类似物的发现[J]. 生命有机化学, 2014, 34(7): 1376-1381. |
| XI Lihua, ZHANG Mei, PAN Haixue, et al. Production of a trioxacarcin analogue by engineering of its biosynthetic pathway [J]. Chinese Journal of Organic Chemistry, 2014, 34(7): 1376-1381. | |
| 38 | MARTIN V J J, PITERA D J, WITHERS S T, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids [J]. Nature Biotechnology, 2003, 21(7): 796-802. |
| 39 | 范楚珧, 刘龙英, 沈玥, 等. 吗啡的合成生物学研究和工业化生产[J]. 科学通报, 2016, 61: 1436-1444. |
| FAN Chuyao, LIU Longying, SHEN Yue, et al. Progress of biosynthesis of morphine and its industrial manufacture [J]. Chinese Science Bulletin, 2016, 61: 1436-1444. | |
| 40 | NAKAGAWA A, MINAMI H, KIM Ju-Sung, et al. A bacterial platform for fermentative production of plant alkaloids [J]. Nature Communications, 2011, 2(1): 1-9. |
| 41 | NEUMANN H, NEUMANN-STAUBITZ P. Synthetic biology approaches in drug discovery and pharmaceutical biotechnology [J]. Applied Microbiology and Biotechnology, 2010, 87(1): 75-86. |
| 42 | ENGELS B, DAHM P, JENNEWEIN S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production [J]. Metabolic Engineering, 2008, 10(3/4): 201-206. |
| 43 | KIRBY J, KEASLING J D. Metabolic engineering of microorganisms for isoprenoid production [J]. Natural Product Reports, 2008, 25(4): 656-661. |
| 44 | WALTHER T, CALVAYRAC F, MALBERT Y, et al. Construction of a synthetic metabolic pathway for the production of 2,4-dihydroxybutyric acid from homoserine [J]. Metabolic Engineering, 2018, 45: 237-245. |
| 45 | WEI Liang, WANG Qian, XU Ning, et al. Combining protein and metabolic engineering strategies for high level production of O-acetylhomoserine in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8: 1153-1167. |
| 46 | WILLIAMS T L, YIN Yuhui W, CARTER C W. Selective inhibition of bacterial tryptophanyl-tRNA synthetases by indolmycin is mechanism-based [J]. Journal of Biological Chemistry, 2016, 291(1): 255-265. |
| 47 | DU Yiling, HIGGINS M A, ZHAO Guiyun, et al. Convergent biosynthetic transformations to a bacterial specialized metabolite [J]. Nature Chemical Biology, 2019, 15(11): 1043-1048. |
| 48 | DANGEL V, WESTRICH L, SMITH M C M, et al. Use of an inducible promoter for antibiotic production in a heterologous host [J]. Applied Microbiology and Biotechnology, 2010, 87(1): 261-269. |
| 49 | YAN Fu, BURGARD C, POPOFF A, et al. Synthetic biology approaches and combinatorial biosynthesis towards heterologous lipopeptide production [J]. Chemical Science, 2018, 9(38): 7510-7519. |
| 50 | KOMATSU M, KOMATSU K, KOIWAI H, et al. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites [J]. ACS Synthetic Biology, 2013, 2(7): 384-396. |
| 51 | LUO Yunzi, HUANG Hua, LIANG Jing, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster [J]. Nature Communications, 2013, 4: 94-105. |
| 52 | TAN Gaoyi, DENG Kunhua, LIU Xinhua, et al. Heterologous biosynthesis of spinosad: an omics-guided large polyketide synthase gene cluster reconstitution in Streptomyces [J]. ACS Synthetic Biology, 2017, 6(6): 995-1005. |
| 53 | D'ISCHIA M, WAKAMATSU K, CICOIRA F, et al. Melanins and melanogenesis: from pigment cells to human health and technological applications [J]. Pigment Cell & Melanoma Research, 2015, 28(5): 520-544. |
| 54 | KIM Young Jo, KHETAN A, WU Wei, et al. Evidence of porphyrin-like structures in natural melanin pigments using electrochemical fingerprinting [J]. Advanced Materials, 2016, 28(16): 3173-3180. |
| 55 | WANG Zheng, TSCHIRHART T, SCHULTZHAUS Z, et al. Characterization and application of melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis [J]. Applied and Environmental Microbiology, 2020, 86(5): e02749-19. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | ZHONG Quanzhou, SHAN Yiyi, PEI Qingyun, JIN Yanyun, WANG Yihan, MENG Luyuan, WANG Xinyun, ZHANG Yuxin, LIU Kunyuan, WANG Huizhong, FENG Shangguo. Research progress in the production of α-arbutin through biosynthesis [J]. Synthetic Biology Journal, 2025, 6(1): 118-135. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [11] | LIU Yining, PU Wei, YANG Jinxing, WANG Yu. Recent advances in the biosynthesis of ω-amino acids and lactams [J]. Synthetic Biology Journal, 2024, 5(6): 1350-1366. |
| [12] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [13] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [14] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [15] | LIU Zijian, MU Baiyang, DUAN Zhiqiang, WANG Xuan, LU Xiaojie. Advances in the development of DNA-compatible chemistries [J]. Synthetic Biology Journal, 2024, 5(5): 1102-1124. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||