Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (6): 1367-1385.DOI: 10.12211/2096-8280.2024-014
• Invited Review • Previous Articles Next Articles
Research progress of diols production by microbes
ZHU Fanghuan1, CEN Xuecong1, CHEN Zhen1,2
- 1.Key Laboratory of Industrial Biocatalysis (Ministry of Education),Department of Chemical Engineering,Tsinghua University,Beijing 100084,China
2.Center for Synthetic and Systems Biology,Tsinghua University,Beijing 100084,China
-
Received:2024-02-04Revised:2024-05-08Online:2025-01-10Published:2024-12-31 -
Contact:CHEN Zhen
微生物合成二元醇研究进展
竺方欢1, 岑雪聪1, 陈振1,2
- 1.清华大学化学工程系,工业生物催化教育部重点实验室,北京 100084
2.清华大学合成与系统生物学中心,北京 100084
-
通讯作者:陈振 -
作者简介:竺方欢 (1996—),女,博士研究生。研究方向为二元醇的绿色生物制造。E-mail:zfh21@mails.tsinghua.edu.cn岑雪聪 (1996—),女,博士研究生。研究方向为二元醇的绿色生物制造。 E-mail:cxc18@mails.tsinghua.edu.cn陈振 (1983—),男,副教授,博士生导师。研究方向为材料、化学品及生物医药的绿色生物制造。E-mail:zhenchen2013@tsinghua.edu.cn -
基金资助:国家重点研发计划(2021YFC2100900);国家自然科学基金(22078172)
CLC Number:
Cite this article
ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes[J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385.
竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-014
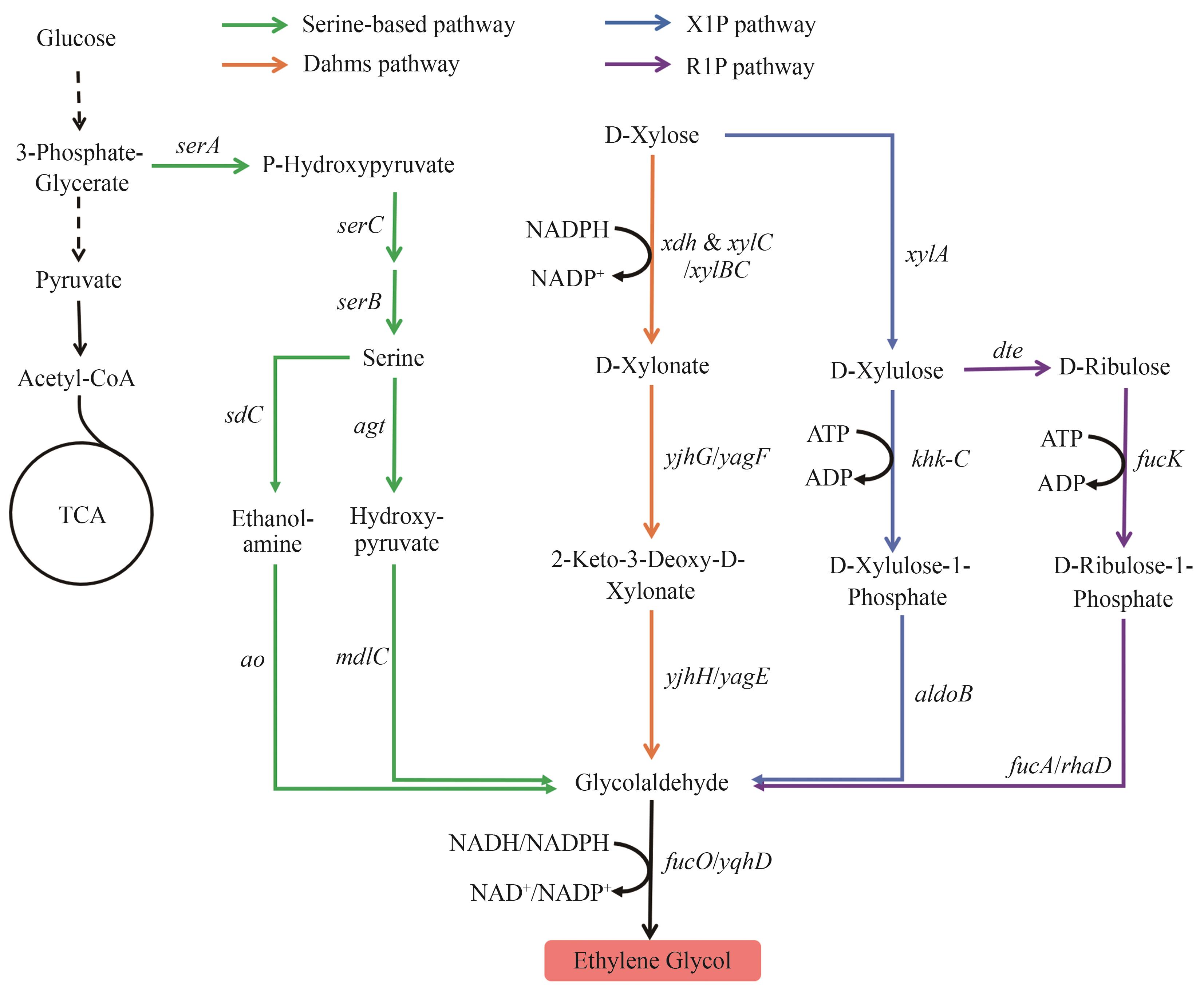
Fig. 1 Biosynthetic pathways of EGX1P—Xylulose-1-phosphate; R1P—Ribulose-1-phosphate The enzymes encoded by the genes: serA—phosphoglycerate dehydrogenase; serC—phosphoserine aminotransferase; serB—phosphoserine phosphatase; sdC—L-serine decarboxylase; ao—amine oxidase; agt—serine-glyoxylate aminotransferase; mdlC—benzoylformate decarboxylase; fucO/yqhD—alcohol dehydrogenase; xdh—D-xylose dehydrogenase; xylB—xylulokinase; xylC—xylonolactonase; yjhG/yagF—D-xylonate dehydrogenase; yjhH/yagE—2-keto-3-deoxy-D-pentose aldolase; xylA—D-xylose isomerase; khk-C—D-xylulose kinase; aldoB—D-xylulose-1-phosphate aldolase; dte—D-tagatose epimerase; fucK—fuculokinase; fucA/rhaD—D-ribulose 1-phosphate aldolase
| 产物 | 合成路径 | 菌株 | 底物 | 基因改造策略 | 产量 /(g/L) | 得率 /(g/g) | 生产效率 /[g/(L·h)] | 理论转化率 /(g/g) | 参考文献 |
|---|---|---|---|---|---|---|---|---|---|
| 乙二醇(EG) | Dahms路径 | Escherichia coli | D-木糖 | (1)下调xylB表达 (2)过表达yqhD | 108.2 | 0.36 | 2.25 | 0.41 | [ |
| Dahms路径 | Escherichia coli | D-木糖 | (1)敲除arcA和aldA (2)过表达yjhH, xdh, xylC, fucO和yjhG | 72.0 | 0.40 | 1.38 | [ | ||
| R1P路径 | Escherichia coli | D-木糖 | (1)过表达fucK, fucO和fucA (2)敲除ald和xylB | 40 | 0.35 | 0.58 | 0.41 | [ | |
| X1P路径 | Escherichia coli | D-木糖 | (1)过表达khk-C, aldoB和fucO (2)敲除xylB和aldA | 20 | 0.38 | 0.37 | 0.41 | [ | |
| X1P路径 | Saccharomyces cerevisiae F251 | D-木糖 | (1)过表达pfk1和pfk2 (2)敲除xks1 | 4.05 | 0.12 | 0.06 | [ | ||
| 丝氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达aao, sdc, serABC和fucO (2)敲除aldA | 4.1 | 0.14 | — | 0.7 | [ | |
| 丝氨酸路径 | Corynebacterium glutamicum | 葡萄糖 | (1)过表达sgt, mdlC, sdc, AO和yqhD (2)插入serACB (3)敲除pabABC和sdaA | 3.5 | 0.09 | — | [ | ||
1,2-丙二醇 (1,2-PDO) | 丙酮醛途径 | K. pneumoniae | 甘油 | (1)过表达mgsA和yqhD (2)敲除tpiA | 9.3 | 0.20 | 0.06 | 0.56 | [ |
| 丙酮醛途径 | Escherichia coli | 葡萄糖 | (1)过表达mgsA, gldA, fdh1和fucO (2)敲除zwf, tpiA, adhE, gloA和ldhA | 5.13 | 0.48 | — | [ | ||
| 乳酸途径 | Escherichia coli | 葡萄糖 | (1)敲除adhE, dld, lldD, frdA, pflB, mgsA, aldA和arcA | 17.3 | 0.18 | 0.72 | 0.56 | [ | |
1,3-丙二醇 (1,3-PDO) | 甘油途径 | Escherichia coli | 葡萄糖 | — | 135 | 3.50 | — | 0.59 | [ |
| 甘油途径 | Corynebacterium glutamicum | 葡萄糖 | (1)敲除ald, pyk, adh, poxB, ldhA, ppc和zwf (2)过表达hdpA-gldA, gpd1, gpp2, yqhD, 和pduCEDGH (3)下调表达gapA | 110.4 | 0.42 | 2.30 | 0.59 | [ | |
| 甘油途径 | Vibrio natriegens | 甘油 | (1)敲除pta-ackA, arcA, adhE, aldB, ldh, pfl, sthA, glpR, aldA和frdABCD (2)过表达pntAB和phaP | 69.5 | 0.51 | 2.90 | 0.67 | [ | |
| 高丝氨酸途径 | Escherichia coli | 葡萄糖 | (1)敲除thrB (2)过表达yqhD, lysC, serCR42W/R77W, metL和pdc | 3.03 | — | 0.05 | 0.55 | [ | |
| 丙二酰辅酶A途径 | Escherichia coli | 葡萄糖 | (1)过表达mcrC, pduP, mcrN, yqhD和prpE | 7.98 | 0.15 | 0.22 | 0.54 | [ | |
| β-丙氨酸途径 | Escherichia coli | 葡萄糖 | (1)敲除lysC (2)过表达ppc (3)下调表达gltA | 11.21 | — | 0.10 | 0.61 | [ | |
1,3-丁二醇 (1,3-BDO) | 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB和bld | 15.75 | 0.19 | 0.16 | 0.53 | [ |
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达 bldL273T, yqhD, phaAB和 pntAB (2)敲除ldh, pta, ackA, adhE | 13.40 | 0.30 | 0.42 | [ | ||
| 3-羟基丁酸还原路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB, yqhD, pntAB, car和sfp (2)敲除ldh, pta, ackA, adhE | 0.40 | 0.02 | — | [ | ||
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达 pk, glpX, thl, hbd, tesB和car (2)敲除 zwf, edd, pfkA和pfkB | 22.66 | 0.40 | 0.32 | 0.56 | [ | |
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB, bld和yqhD (2)敲除adhE, poxB, ldhA, pta-ackA, atoB, tesB和yciA | 23.10 | 0.26 | 0.64 | [ | ||
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB, bld, yjgB和zwf (2)敲除adhE, poxB, ldhA, yciA, pdhR, pgi和gntR | 71.10 | 0.34 | 1.55 | [ | ||
| DERA-AKR 路径 | Escherichia coli | 葡萄糖 | (1)过表达 AKR, DERA和 PDC (2)敲除pta, yjgB, adhE, ldhA, pflB, adhP, yqhD, eutG, ilvB, and poxB; | 2.40 | 0.06 | — | 0.55 | [ | |
1,4-丁二醇 (1,4-BDO) | 琥珀酰辅酶A路径 | Escherichia coli | 葡萄糖 | — | >125 | 0.40 | >3.5 | 0.5 | [ |
| 谷氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达gadB, gabT, yqhD, car, ppc, gltAR163L | 1.41 | 0.07 | 0.03 | 0.58 | [ | |
| 非磷酸化路径 | Escherichia coli | 葡萄糖、木糖 | (1)敲除yagE, xylA和yjhH (2)过表达KvidV461I, xylBCDX和yqhD | 12 | 0.26 (木糖) | 0.40 | 0.41 | [ | |
1,2-丁二醇 (1,2-BDO) | 苏氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达pyc, thrABC, ilvA, L-ldh, car, yqhD (2)敲除lldd, dld | 0.15 | — | — | 0.48 | [ |
2,3-丁二醇 (2,3-BDO) | 丙酮酸路径 | Saccharomyces cerevisiae | 葡萄糖 | (1)过表达BDH1 (2)下调PDC1, PDC6, 和AHD1 | 178 | 0.34 | 2.64 | 0.5 | [ |
| Corynebacterium glutamicum | 葡萄糖 | (1)过表达budABC和acs (2)敲除ldhA, adhE, frdA和pta | 144.9 | 0.43 | 1.10 | [ | |||
1,5-戊二醇 (1,5-PDO) | 赖氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达 lysCQ298G, asd,ddh, dapA,patA, patD, cadA, gabT, yahk, car, sfp, yqhD (2)敲除gabD, gdhA | 9.25 | 0.16 | 0.05 | 0.40 | [ |
| 赖氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达 lysC, dapA, davB, davA, gabT, yqhD, car和sfp (2)敲除iclR | 0.97 | 0.05 | — | 0.38 | [ | |
| 赖氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达 lysCQ298G, asd,ddh, dapA, davB, davA, gabT, yqhD, abfT, bldL273T 和pntAB | 0.12 | 0.006 | — | 0.3 | [ |
Table 1 Typical pathways and metabolic engineering modification strategies for biosynthesis of diols
| 产物 | 合成路径 | 菌株 | 底物 | 基因改造策略 | 产量 /(g/L) | 得率 /(g/g) | 生产效率 /[g/(L·h)] | 理论转化率 /(g/g) | 参考文献 |
|---|---|---|---|---|---|---|---|---|---|
| 乙二醇(EG) | Dahms路径 | Escherichia coli | D-木糖 | (1)下调xylB表达 (2)过表达yqhD | 108.2 | 0.36 | 2.25 | 0.41 | [ |
| Dahms路径 | Escherichia coli | D-木糖 | (1)敲除arcA和aldA (2)过表达yjhH, xdh, xylC, fucO和yjhG | 72.0 | 0.40 | 1.38 | [ | ||
| R1P路径 | Escherichia coli | D-木糖 | (1)过表达fucK, fucO和fucA (2)敲除ald和xylB | 40 | 0.35 | 0.58 | 0.41 | [ | |
| X1P路径 | Escherichia coli | D-木糖 | (1)过表达khk-C, aldoB和fucO (2)敲除xylB和aldA | 20 | 0.38 | 0.37 | 0.41 | [ | |
| X1P路径 | Saccharomyces cerevisiae F251 | D-木糖 | (1)过表达pfk1和pfk2 (2)敲除xks1 | 4.05 | 0.12 | 0.06 | [ | ||
| 丝氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达aao, sdc, serABC和fucO (2)敲除aldA | 4.1 | 0.14 | — | 0.7 | [ | |
| 丝氨酸路径 | Corynebacterium glutamicum | 葡萄糖 | (1)过表达sgt, mdlC, sdc, AO和yqhD (2)插入serACB (3)敲除pabABC和sdaA | 3.5 | 0.09 | — | [ | ||
1,2-丙二醇 (1,2-PDO) | 丙酮醛途径 | K. pneumoniae | 甘油 | (1)过表达mgsA和yqhD (2)敲除tpiA | 9.3 | 0.20 | 0.06 | 0.56 | [ |
| 丙酮醛途径 | Escherichia coli | 葡萄糖 | (1)过表达mgsA, gldA, fdh1和fucO (2)敲除zwf, tpiA, adhE, gloA和ldhA | 5.13 | 0.48 | — | [ | ||
| 乳酸途径 | Escherichia coli | 葡萄糖 | (1)敲除adhE, dld, lldD, frdA, pflB, mgsA, aldA和arcA | 17.3 | 0.18 | 0.72 | 0.56 | [ | |
1,3-丙二醇 (1,3-PDO) | 甘油途径 | Escherichia coli | 葡萄糖 | — | 135 | 3.50 | — | 0.59 | [ |
| 甘油途径 | Corynebacterium glutamicum | 葡萄糖 | (1)敲除ald, pyk, adh, poxB, ldhA, ppc和zwf (2)过表达hdpA-gldA, gpd1, gpp2, yqhD, 和pduCEDGH (3)下调表达gapA | 110.4 | 0.42 | 2.30 | 0.59 | [ | |
| 甘油途径 | Vibrio natriegens | 甘油 | (1)敲除pta-ackA, arcA, adhE, aldB, ldh, pfl, sthA, glpR, aldA和frdABCD (2)过表达pntAB和phaP | 69.5 | 0.51 | 2.90 | 0.67 | [ | |
| 高丝氨酸途径 | Escherichia coli | 葡萄糖 | (1)敲除thrB (2)过表达yqhD, lysC, serCR42W/R77W, metL和pdc | 3.03 | — | 0.05 | 0.55 | [ | |
| 丙二酰辅酶A途径 | Escherichia coli | 葡萄糖 | (1)过表达mcrC, pduP, mcrN, yqhD和prpE | 7.98 | 0.15 | 0.22 | 0.54 | [ | |
| β-丙氨酸途径 | Escherichia coli | 葡萄糖 | (1)敲除lysC (2)过表达ppc (3)下调表达gltA | 11.21 | — | 0.10 | 0.61 | [ | |
1,3-丁二醇 (1,3-BDO) | 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB和bld | 15.75 | 0.19 | 0.16 | 0.53 | [ |
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达 bldL273T, yqhD, phaAB和 pntAB (2)敲除ldh, pta, ackA, adhE | 13.40 | 0.30 | 0.42 | [ | ||
| 3-羟基丁酸还原路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB, yqhD, pntAB, car和sfp (2)敲除ldh, pta, ackA, adhE | 0.40 | 0.02 | — | [ | ||
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达 pk, glpX, thl, hbd, tesB和car (2)敲除 zwf, edd, pfkA和pfkB | 22.66 | 0.40 | 0.32 | 0.56 | [ | |
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB, bld和yqhD (2)敲除adhE, poxB, ldhA, pta-ackA, atoB, tesB和yciA | 23.10 | 0.26 | 0.64 | [ | ||
| 3-羟基丁酰辅酶A路径 | Escherichia coli | 葡萄糖 | (1)过表达phaAB, bld, yjgB和zwf (2)敲除adhE, poxB, ldhA, yciA, pdhR, pgi和gntR | 71.10 | 0.34 | 1.55 | [ | ||
| DERA-AKR 路径 | Escherichia coli | 葡萄糖 | (1)过表达 AKR, DERA和 PDC (2)敲除pta, yjgB, adhE, ldhA, pflB, adhP, yqhD, eutG, ilvB, and poxB; | 2.40 | 0.06 | — | 0.55 | [ | |
1,4-丁二醇 (1,4-BDO) | 琥珀酰辅酶A路径 | Escherichia coli | 葡萄糖 | — | >125 | 0.40 | >3.5 | 0.5 | [ |
| 谷氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达gadB, gabT, yqhD, car, ppc, gltAR163L | 1.41 | 0.07 | 0.03 | 0.58 | [ | |
| 非磷酸化路径 | Escherichia coli | 葡萄糖、木糖 | (1)敲除yagE, xylA和yjhH (2)过表达KvidV461I, xylBCDX和yqhD | 12 | 0.26 (木糖) | 0.40 | 0.41 | [ | |
1,2-丁二醇 (1,2-BDO) | 苏氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达pyc, thrABC, ilvA, L-ldh, car, yqhD (2)敲除lldd, dld | 0.15 | — | — | 0.48 | [ |
2,3-丁二醇 (2,3-BDO) | 丙酮酸路径 | Saccharomyces cerevisiae | 葡萄糖 | (1)过表达BDH1 (2)下调PDC1, PDC6, 和AHD1 | 178 | 0.34 | 2.64 | 0.5 | [ |
| Corynebacterium glutamicum | 葡萄糖 | (1)过表达budABC和acs (2)敲除ldhA, adhE, frdA和pta | 144.9 | 0.43 | 1.10 | [ | |||
1,5-戊二醇 (1,5-PDO) | 赖氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达 lysCQ298G, asd,ddh, dapA,patA, patD, cadA, gabT, yahk, car, sfp, yqhD (2)敲除gabD, gdhA | 9.25 | 0.16 | 0.05 | 0.40 | [ |
| 赖氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达 lysC, dapA, davB, davA, gabT, yqhD, car和sfp (2)敲除iclR | 0.97 | 0.05 | — | 0.38 | [ | |
| 赖氨酸路径 | Escherichia coli | 葡萄糖 | (1)过表达 lysCQ298G, asd,ddh, dapA, davB, davA, gabT, yqhD, abfT, bldL273T 和pntAB | 0.12 | 0.006 | — | 0.3 | [ |
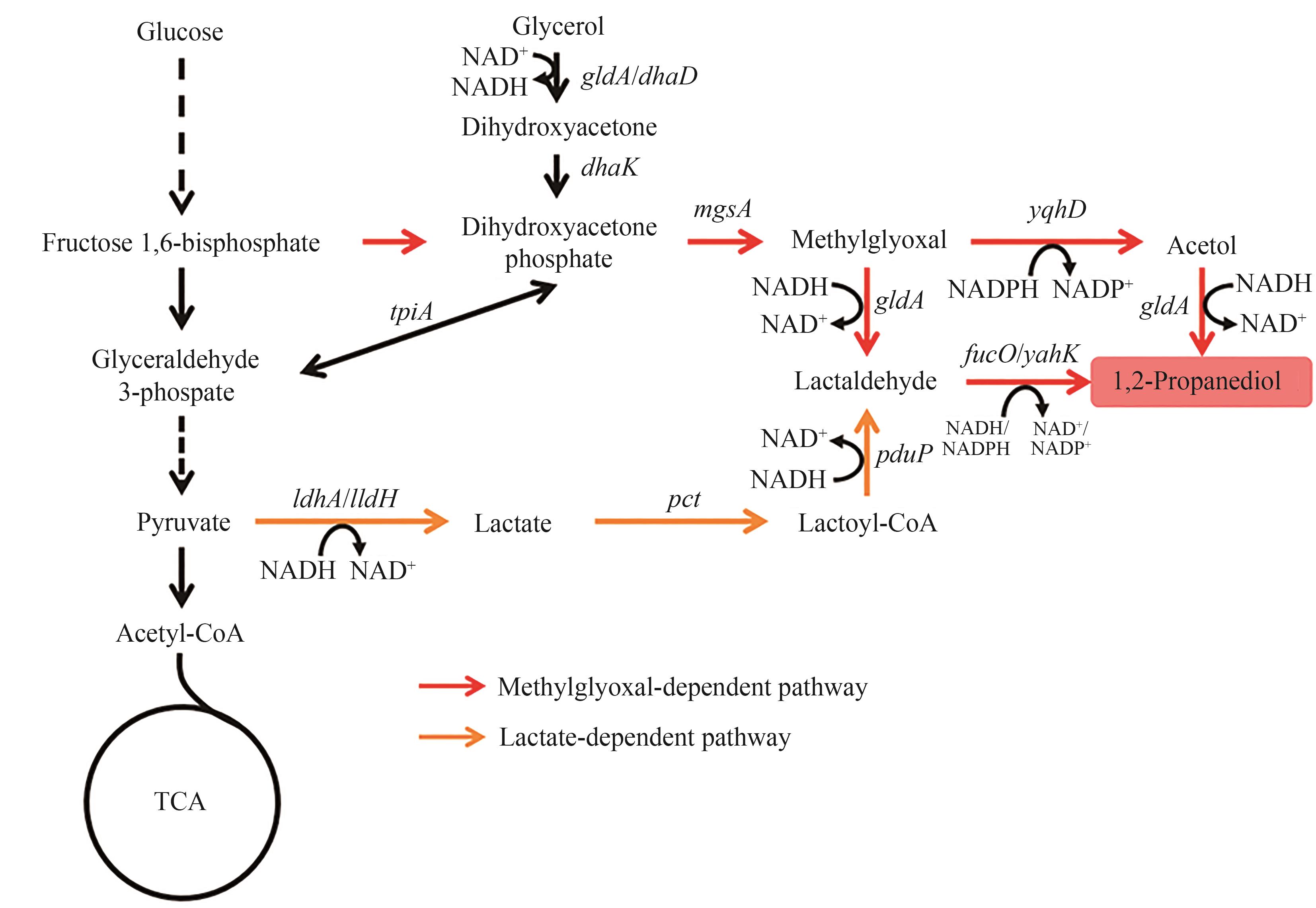
Fig. 2 Biosynthetic pathways of 1,2-PDOThe enzymes encoded by the genes: gldA/dhaD—glycerol dehydrogenase; dhaK—PEP-dependent dihydroxyacetone kinase; mgsA—methylglyoxal synthase; yqhD—alcohol dehydrogenase; yahK—alcohol dehydrogenase; fucO—alcohol dehydrogenase; ldhA/lldH—lactate dehydrogenase; pct—propionate CoA-transferase; pduP, aldehyde dehydrogenase
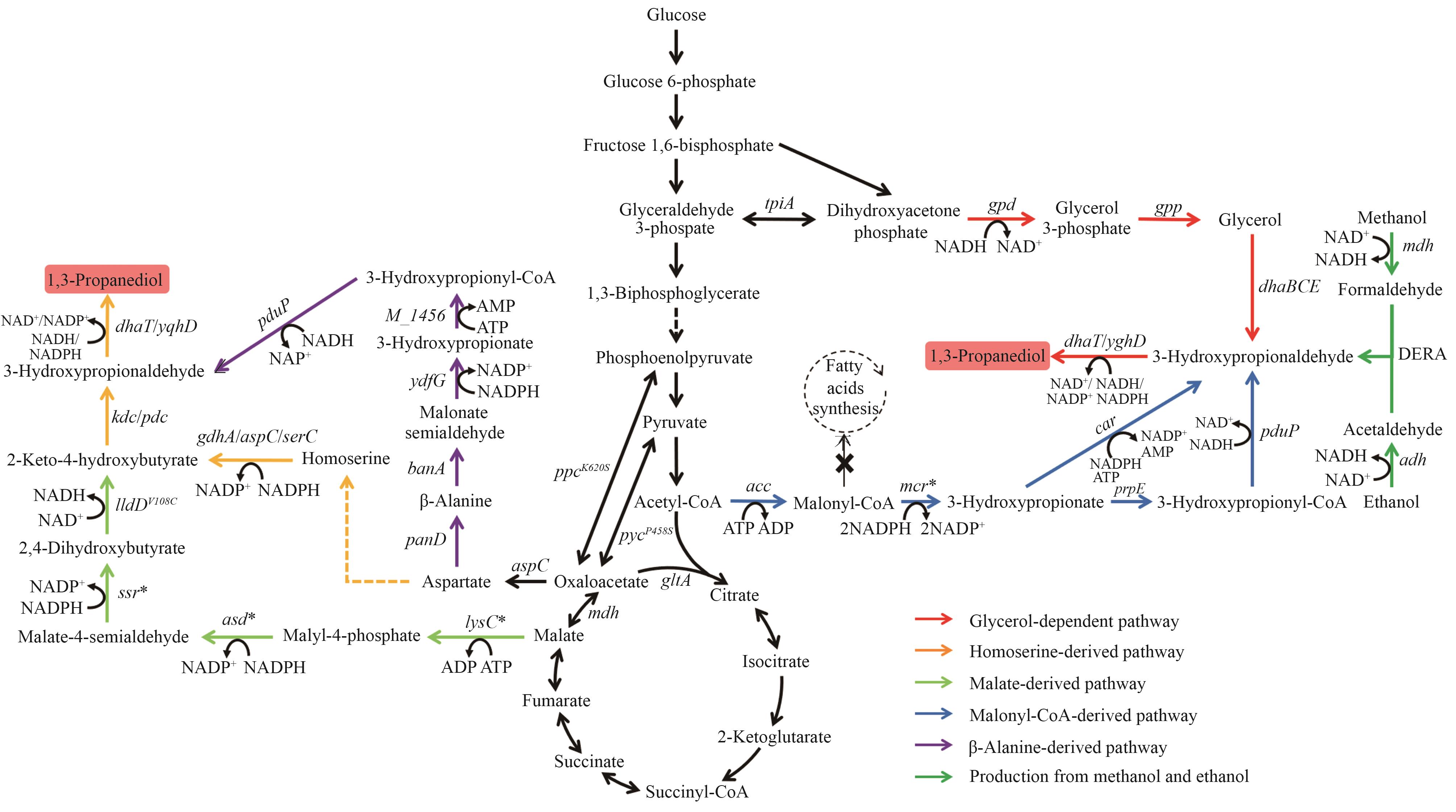
Fig. 3 Biosynthetic pathways of 1,3-PDOThe enzymes encoded by the genes: tpiA—triose-phosphate isomerase; gpd—glycerol-3-phosphate dehydrogenase; gpp—glycerol-3-phosphate phosphatase; dhaBCE—vitamin B12-dependent glycerol dehydratase; dhaT/yqhD—alcohol dehydrogenase; acc—acetyl-CoA carboxylase; mcr—malonyl-CoA reductase; prpE—3-hydroxypropionyl-CoA synthetase; pduP—aldehyde dehydrogenase; car—carboxylic acid reductase; mdh—methanol dehydrogenase; adh—alcohol dehydrogenase; DERA—deoxyribose-5-phosphate aldolase; aspC—aspartate transaminase; panD—aspartate decarboxylase; bauA—β-alanine-pyruvate aminotransferase; ydfG—3-hydroxy acid dehydrogenase; M_1456—3-hydroxypropionyl-coenzyme A synthetase; lysC—malate kinase; asd—malate semialdehyde dehydrogenase; ssr—malate semialdehyde reductase; lldD—lactate dehydrogenase; kdc—ketoacid decarboxylase; pdc—pyruvate decarboxylase; gdhA—glutamate dehydrogenase; serC—phosphoserine aminotransferase
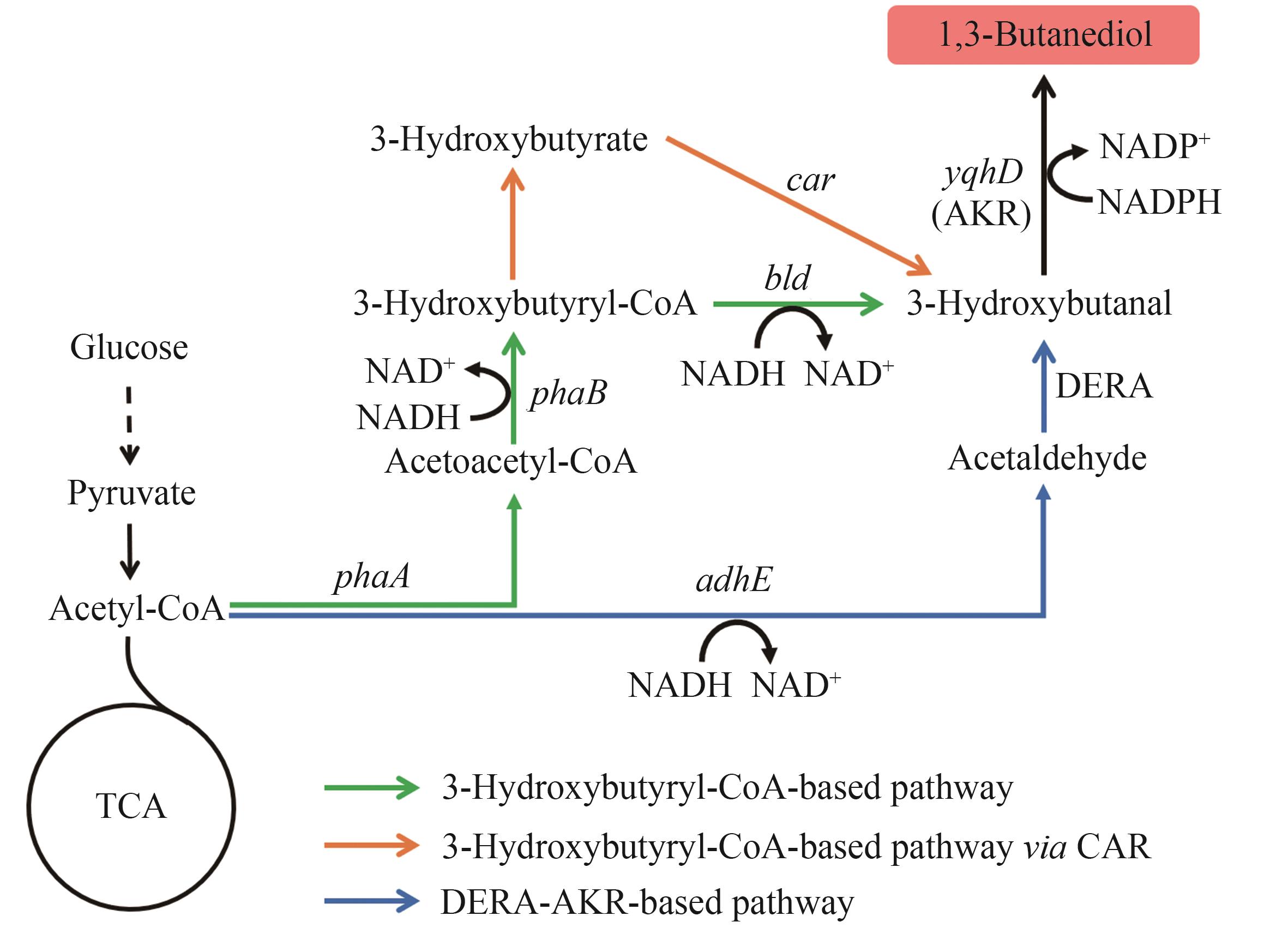
Fig. 4 Biosynthetic pathways of 1,3-BDOThe enzymes encoded by the genes: adhE—alcohol dehydrogenase; DERA—deoxyribose-5-phosphate aldolase; yqhD—alcohol dehydrogenase; phaA—acetyl-CoA acetyltransferase; phaB—acetoacetyl-CoA reductase; bld—3-hydroxybutyryl-CoA dehydrogenase; car—carboxylic acid reductase
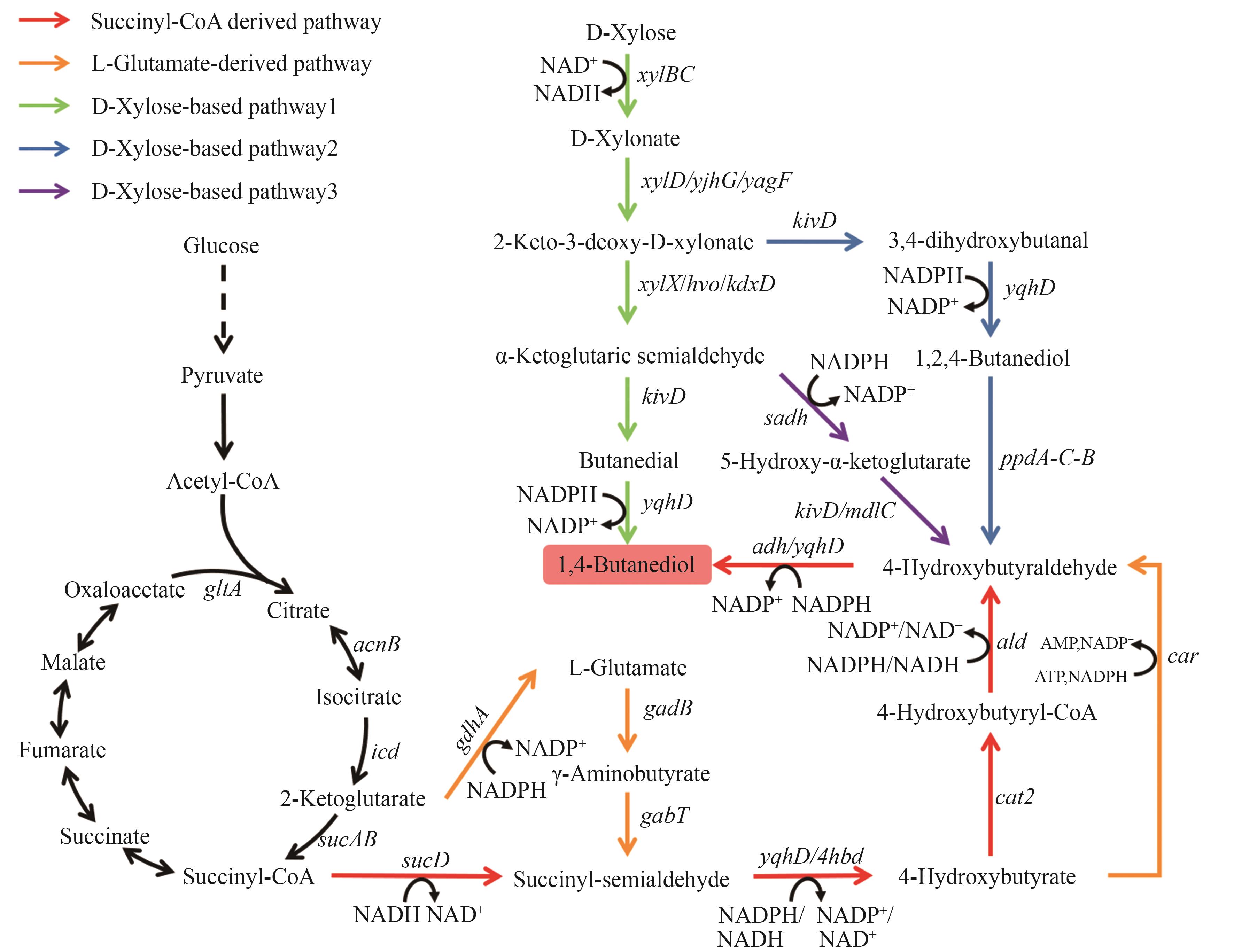
Fig. 5 Biosynthetic pathways of 1,4-BDOThe enzymes encoded by the genes: gdhA—glutamate dehydrogenase; gadB—glutamate decarboxylase; gabT—aminotransferase; sucD—succinate semialdehyde dehydrogenase; yqhD/4hbd/adh—alcohol dehydrogenase; cat2—4-hydroxybutyrate-CoA transferase; ald—aldehyde dehydrogenase; car—carboxylic acid reductase; xylBC—D-xylose dehydrogenase; xylD/yjhG/yagF—D-xylonate dehydratase; xylX/hvo/kdxD—2-keto-3-deoxy-D-xylonate dehydratase; sadh—alcohol dehydrogenase; kivD/mdlC—decarboxylase
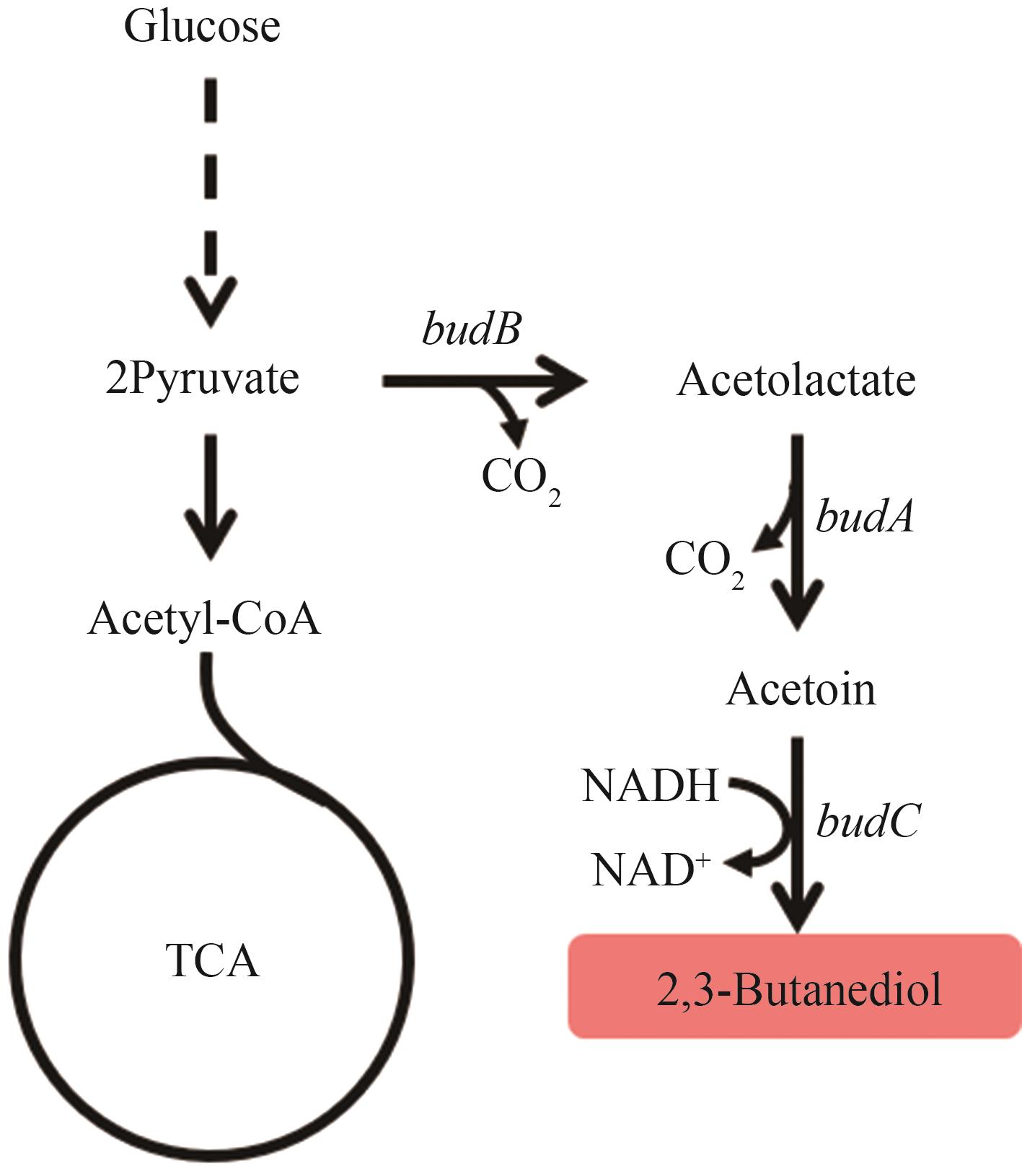
Fig. 6 Biosynthetic pathways of 2,3-BDOThe enzymes encoded by the genes: budB—acetolactate synthase; budA—acetolactate decarboxylase; budC—2,3-butanediol dehydrogenase
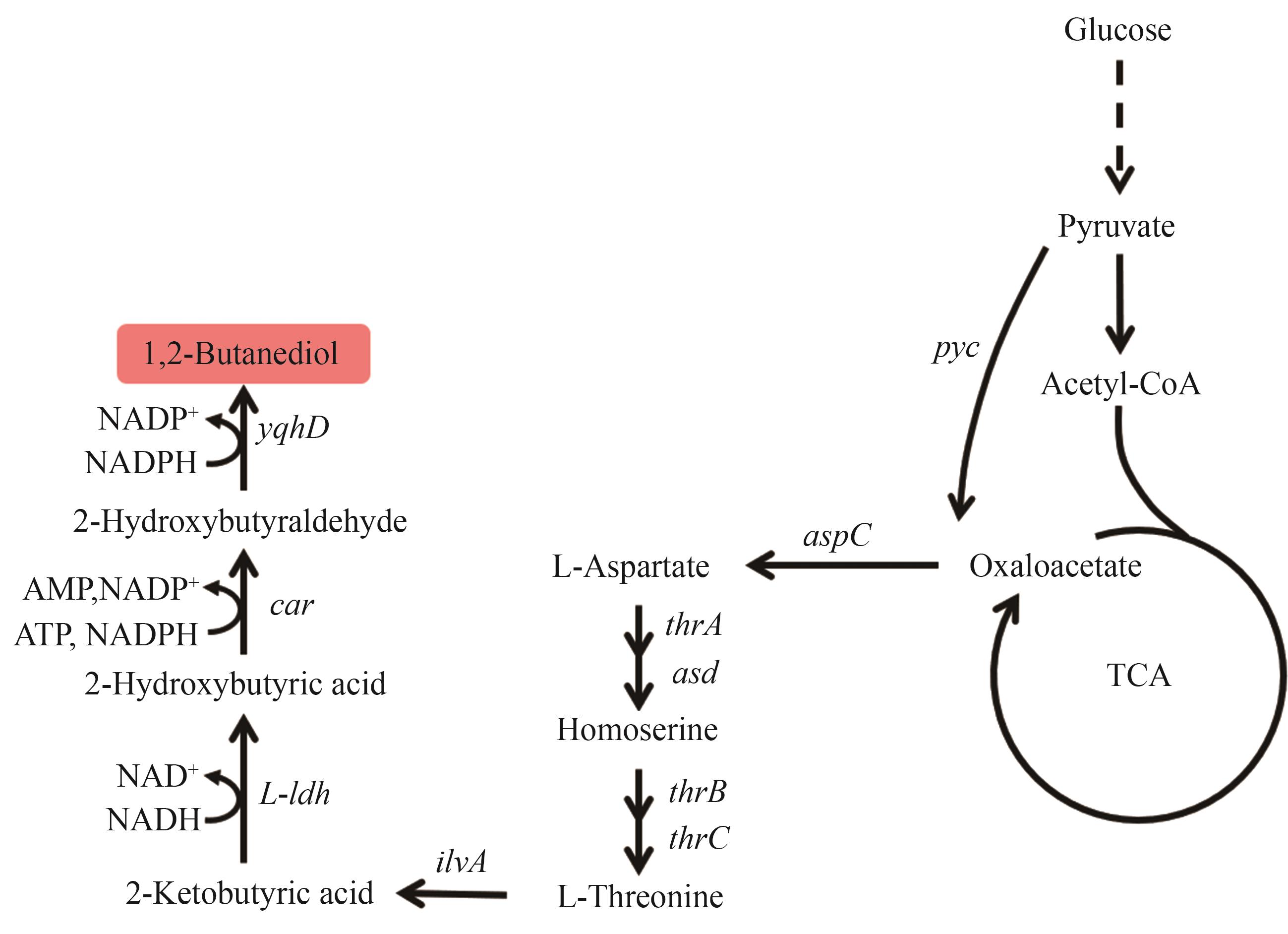
Fig. 7 Biosynthetic pathway of 1,2-BDOThe enzymes encoded by the genes: pyc—pyruvate carboxylase; aspC—aspartate transaminase; thrA—homoserine dehydrogenase; asd—aspartate semialdehyde dehydrogenase; thrB—homoserine kinase; thrC—threonine synthase; ilvA—L-threonine dehydratase; L-ldh—L-lactate dehydrogenase; car—carboxylic acid reductase; yqhD—alcohol dehydrogenase
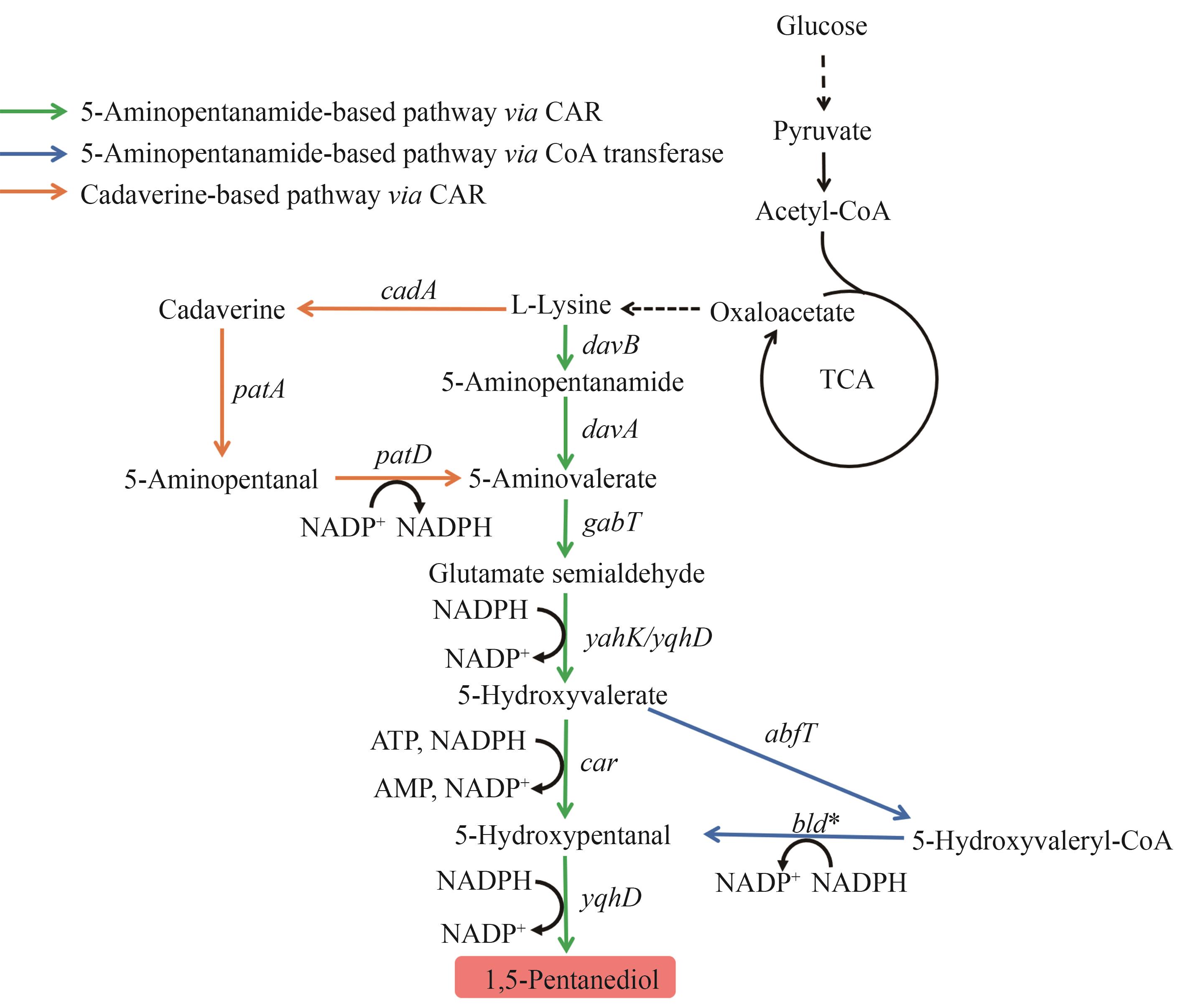
Fig. 8 Biosynthetic pathways of 1,5-PDOThe enzymes encoded by the genes: davB—lysine monooxygenase; davA—5-aminovaleramidase; gabT—4-aminobutyrate aminotransferase; yahK/yqhD—alcohol dehydrogenase; car—carboxylic acid reductase; abfT—5-hydroxyvalerate-CoA transferase; bld—aldehyde dehydrogenase; cadA—lysine decarboxylase; patA—putrescine aminotransferase; patD—alcohol dehydrogenase
| 1 | CHOI S, SONG C W, SHIN J H, et al. Biorefineries for the production of top building block chemicals and their derivatives[J]. Metabolic Engineering, 2015, 28: 223-239. |
| 2 | LIU Y F, WANG W, ZENG A P. Biosynthesizing structurally diverse diols via a general route combining oxidative and reductive formations of OH-groups[J]. Nature Communications, 2022, 13(1): 1595. |
| 3 | LIU Y, CEN X C, LIU D H, et al. Metabolic engineering of Escherichia coli for high-yield production of (R)-1,3-butanediol[J]. ACS Synthetic Biology, 2021, 10(8): 1946-1955. |
| 4 | ZHANG Y, LIU D H, CHEN Z. Production of C2-C4 diols from renewable bioresources: new metabolic pathways and metabolic engineering strategies[J]. Biotechnology for Biofuels, 2017, 10: 299. |
| 5 | CEN X C, LIU Y J, ZHU F H, et al. Metabolic engineering of Escherichia coli for high production of 1,5-pentanediol via a cadaverine-derived pathway[J]. Metabolic Engineering, 2022, 74: 168-177. |
| 6 | CEN X C, DONG Y, LIU D H, et al. New pathways and metabolic engineering strategies for microbial synthesis of diols[J]. Current Opinion in Biotechnology, 2022, 78: 102845. |
| 7 | VIVEK N, HAZEENA S H, ALPHY M P, et al. Recent advances in microbial biosynthesis of C3-C5 diols: genetics and process engineering approaches[J]. Bioresource Technology, 2021, 322: 124527. |
| 8 | WU T, LIU Y M, LIU J S, et al. Metabolic engineering and regulation of diol biosynthesis from renewable biomass in Escherichia coli [J]. Biomolecules, 2022, 12(5): 715. |
| 9 | 陈勇,李凤梅,韩荣伟, 等. 酵母不对称催化制备R-1,2丙二醇[J].生物加工过程, 2009, 7(4): 61-64. |
| CHEN Y, LI F M, HAN R W, et al. Preparation of (R)-1,2-propanediol through asymmetric reduction with bakers yeast[J]. Chinese Journal of Bioprocess Engineering, 2009, 7(4): 61-64. | |
| 10 | MATSUYAMA A, YAMAMOTO H, KAWADA N, et al. Industrial production of (R)-1,3-butanediol by new biocatalysts[J]. Journal of Molecular Catalysis B: Enzymatic, 2001, 11(4/5/6): 513-521. |
| 11 | ZHU F H, LIU D H, CHEN Z. Recent advances in biological production of 1,3-propanediol: new routes and engineering strategies[J]. Green Chemistry, 2022, 24(4): 1390-1403. |
| 12 | BURGARD A, BURK M J, OSTERHOUT R, et al. Development of a commercial scale process for production of 1,4-butanediol from sugar[J]. Current Opinion in Biotechnology, 2016, 42: 118-125. |
| 13 | SALUSJÄRVI L, HAVUKAINEN S, KOIVISTOINEN O, et al. Biotechnological production of glycolic acid and ethylene glycol: current state and perspectives[J]. Applied Microbiology and Biotechnology, 2019, 103(6): 2525-2535. |
| 14 | YUE H R, ZHAO Y J, MA X B, et al. Ethylene glycol: properties, synthesis, and applications[J]. Chemical Society Reviews, 2012, 41(11): 4218-4244. |
| 15 | CHAE T U, CHOI S Y, RYU J Y, et al. Production of ethylene glycol from xylose by metabolically engineered Escherichia coli [J]. AIChE Journal, 2018, 64(12): 4193-4200. |
| 16 | WANG Y H, XIAN M, FENG X J, et al. Biosynthesis of ethylene glycol from D-xylose in recombinant Escherichia coli [J]. Bioengineered, 2018, 9(1): 233-241. |
| 17 | PEREIRA B, LI Z J, DE MEY M, et al. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate[J]. Metabolic Engineering, 2016, 34: 80-87. |
| 18 | URANUKUL B, WOOLSTON B M, FINK G R, et al. Biosynthesis of monoethylene glycol in Saccharomyces cerevisiae utilizing native glycolytic enzymes[J]. Metabolic Engineering, 2019, 51: 20-31. |
| 19 | ALKIM C, CAM Y, TRICHEZ D, et al. Optimization of ethylene glycol production from (D)-xylose via a synthetic pathway implemented in Escherichia coli [J]. Microbial Cell Factories, 2015, 14: 127. |
| 20 | PEREIRA B, ZHANG H R, DE MEY M, et al. Engineering a novel biosynthetic pathway in Escherichia coli for production of renewable ethylene glycol[J]. Biotechnology and Bioengineering, 2016, 113(2): 376-383. |
| 21 | CHEN Z, HUANG J H, WU Y, et al. Metabolic engineering of Corynebacterium glutamicum for the de novo production of ethylene glycol from glucose[J]. Metabolic Engineering, 2016, 33: 12-18. |
| 22 | SUN S Q, SHU L, LU X Y, et al. 1,2-Propanediol production from glycerol via an endogenous pathway of Klebsiella pneumoniae [J]. Applied Microbiology and Biotechnology, 2021, 105(23): 9003-9016. |
| 23 | JAIN R, SUN X X, YUAN Q P, et al. Systematically engineering Escherichia coli for enhanced production of 1,2-propanediol and 1-propanol[J]. ACS Synthetic Biology, 2015, 4(6): 746-756. |
| 24 | NIU W, KRAMER L, MUELLER J, et al. Metabolic engineering of Escherichia coli for the de novo stereospecific biosynthesis of 1,2-propanediol through lactic acid[J]. Metabolic Engineering Communications, 2019, 8: e00082. |
| 25 | KURIAN J V. A new polymer platform for the future—sorona® from corn derived 1,3-propanediol[J]. Journal of Polymers and the Environment, 2005, 13(2): 159-167. |
| 26 | LI Z H, DONG Y F, LIU Y, et al. Systems metabolic engineering of Corynebacterium glutamicum for high-level production of 1,3-propanediol from glucose and xylose[J]. Metabolic Engineering, 2022, 70: 79-88. |
| 27 | ZHANG Y, SUN Q, LIU Y, et al. Development of a plasmid stabilization system in Vibrio natriegens for the high production of 1,3-propanediol and 3-hydroxypropionate[J]. Bioresources and Bioprocessing, 2021, 8(1): 125. |
| 28 | ZHANG Y J, MA C W, DISCHERT W, et al. Engineering of phosphoserine aminotransferase increases the conversion of L-homoserine to 4-hydroxy-2-ketobutyrate in a glycerol-independent pathway of 1,3-propanediol production from glucose[J]. Biotechnology Journal, 2019, 14(9): e1900003. |
| 29 | LI Z H, WU Z Y, CEN X C, et al. Efficient production of 1,3-propanediol from diverse carbohydrates via a non-natural pathway using 3-hydroxypropionic acid as an intermediate[J]. ACS Synthetic Biology, 2021, 10(3): 478-486. |
| 30 | LI M D, ZHANG Y, LI J C, et al. Biosynthesis of 1,3-propanediol via a new pathway from glucose in Escherichia coli [J]. ACS Synthetic Biology, 2023, 12(7): 2083-2093. |
| 31 | KATAOKA N, VANGNAI A S, UEDA H, et al. Enhancement of (R)-1,3-butanediol production by engineered Escherichia coli using a bioreactor system with strict regulation of overall oxygen transfer coefficient and pH[J]. Bioscience, Biotechnology, and Biochemistry, 2014, 78(4): 695-700. |
| 32 | WANG J, ZHANG R H, ZHANG J L, et al. Tunable hybrid carbon metabolism coordination for the carbon-efficient biosynthesis of 1,3-butanediol in Escherichia coli [J]. Green Chemistry, 2021, 23(21): 8694-8706. |
| 33 | ISLAM T, NGUYEN-VO T P, GAUR V K, et al. Metabolic engineering of Escherichia coli for biological production of 1,3-butanediol[J]. Bioresource Technology, 2023, 376: 128911. |
| 34 | ISLAM T, NGUYEN-VO T P, CHO S, et al. Metabolic engineering of Escherichia coli for enhanced production of 1,3-butanediol from glucose[J]. Bioresource Technology, 2023, 389: 129814. |
| 35 | KIM T, FLICK R, BRUNZELLE J, et al. Novel Aldo-Keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol: structural and biochemical studies[J]. Applied and Environmental Microbiology, 2017, 83(7): e03172-16. |
| 36 | WANG J, LI C Y, ZOU Y S, et al. Bacterial synthesis of C3-C5 diols via extending amino acid catabolism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(32): 19159-19167. |
| 37 | TAI Y S, XIONG M Y, JAMBUNATHAN P, et al. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products[J]. Nature Chemical Biology, 2016, 12(4): 247-253. |
| 38 | QIN N, ZHU F H, LIU Y Y, et al. Metabolic engineering of Escherichia coli for de novo production of 1,2-butanediol[J]. ACS Synthetic Biology, 2024, 13(1): 351-357. |
| 39 | LEE Y G, SEO J H. Production of 2,3-butanediol from glucose and cassava hydrolysates by metabolically engineered industrial polyploid Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2019, 12: 204. |
| 40 | KOU M Y, CUI Z Z, FU J, et al. Metabolic engineering of Corynebacterium glutamicum for efficient production of optically pure (2R,3R)-2,3-butanediol[J]. Microbial Cell Factories, 2022, 21(1): 150. |
| 41 | CEN X C, LIU Y, CHEN B, et al. Metabolic engineering of Escherichia coli for de novo production of 1,5-pentanediol from glucose[J]. ACS Synthetic Biology, 2021, 10(1): 192-203. |
| 42 | TAO Y M, BU C Y, ZOU L H, et al. A comprehensive review on microbial production of 1,2-propanediol: micro-organisms, metabolic pathways, and metabolic engineering[J]. Biotechnology for Biofuels, 2021, 14(1): 216. |
| 43 | MARINAS A, BRUIJNINCX P, FTOUNI J, et al. Sustainability metrics for a fossil- and renewable-based route for 1,2-propanediol production: a comparison[J]. Catalysis Today, 2015, 239: 31-37. |
| 44 | INGVADOTTIR E M, SCULLY S M, ORLYGSSON J. Production of (S)-1,2-propanediol from L-rhamnose using the moderately thermophilic Clostridium strain AK1[J]. Anaerobe, 2018, 54: 26-30. |
| 45 | BADÍA J, ROS J, AGUILAR J. Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae [J]. Journal of Bacteriology, 1985, 161(1): 435-437. |
| 46 | JUN S A, MOON C, KANG C H, et al. Microbial fed-batch production of 1,3-propanediol using raw glycerol with suspended and immobilized Klebsiella pneumoniae [J]. Applied Biochemistry and Biotechnology, 2010, 161(1-8): 491-501. |
| 47 | LIU H J, XU Y Z, ZHENG Z M, et al. 1, 3-Propanediol and its copolymers: research, development and industrialization[J]. Biotechnology Journal, 2010, 5(11): 1137-1148. |
| 48 | YANG M M, YUN J H, ZHANG H H, et al. Genetically engineered strains: application and advances for 1,3-propanediol production from glycerol[J]. Food Technology and Biotechnology, 2018, 56(1): 3-15. |
| 49 | OH B R, LEE S M, HEO S Y, et al. Efficient production of 1,3-propanediol from crude glycerol by repeated fed-batch fermentation strategy of a lactate and 2,3-butanediol deficient mutant of Klebsiella pneumoniae [J]. Microbial Cell Factories, 2018, 17(1): 92. |
| 50 | ZHOU S, HUANG Y H, MAO X L, et al. Impact of acetolactate synthase inactivation on 1,3-propanediol fermentation by Klebsiella pneumoniae [J]. PLoS One, 2019, 14(4): e0200978. |
| 51 | CHEN Z, GENG F, ZENG A P. Protein design and engineering of a de novo pathway for microbial production of 1,3-propanediol from glucose[J]. Biotechnology Journal, 2015, 10(2): 284-289. |
| 52 | ZHONG W Q, ZHANG Y, WU W J, et al. Metabolic engineering of a homoserine-derived non-natural pathway for the de novo production of 1,3-propanediol from glucose[J]. ACS Synthetic Biology, 2019, 8(3): 587-595. |
| 53 | WALTHER T, TOPHAM C M, IRAGUE R, et al. Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor 2,4-dihydroxybutyric acid[J]. Nature Communications, 2017, 8: 15828. |
| 54 | FRAZÃO C J R, TRICHEZ D, SERRANO-BATAILLE H, et al. Construction of a synthetic pathway for the production of 1,3-propanediol from glucose[J]. Scientific Reports, 2019, 9(1): 11576. |
| 55 | ZHANG C J, SHARMA S, MA C W, et al. Strain evolution and novel downstream processing with integrated catalysis enable highly efficient coproduction of 1,3-propanediol and organic acid esters from crude glycerol[J]. Biotechnology and Bioengineering, 2022, 119(6): 1450-1466. |
| 56 | MENG H, WANG C, YUAN Q P, et al. An aldolase-based new pathway for bioconversion of formaldehyde and ethanol into 1,3-propanediol in Escherichia coli [J]. ACS Synthetic Biology, 2021, 10(4): 799-809. |
| 57 | TROTTER C L, BABU G S, WALLACE S. Engineering biology for sustainable 1,4-butanediol synthesis[J]. Trends in Biotechnology, 2023, 41(3): 286-288. |
| 58 | ZHU Y, YANG J M, MEI F, et al. Bio-based 1,4-butanediol and tetrahydrofuran synthesis: perspective[J]. Green Chemistry, 2022, 24(17): 6450-6466. |
| 59 | YIM H, HASELBECK R, NIU W, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol[J]. Nature Chemical Biology, 2011, 7(7): 445-452. |
| 60 | BARTON N R, BURGARD A P, BURK M J, et al. An integrated biotechnology platform for developing sustainable chemical processes[J]. Journal of Industrial Microbiology & Biotechnology, 2015, 42(3): 349-360. |
| 61 | LIU H W, LU T. Autonomous production of 1, 4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli [J]. Metabolic Engineering, 2015, 29: 135-141. |
| 62 | WANG J, JAIN R, SHEN X L, et al. Rational engineering of diol dehydratase enables 1,4-butanediol biosynthesis from xylose[J]. Metabolic Engineering, 2017, 40: 148-156. |
| 63 | LIU X, FABOS V, TAYLOR S, et al. One-step production of 1,3-butadiene from 2,3-butanediol dehydration[J]. Chemistry, 2016, 22(35): 12290-12294. |
| 64 | BAEK H S, WOO B Y, YOO S J, et al. Composition containing meso-2,3-butanediol: US 10525017[P/OL]. 2020-01-07[2024-01-01]. . |
| 65 | OTAGIRI M, UI S, TAKUSAGAWA Y, et al. Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases[J]. FEBS Letters, 2010, 584(1): 219-223. |
| 66 | YU B, SUN J B, BOMMAREDDY R R, et al. Novel (2R,3R)-2,3-butanediol dehydrogenase from potential industrial strain Paenibacillus polymyxa ATCC 12321[J]. Applied and Environmental Microbiology, 2011, 77(12): 4230-4233. |
| 67 | LEE J W, LEE Y G, JIN Y S, et al. Metabolic engineering of non-pathogenic microorganisms for 2,3-butanediol production[J]. Applied Microbiology and Biotechnology, 2021, 105(14/15): 5751-5767. |
| 68 | MIZOBATA A, MITSUI R, YAMADA R, et al. Improvement of 2,3-butanediol tolerance in Saccharomyces cerevisiae by using a novel mutagenesis strategy[J]. Journal of Bioscience and Bioengineering, 2021, 131(3): 283-289. |
| 69 | NOVAK K, KUTSCHA R, PFLÜGL S. Microbial upgrading of acetate into 2,3-butanediol and acetoin by E. coli W[J]. Biotechnology for Biofuels, 2020, 13: 177. |
| 70 | REHMAN S, LENG L, ZHUANG H C, et al. Genomic insights to facilitate the construction of a high-xylose-utilization Enterococcus faecalis OPS2 for 2,3-BDO production[J]. Chemical Engineering Journal, 2022, 448: 137617. |
| 71 | LIN H, XU J Y, SUN W L, et al. Efficient 1-hydroxy-2-butanone production from 1,2-butanediol by whole cells of engineered E. coli [J]. Catalysts, 2021, 11(10): 1184. |
| 72 | LU H Y, DIAZ D J, CZARNECKI N J, et al. Machine learning-aided engineering of hydrolases for PET depolymerization[J]. Nature, 2022, 604(7907): 662-667. |
| 73 | JIANG S, WANG R R, WANG D H, et al. Metabolic reprogramming and biosensor-assisted mutagenesis screening for high-level production of L-arginine in Escherichia coli [J]. Metabolic Engineering, 2023, 76: 146-157. |
| 74 | LIN J L, WAGNER J M, ALPER H S. Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications[J]. Biotechnology Advances, 2017, 35(8): 950-970. |
| 75 | CHAE T U, CHOI S Y, KIM J W, et al. Recent advances in systems metabolic engineering tools and strategies[J]. Current Opinion in Biotechnology, 2017, 47: 67-82. |
| 76 | ZHANG Y W, YANG J W, YANG S Q, et al. Programming cells by multicopy chromosomal integration using CRISPR-associated transposases[J]. The CRISPR Journal, 2021, 4(3): 350-359. |
| 77 | DONG Y F, ZHANG Y, LIU D H, et al. Strain and process engineering toward continuous industrial fermentation[J]. Frontiers of Chemical Science and Engineering, 2023, 17(10): 1336-1353. |
| 78 | JIANG L L, LIU H F, MU Y, et al. High tolerance to glycerol and high production of 1,3-propanediol in batch fermentations by microbial consortium from marine sludge[J]. Engineering in Life Sciences, 2017, 17(6): 635-644. |
| 79 | LEE S Y, KIM H U, CHAE T U, et al. A comprehensive metabolic map for production of bio-based chemicals[J]. Nature Catalysis, 2019, 2: 18-33. |
| 80 | UTESCH T, SABRA W, PRESCHER C, et al. Enhanced electron transfer of different mediators for strictly opposite shifting of metabolism in Clostridium pasteurianum grown on glycerol in a new electrochemical bioreactor[J]. Biotechnology and Bioengineering, 2019, 116(7): 1627-1643. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | ZHONG Quanzhou, SHAN Yiyi, PEI Qingyun, JIN Yanyun, WANG Yihan, MENG Luyuan, WANG Xinyun, ZHANG Yuxin, LIU Kunyuan, WANG Huizhong, FENG Shangguo. Research progress in the production of α-arbutin through biosynthesis [J]. Synthetic Biology Journal, 2025, 6(1): 118-135. |
| [3] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [4] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [5] | LIU Yining, PU Wei, YANG Jinxing, WANG Yu. Recent advances in the biosynthesis of ω-amino acids and lactams [J]. Synthetic Biology Journal, 2024, 5(6): 1350-1366. |
| [6] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [7] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [8] | LIU Zijian, MU Baiyang, DUAN Zhiqiang, WANG Xuan, LU Xiaojie. Advances in the development of DNA-compatible chemistries [J]. Synthetic Biology Journal, 2024, 5(5): 1102-1124. |
| [9] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [10] | ZHANG Shouqi, WANG Tao, KONG Yao, ZOU Jiasheng, LIU Yuanning, XU Zhengren. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [11] | XIE Xiangqian, GUO Wen, WANG Huan, LI Jin. Biosynthesis and chemical synthesis of ribosomally synthesized and post-translationally modified peptides containing aminovinyl cysteine [J]. Synthetic Biology Journal, 2024, 5(5): 981-996. |
| [12] | TANG Zhijun, HU Youcai, LIU Wen. Enzymatic (4+2)- and (2+2)-cycloaddition reactions: fundamentals and applications of regio- and stereoselectivity [J]. Synthetic Biology Journal, 2024, 5(3): 401-407. |
| [13] | ZHANG Jun, JIN Shixue, YUN Qian, QU Xudong. Biosynthesis of the unnatural extender units with polyketides and their structural modifications for applications in medicines [J]. Synthetic Biology Journal, 2024, 5(3): 561-570. |
| [14] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [15] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||