Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (1): 18-44.DOI: 10.12211/2096-8280.2023-040
• Invited Review • Previous Articles Next Articles
Advances in microbial production of liquid biofuels
GUO Shuyuan1,2( ), ZHANG Qiannan1,2, Gulikezi· MAIMAITIREXIATI1,2, YANG Yiqun1,2, YU Tao1,2
), ZHANG Qiannan1,2, Gulikezi· MAIMAITIREXIATI1,2, YANG Yiqun1,2, YU Tao1,2
- 1.Center for Synthetic Biochemistry,Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences (CAS),Shenzhen 518055,Guangdong,China
2.CAS key laboratory of Quantitative Engineering Biology,Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences (CAS),Shenzhen 518055,Guangdong,China
-
Received:2023-06-13Revised:2024-01-30Online:2025-03-12Published:2025-02-28 -
Contact:GUO Shuyuan, YU Tao
液体生物燃料合成与炼制的研究进展
郭姝媛1,2( ), 张倩楠1,2, 姑丽克孜·买买提热夏提1,2, 杨一群1,2, 于涛1,2
), 张倩楠1,2, 姑丽克孜·买买提热夏提1,2, 杨一群1,2, 于涛1,2
- 1.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,合成生物化学研究中心,广东 深圳 518055
2.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点实验室,广东 深圳 518055
-
通讯作者:郭姝媛,于涛 -
作者简介:郭姝媛 (1991—),女,博士,助理研究员。研究方向为甲醇生物转化及产物合成。E-mail:sy.guo@siat.ac.cn于涛 (1986—),男,博士,研究员。研究方向为酿酒酵母的合成生物学。E-mail:tao.yu@siat.ac.cn -
基金资助:国家重点研发计划(2021YFA0911000);广东省重点区域研究与发展计划项目(2022B1111080005);国家自然科学基金(NSFC32071416);深圳合成生物学创新研究院科研基金(JCHZ20200003);深圳市科技计划(ZDSYS20210623091810032);中国科学院战略重点研究项目(XDB0480000);招商局集团先进技术研究院有限公司(基于电催化CO2转化与生物炼制的绿色制造项目);中海石油化学股份有限公司和海洋石油富岛有限公司“碳中和与粮食安全交叉创新联合实验室”项目;深圳先进院跨所联合攻关青年团队项目(电驱动CO2转化与生物炼制规模化示范)
CLC Number:
Cite this article
GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels[J]. Synthetic Biology Journal, 2025, 6(1): 18-44.
郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-040
| 生物燃料 | 宿主 | 主要底物 | 发酵培养基 | 产物和产量 | 主要途径 | 改造策略及相关基因 | 备注 | 参考文献 |
|---|---|---|---|---|---|---|---|---|
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 葡萄糖 | Luria-Bertani (LB) | 高级醇混合物(1.8 g/L) | 逆β氧化途径 | 表达酰基CoA还原酶(TER),硫解酶(FadA),羟基酰CoA还原酶(FADB) | ①1 L生物反应器 ②产物:丁醇、己醇、辛醇、癸醇、十二醇、十四醇、十六醇 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 葡萄糖 | 无机盐基础培养基,多种氨基酸 | 丁醇(14 g/L);3-酮丁酸(500 mg/L);脂肪酸(7 g/L) | 逆β氧化途径 | ①丁醇合成:ΔyqhD,ΔeutE,表达酰基转移酶(YQEF),丙二醇氧化还原酶(FUCO) ②羧酸(C>4)合成:ΔfadB,ΔydiO,表达硫酯酶(TESA,TESB),脂肪酰转移酶(YAQF) ③脂肪酸合成:ΔyqhD,ΔfucO,ΔfadD,表达硫酯酶(TESA,TESB,FADM,YCIA) ④长链醇(C>4):表达酰基CoA还原酶,醇脱氢酶(YIAY,BETA,EUTG) | ①生产高级醇(C>4)和脂肪酸(C>10)具有更高的效率 ②丁醇产率:0.33 g/g葡萄糖 ③脂肪酸产率:0.28 g/g葡萄糖 ④不同硫酯酶的使用可以产生不同碳链的脂肪酸 ⑤利用生物反应器生产丁醇和脂肪酸 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 葡萄糖 | Terrifc broth(TB) | 3-羟基丁酸(29.8g/L) | 逆β氧化途径 | ①多元重组酶调控系统 ②表达3-羟基丁酸酰基CoA脱氢酶(HBD),3-羟基丁酸酰基脱水酶(CRT),烯酰CoA还原酶(TER),酰基CoA酯化酶(TESB) ③表达RPOS,σ-38 | ①建立二元或多元重组酶依赖的开关调控系统用于延长菌株的复制周期 ②通过提高菌株的复制周期提高物质产量 ③5 L生物反应器 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠 杆菌 | 葡萄糖 | TB | 1-丁醇(30 g/L) | 逆β氧化途径 | ①表达烯酰CoA还原酶(TER) ②打断NADH竞争利用途径:ΔldhA,ΔadhE,ΔfrdBC ③Δpta ④表达甲酸脱氢酶(FDH) | ①厌氧发酵 ②产率:70%~88% | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠 杆菌 | 葡萄糖 | 无机盐基础培养基,多种氨基酸 | 异丁醇(23 mmol/L);1-丁醇(0.6 mmol/L) | 酮酸途径 | ①表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) ②表达缬氨酸或亮氨酸合成途径 | ①以苏氨酸、缬氨酸、异亮氨酸、亮氨酸等氨基酸生物合成途径为基础 ②供应不同底物可以产生不同化合物,如2-甲基-1-丁醇、3-甲基-1-丁醇或2-苯乙醇 | [ |
| 生物高级醇、酮,短链酸类物质 | 酿酒 酵母 | 葡萄糖 | 酵母合成培养基(SC),硫酸铜 | 异丁醇(263.2 mg/L) | 酮酸途径 | ①表达α-乙酰乳酸合酶(ALSS,Bacillus subtilis),酮醇酸还原异构酶(ILV5),二羟基酸脱氢酶(ILV3) ②表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) | ①通过引入铜诱导启动子CUP1缓解中间产物乙酰乳酸毒性 ②Delta位点多拷贝整合 ③以缬氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基、酵母合成培养基 | 异丁醇 [(635±23)mg/L];异戊醇 [(95±12)mg/L];2-甲基-1-丁醇[(118±28)mg/L] | 酮酸途径 | ①表达α-乙酰乳酸合酶(ALSS),酮醇酸还原异构酶(ILV5),二羟基酸脱氢酶(ILV3) ②表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) | ①线粒体靶向表达 ②以缬氨酸、亮氨酸、异亮氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 酿酒 酵母 | 葡萄糖 | YP | 异丁醇(2.09 g/L) | 酮酸途径 | ①乙酰乳酸合成酶(ALSS),乙酰羟基酸还原异构酶(ILV5),二醇酸脱水酶(ILV3) ②ΔILV2 ③敲除合成副产物的基因 | ①以缬氨酸合成途径为基础 ②转化率:59.55 mg/g葡萄糖 | [ |
| 生物高级醇、酮,短链酸类物质 | 谷氨酸棒状杆菌 | 葡萄糖 | CGXⅡ培养基 | 2-甲基-1-丁醇(0.37 g/L);3-甲基-1-丁醇(2.76 g/L) | 酮酸途径 | 表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) | 以缬氨酸和异亮氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 毕赤酵母 | 甘油 | 无机盐基础培养基 | 异戊醇;3-甲基-1-丁醇[(191.0±9.6) mg/L] | 酮酸途径 | ①表达乙酰乳酸合成酶(ILV2),乙酰羟基酸还原异构酶(ILV5),二醇酸脱水酶(ILV3),酮酸脱羧酶(KDC),乙醇脱氢酶(ADH) ②下调丙酮酸脱羧酶(PDC) | 通过表达缬氨酸和亮氨酸合成途径增加中间产物2-酮异戊酸的产量 | [ |
| 生物高级醇、酮,短链酸类物质 | 毕赤 酵母 | 葡萄糖/甘油 | 无机盐基础培养基 | 异丁醇(2.22 g/L);乙酸异丁酯(24 mg/L) | 酮酸途径 | ①表达缬氨酸合成途径(ILV2,ILV5,ILV3),酮酸脱羧酶(KDC),醇脱氢酶(ADH) ②表达醇氧酰基转移酶用于乙酸异丁酯合成(ATF) | 以缬氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 黄色 短杆菌(Breviba-cterium flavum) | 葡萄糖 | 无机盐培养基,酵母提取物 | 异丁醇(5362 mg/L);2-甲基-1-丁醇(1945 mg/L);3-甲基-1-丁醇(785.34 mg/L) | 酮酸途径 | ①表达酮酸脱羧酶(KDC),酮基异戊酸脱羧酶(KIVD),醇脱氢酶(ADH) ②苯丙酮酸脱羧酶(ARO10) | ①诱变结合高通量筛选 ②以亮氨酸、异亮氨酸、缬氨酸为基础合成 | [ |
| 生物高级醇、酮,短链酸类物质 | 枯草芽孢杆菌 | 葡萄糖 | LB和无机盐混合培养基 | 异丁醇(2.62 g/L);乙醇(1.2 g/L);苯乙醇(1.06 g/L) | 酮酸途径 | 乙酰乳酸合酶(ALSS),酮酸还原异构酶(ILVC),二羟酸脱水酶(ILVD),酮酸脱羧酶(KDC),醇脱氢酶(ADH) | ①以缬氨酸合成途径为基础 ②丙酮酸和磷酸烯醇式丙酮酸为乙醇和苯乙醇的前体物质 ③1 L摇瓶发酵 | [ |
| 生物高级醇、酮,短链酸类物质 | 解脂耶氏酵母(Yarrowia lipolytica) | 甘油 | YP | 异丙醇(1.94 g/L) | — | 表达丙酮酰CoA合成酶(nphT7),表达异丙醇合成酶 | ①利用该酵母生长异丙醇的最高滴度 | [ |
| ②纯甘油作为碳源可产生1.94 g/L 异丙醇;利用原油作为碳源可产生1.6 g/L异丙醇 | ||||||||
| ③5 L生物反应器 | ||||||||
| 生物高级醇、酮,短链酸类物质 | 链霉菌(Strepto-myces albus) | 葡萄糖,木糖 | — | 短链酮(C5~C7) | 聚酮合成途径 | 表达聚酮合成酶(PKS) | ①利用多结构域融合酶合成燃料 | [ |
| ②C6~C7乙基酮:>1 g/L;C5~C6甲基酮:250 mg/L | ||||||||
| ③原料为玉米秸秆 | ||||||||
| ④2 L生物反应器 | ||||||||
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌Re2133(Cupria-vidus necator) | 果糖 | 无机盐基础培养基 | 异丙醇(3.44g/L) | — | ①表达酮硫解酶(THL),CoA转移酶(CTF),乙酰乙酸脱羧酶(ADC),醇脱氢酶(ADH) | — | [ |
| ②ΔphaB,ΔphaC | ||||||||
| 萜类物质 | 紫色非硫光合细菌(Rhodobac-tercapsula-tus) | 葡萄糖 | 无机盐基础培养基,酵母提取物 | 红没药烯(1 g/L) | 类异戊二烯途径 | ①筛选红没药烯合成酶表达启动子 ②Δzwf1 ③增加NADPH:ΔgltBD,ΔphbC ④敲除FBB竞争途径 ⑤表达异源MVA途径;乙酰CoA酰基转移酶(ATOB),HMG-CoA合成酶(HMGCS),HMG-CoA还原酶(HMGCR),甲羟戊酸激酶(MK),磷酸甲羟戊酸激酶(PMK),甲羟戊酸二磷酸脱羧酶(PMD),异戊二烯二磷酸异构酶(IDI),法呢基二磷酸合酶(ISPA) | ①摇瓶产量1 g/L ②5 L生物反应器中,产量:9.8 g/L,产率>0.196 g/g葡萄糖 | [ |
| 萜类物质 | 大肠 杆菌 | 葡萄糖 | EZ-Rich,YP | 红没药烯(900 mg/L) | 类异戊二烯途径 | ①表达没药烯合成酶(TPS, Abies grandis) | — | [ |
| ②表达MVA途径:乙酰CoA酰基转移酶(ATOB),HMG-CoA合成酶(HMGCS),HMG-CoA还原酶(HMGCR),甲羟戊酸激酶(MK),磷酸甲羟戊酸激酶(PMK),甲羟戊酸二磷酸脱羧酶(PMD),异戊二烯二磷酸异构酶(IDI),法呢基二磷酸合酶(ISPA) | ||||||||
| 萜类物质 | 大肠 杆菌 | 葡萄糖 | 无机盐基础培养基 | 异戊二烯[(587±47) mg/L] | 类异戊二烯途径 | 表达MVA途径:乙酰乙酰辅酶A硫代酶(MVAE),合成酶(MVAS),激酶(MVK),磷酸甲羟戊酸激酶(PMK),二磷酸甲羟戊酸脱羧酶(MVAD),异戊烯基二磷酸异构酶(IDI),异戊二烯合酶(ISPS) | [ | |
| 萜类物质 | 大肠 杆菌 | 葡萄糖 | EZ-Rich | 柠烯(435 mg/L) | 类异戊二烯途径 | ①表达柠烯合成酶(LS),细胞色素P450 | MVA途径为基础 | [ |
| ②表达MVA途径:乙酰CoA酰基转移酶(ATOB),HMG-CoA合成酶(HMGCS),HMG-CoA还原酶(HMGCR),甲羟戊酸激酶(MK),磷酸甲羟戊酸激酶(PMK),甲羟戊酸二磷酸脱羧酶(PMD) | ||||||||
| ③香叶基焦磷酸合成酶(GPPS) | ||||||||
| 萜类物质 | 酿酒 酵母 | 葡萄糖,蔗糖 | 无机盐培养基 | 法呢烯(130 g/L) | 类异戊二烯途径 | ①表达磷酸转酮酶(XPK),磷脂酰转移酶(PTA),乙醛脱氢酶(ADA),HMG-CoA还原酶(HMGCR),法呢烯合成酶(FS) ②Δacs2,Δacs1,Δacs6,Δhr2 | ①首次在酿酒酵母中高效合成法呢烯 ②产率:17.3% g/g 葡萄糖 | [ |
| 萜类物质 | 解脂耶氏酵母(Yarrowialipolytica) | 葡萄糖 | YP | β-法呢烯(22.8 g/L) | 类异戊二烯途径 | ①表达MVA途径:HMG-CoA还原酶(HMGCR),法呢基二磷酸合成酶(ERG20),法呢烯合成酶(FS) ②∆DGA1,∆DGA2 | ①以MVA途径为基础 ②2 L生物反应器 | [ |
| 萜类物质 | 解脂耶 氏酵母(Yarrowialipolytica) | 葡萄糖 | YP | α-法呢烯(25.55 g/L) | 类异戊二烯途径 | ①表达MVA途径:乙酰CoA酰基转移酶(ATOB),HMG-CoA还原酶(HMGCR) ②法呢基二磷酸合成酶(ERG20),法呢烯合成酶(FS) | ①以MVA途径为基础 ②1 L生物反应器 | [ |
| 脂肪酸及其衍生物 | 大肠 杆菌 | 甘油 | 无机盐基础培养基,酵母提取物 | 游离脂肪酸(30 g/L) | — | ihfAL- -aidB+ - ryfAM--gadAH- | ①利用CRISPRi高通量筛选结合组学分析探究提高脂肪酸产量的靶基因 ②5 L生物反应器 | [ |
脂肪酸及其 衍生物 | 大肠 杆菌 | 葡萄糖 | 无机盐基础培养基,多种氨基酸 | 脂肪酸异丙酯(203.4 mg/L) | 脂肪酸合成途径,逆β氧化途径 | ①表达酰基CoA-酰基转移酶(ATOB),乙酰乙酰 CoA转移酶(ATOAD),乙酰乙酸脱羧酶(ADC),乙醇脱氢酶(ADH) ②硫酯酶(TESA),脂酰辅酶A合成酶(FADD),酰基转移酶(DGAT) | 逆β氧化途径和脂肪酸合成途径共同作用 | [ |
脂肪酸及其 衍生物 | 大肠 杆菌 | 甘油 | 无机盐基础培养基,酵母提取物 | 脂肪酸短链酯(1 g/L) | 酮酸途径,脂肪酸合成途径 | ①表达酮酸脱羧酶(ARO10),乙醇脱氢酶(ADH),酰基转酯酶(DGAT) ②表达硫酯酶(TESA),脂酰辅酶A合成酶(FADD) ③ΔfadE | ①酮酸途径合成短链醇,脂肪酸合成途径提供乙酰CoA,随后酯化形成终产物 ②6 L生物反应器 | [ |
脂肪酸及其 衍生物 | 大肠 杆菌 | 甘油 | 无机盐基础培养基,酵母提取物,胰蛋白胨 | 脂肪酸乙酯(813 mg/L) | 脂肪酸合成途径 | 表达酰基辅酶A:二酰基甘油酰基转移酶(ATFA) | 5 L生物反应器 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | YP | C12~C18脂肪醇(6 g/L) | 脂肪酸合成途径 | ①Δhfd1,Δadh6,Δgdh1,Δdga1 ②表达脂肪酸还原酶(FAR,Mus musculus),乙酰CoA羧化酶(ACC1),脂肪酸合成酶(FAS),脂肪酸去饱和酶(OLE1) | ①木质纤维素作为原材料 ②产率:葡萄糖最大理论转化率的20% ③2 L生物反应器 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖,半乳糖 | 无机盐基础培养基 | 超长链脂肪酸(83.5 mg/L) | 脂肪酸合成途径 | ①表达脂肪酸合成酶(FAS) ②ΔElo3,Δgal1 ③表达脂肪酸还原酶(FAR) ④表达乙酰CoA羧化酶(ACC1),延长酶(ELO1,ELO2) | ①C22脂肪酸及脂肪醇为主 ②不同链长脂肪酸表达不同的延长酶 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基 | 脂肪酸(33.4 g/L) | 脂肪酸合成途径 | ①增强乙酰CoA供应:表达丙酮酸羧化酶(PYC1),乙酰CoA羧化酶(ACC1),线粒体丙酮酸载体(MPC),柠檬酸合成酶(CIT1),柠檬酸裂解酶(ACL),胞质异柠檬酸脱氢酶(IDP2),柠檬酸穿梭蛋白(YHM2) ②加强PPP途径,降低葡萄糖磷酸异构酶(PGI1) ③降低异柠檬酸脱氢酶1(IDH1) ④Δpdc(丙酮酸脱羧酶) ⑤pyk突变 | ①葡萄糖生产脂肪酸的最高产量 ②1 L生物反应器 ③挖掘进化的关键基因并通过反向工程验证,阐明高产油机制 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基 | 中链脂肪酸[C6~C12:(1.39±0.05) g/L] | 脂肪酸合成途径 | ①工程化改造脂肪酸合成酶(FAS) ②Δhfd1 ③膜转运蛋白(TOP1)易错PCR结合进化筛选 ④菌株进化结合代谢重塑 | ①摇瓶发酵 ②产率:18.9%±0.6% | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基 | 脂肪酸(20 g/L) | 脂肪酸合成途径 | ①Δpgi,Δpdc1,Δpdc5,Δpdc6 ②增加胞质 NADH:表达谷氨酸脱氢酶(GDH1, GDH2) ③表达琥珀酸生成途径:延胡索酸酶(FUM1),苹果酸酶(tMDH3),丙酮酸羧化酶(PYC2),富马酸还原酶(FDR1) ④下调PFK1,Δpfk2 ⑤捕获胞质NADH进入呼吸链:表达NADH脱氢酶(NDE1,NDE2) ⑥表达脂肪酸合成途径:脂肪酸合成酶(FAS),硫酯酶(TESA),乙酰CoA羧化酶(ACC1) ⑦Δfaa1,Δfaa4,Δpox1 ⑧过表达PPP途径:6-磷酸葡萄糖脱氢酶(ZWF1),磷酸葡糖酸脱氢酶(GND1),转酮酶(TKL1),转醛酶(TAL1) ⑨利用不同启动子表达不同来源的果糖1,6-二磷酸酶(FBP) ⑩表达NOG途径:磷酸转酮酶(XFPK),磷脂酰转移酶(PTA) | ①利用合成的能量系统代替TCA进行能量供应,用于脂肪酸生成 ②产率:0.134 g/g 葡萄糖,40%的产率为已知报道的最高 | [ |
脂肪酸及其 衍生物 | 斯达油脂酵母(Lipomyces starkeyi),解脂耶氏酵母(Yarrowialipolytica) | 葡萄糖,木糖 | 无机盐基础培养基 | 脂肪醇 | 脂肪酸合成途径 | 表达脂肪酰辅酶A还原酶(FAR,Marinobactor aquaeolei VT8) | ①十六烷醇(C16∶0)和十八醇(C18∶0)占主导地位 ②不同底物所得的脂肪醇产量不同 | [ |
Table 1 Engineered microbial chassis to synthetic advanced biofuels
| 生物燃料 | 宿主 | 主要底物 | 发酵培养基 | 产物和产量 | 主要途径 | 改造策略及相关基因 | 备注 | 参考文献 |
|---|---|---|---|---|---|---|---|---|
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 葡萄糖 | Luria-Bertani (LB) | 高级醇混合物(1.8 g/L) | 逆β氧化途径 | 表达酰基CoA还原酶(TER),硫解酶(FadA),羟基酰CoA还原酶(FADB) | ①1 L生物反应器 ②产物:丁醇、己醇、辛醇、癸醇、十二醇、十四醇、十六醇 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 葡萄糖 | 无机盐基础培养基,多种氨基酸 | 丁醇(14 g/L);3-酮丁酸(500 mg/L);脂肪酸(7 g/L) | 逆β氧化途径 | ①丁醇合成:ΔyqhD,ΔeutE,表达酰基转移酶(YQEF),丙二醇氧化还原酶(FUCO) ②羧酸(C>4)合成:ΔfadB,ΔydiO,表达硫酯酶(TESA,TESB),脂肪酰转移酶(YAQF) ③脂肪酸合成:ΔyqhD,ΔfucO,ΔfadD,表达硫酯酶(TESA,TESB,FADM,YCIA) ④长链醇(C>4):表达酰基CoA还原酶,醇脱氢酶(YIAY,BETA,EUTG) | ①生产高级醇(C>4)和脂肪酸(C>10)具有更高的效率 ②丁醇产率:0.33 g/g葡萄糖 ③脂肪酸产率:0.28 g/g葡萄糖 ④不同硫酯酶的使用可以产生不同碳链的脂肪酸 ⑤利用生物反应器生产丁醇和脂肪酸 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 葡萄糖 | Terrifc broth(TB) | 3-羟基丁酸(29.8g/L) | 逆β氧化途径 | ①多元重组酶调控系统 ②表达3-羟基丁酸酰基CoA脱氢酶(HBD),3-羟基丁酸酰基脱水酶(CRT),烯酰CoA还原酶(TER),酰基CoA酯化酶(TESB) ③表达RPOS,σ-38 | ①建立二元或多元重组酶依赖的开关调控系统用于延长菌株的复制周期 ②通过提高菌株的复制周期提高物质产量 ③5 L生物反应器 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠 杆菌 | 葡萄糖 | TB | 1-丁醇(30 g/L) | 逆β氧化途径 | ①表达烯酰CoA还原酶(TER) ②打断NADH竞争利用途径:ΔldhA,ΔadhE,ΔfrdBC ③Δpta ④表达甲酸脱氢酶(FDH) | ①厌氧发酵 ②产率:70%~88% | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠 杆菌 | 葡萄糖 | 无机盐基础培养基,多种氨基酸 | 异丁醇(23 mmol/L);1-丁醇(0.6 mmol/L) | 酮酸途径 | ①表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) ②表达缬氨酸或亮氨酸合成途径 | ①以苏氨酸、缬氨酸、异亮氨酸、亮氨酸等氨基酸生物合成途径为基础 ②供应不同底物可以产生不同化合物,如2-甲基-1-丁醇、3-甲基-1-丁醇或2-苯乙醇 | [ |
| 生物高级醇、酮,短链酸类物质 | 酿酒 酵母 | 葡萄糖 | 酵母合成培养基(SC),硫酸铜 | 异丁醇(263.2 mg/L) | 酮酸途径 | ①表达α-乙酰乳酸合酶(ALSS,Bacillus subtilis),酮醇酸还原异构酶(ILV5),二羟基酸脱氢酶(ILV3) ②表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) | ①通过引入铜诱导启动子CUP1缓解中间产物乙酰乳酸毒性 ②Delta位点多拷贝整合 ③以缬氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基、酵母合成培养基 | 异丁醇 [(635±23)mg/L];异戊醇 [(95±12)mg/L];2-甲基-1-丁醇[(118±28)mg/L] | 酮酸途径 | ①表达α-乙酰乳酸合酶(ALSS),酮醇酸还原异构酶(ILV5),二羟基酸脱氢酶(ILV3) ②表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) | ①线粒体靶向表达 ②以缬氨酸、亮氨酸、异亮氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 酿酒 酵母 | 葡萄糖 | YP | 异丁醇(2.09 g/L) | 酮酸途径 | ①乙酰乳酸合成酶(ALSS),乙酰羟基酸还原异构酶(ILV5),二醇酸脱水酶(ILV3) ②ΔILV2 ③敲除合成副产物的基因 | ①以缬氨酸合成途径为基础 ②转化率:59.55 mg/g葡萄糖 | [ |
| 生物高级醇、酮,短链酸类物质 | 谷氨酸棒状杆菌 | 葡萄糖 | CGXⅡ培养基 | 2-甲基-1-丁醇(0.37 g/L);3-甲基-1-丁醇(2.76 g/L) | 酮酸途径 | 表达酮酸脱羧酶(KDC),醇脱氢酶(ADH) | 以缬氨酸和异亮氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 毕赤酵母 | 甘油 | 无机盐基础培养基 | 异戊醇;3-甲基-1-丁醇[(191.0±9.6) mg/L] | 酮酸途径 | ①表达乙酰乳酸合成酶(ILV2),乙酰羟基酸还原异构酶(ILV5),二醇酸脱水酶(ILV3),酮酸脱羧酶(KDC),乙醇脱氢酶(ADH) ②下调丙酮酸脱羧酶(PDC) | 通过表达缬氨酸和亮氨酸合成途径增加中间产物2-酮异戊酸的产量 | [ |
| 生物高级醇、酮,短链酸类物质 | 毕赤 酵母 | 葡萄糖/甘油 | 无机盐基础培养基 | 异丁醇(2.22 g/L);乙酸异丁酯(24 mg/L) | 酮酸途径 | ①表达缬氨酸合成途径(ILV2,ILV5,ILV3),酮酸脱羧酶(KDC),醇脱氢酶(ADH) ②表达醇氧酰基转移酶用于乙酸异丁酯合成(ATF) | 以缬氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 黄色 短杆菌(Breviba-cterium flavum) | 葡萄糖 | 无机盐培养基,酵母提取物 | 异丁醇(5362 mg/L);2-甲基-1-丁醇(1945 mg/L);3-甲基-1-丁醇(785.34 mg/L) | 酮酸途径 | ①表达酮酸脱羧酶(KDC),酮基异戊酸脱羧酶(KIVD),醇脱氢酶(ADH) ②苯丙酮酸脱羧酶(ARO10) | ①诱变结合高通量筛选 ②以亮氨酸、异亮氨酸、缬氨酸为基础合成 | [ |
| 生物高级醇、酮,短链酸类物质 | 枯草芽孢杆菌 | 葡萄糖 | LB和无机盐混合培养基 | 异丁醇(2.62 g/L);乙醇(1.2 g/L);苯乙醇(1.06 g/L) | 酮酸途径 | 乙酰乳酸合酶(ALSS),酮酸还原异构酶(ILVC),二羟酸脱水酶(ILVD),酮酸脱羧酶(KDC),醇脱氢酶(ADH) | ①以缬氨酸合成途径为基础 ②丙酮酸和磷酸烯醇式丙酮酸为乙醇和苯乙醇的前体物质 ③1 L摇瓶发酵 | [ |
| 生物高级醇、酮,短链酸类物质 | 解脂耶氏酵母(Yarrowia lipolytica) | 甘油 | YP | 异丙醇(1.94 g/L) | — | 表达丙酮酰CoA合成酶(nphT7),表达异丙醇合成酶 | ①利用该酵母生长异丙醇的最高滴度 | [ |
| ②纯甘油作为碳源可产生1.94 g/L 异丙醇;利用原油作为碳源可产生1.6 g/L异丙醇 | ||||||||
| ③5 L生物反应器 | ||||||||
| 生物高级醇、酮,短链酸类物质 | 链霉菌(Strepto-myces albus) | 葡萄糖,木糖 | — | 短链酮(C5~C7) | 聚酮合成途径 | 表达聚酮合成酶(PKS) | ①利用多结构域融合酶合成燃料 | [ |
| ②C6~C7乙基酮:>1 g/L;C5~C6甲基酮:250 mg/L | ||||||||
| ③原料为玉米秸秆 | ||||||||
| ④2 L生物反应器 | ||||||||
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌Re2133(Cupria-vidus necator) | 果糖 | 无机盐基础培养基 | 异丙醇(3.44g/L) | — | ①表达酮硫解酶(THL),CoA转移酶(CTF),乙酰乙酸脱羧酶(ADC),醇脱氢酶(ADH) | — | [ |
| ②ΔphaB,ΔphaC | ||||||||
| 萜类物质 | 紫色非硫光合细菌(Rhodobac-tercapsula-tus) | 葡萄糖 | 无机盐基础培养基,酵母提取物 | 红没药烯(1 g/L) | 类异戊二烯途径 | ①筛选红没药烯合成酶表达启动子 ②Δzwf1 ③增加NADPH:ΔgltBD,ΔphbC ④敲除FBB竞争途径 ⑤表达异源MVA途径;乙酰CoA酰基转移酶(ATOB),HMG-CoA合成酶(HMGCS),HMG-CoA还原酶(HMGCR),甲羟戊酸激酶(MK),磷酸甲羟戊酸激酶(PMK),甲羟戊酸二磷酸脱羧酶(PMD),异戊二烯二磷酸异构酶(IDI),法呢基二磷酸合酶(ISPA) | ①摇瓶产量1 g/L ②5 L生物反应器中,产量:9.8 g/L,产率>0.196 g/g葡萄糖 | [ |
| 萜类物质 | 大肠 杆菌 | 葡萄糖 | EZ-Rich,YP | 红没药烯(900 mg/L) | 类异戊二烯途径 | ①表达没药烯合成酶(TPS, Abies grandis) | — | [ |
| ②表达MVA途径:乙酰CoA酰基转移酶(ATOB),HMG-CoA合成酶(HMGCS),HMG-CoA还原酶(HMGCR),甲羟戊酸激酶(MK),磷酸甲羟戊酸激酶(PMK),甲羟戊酸二磷酸脱羧酶(PMD),异戊二烯二磷酸异构酶(IDI),法呢基二磷酸合酶(ISPA) | ||||||||
| 萜类物质 | 大肠 杆菌 | 葡萄糖 | 无机盐基础培养基 | 异戊二烯[(587±47) mg/L] | 类异戊二烯途径 | 表达MVA途径:乙酰乙酰辅酶A硫代酶(MVAE),合成酶(MVAS),激酶(MVK),磷酸甲羟戊酸激酶(PMK),二磷酸甲羟戊酸脱羧酶(MVAD),异戊烯基二磷酸异构酶(IDI),异戊二烯合酶(ISPS) | [ | |
| 萜类物质 | 大肠 杆菌 | 葡萄糖 | EZ-Rich | 柠烯(435 mg/L) | 类异戊二烯途径 | ①表达柠烯合成酶(LS),细胞色素P450 | MVA途径为基础 | [ |
| ②表达MVA途径:乙酰CoA酰基转移酶(ATOB),HMG-CoA合成酶(HMGCS),HMG-CoA还原酶(HMGCR),甲羟戊酸激酶(MK),磷酸甲羟戊酸激酶(PMK),甲羟戊酸二磷酸脱羧酶(PMD) | ||||||||
| ③香叶基焦磷酸合成酶(GPPS) | ||||||||
| 萜类物质 | 酿酒 酵母 | 葡萄糖,蔗糖 | 无机盐培养基 | 法呢烯(130 g/L) | 类异戊二烯途径 | ①表达磷酸转酮酶(XPK),磷脂酰转移酶(PTA),乙醛脱氢酶(ADA),HMG-CoA还原酶(HMGCR),法呢烯合成酶(FS) ②Δacs2,Δacs1,Δacs6,Δhr2 | ①首次在酿酒酵母中高效合成法呢烯 ②产率:17.3% g/g 葡萄糖 | [ |
| 萜类物质 | 解脂耶氏酵母(Yarrowialipolytica) | 葡萄糖 | YP | β-法呢烯(22.8 g/L) | 类异戊二烯途径 | ①表达MVA途径:HMG-CoA还原酶(HMGCR),法呢基二磷酸合成酶(ERG20),法呢烯合成酶(FS) ②∆DGA1,∆DGA2 | ①以MVA途径为基础 ②2 L生物反应器 | [ |
| 萜类物质 | 解脂耶 氏酵母(Yarrowialipolytica) | 葡萄糖 | YP | α-法呢烯(25.55 g/L) | 类异戊二烯途径 | ①表达MVA途径:乙酰CoA酰基转移酶(ATOB),HMG-CoA还原酶(HMGCR) ②法呢基二磷酸合成酶(ERG20),法呢烯合成酶(FS) | ①以MVA途径为基础 ②1 L生物反应器 | [ |
| 脂肪酸及其衍生物 | 大肠 杆菌 | 甘油 | 无机盐基础培养基,酵母提取物 | 游离脂肪酸(30 g/L) | — | ihfAL- -aidB+ - ryfAM--gadAH- | ①利用CRISPRi高通量筛选结合组学分析探究提高脂肪酸产量的靶基因 ②5 L生物反应器 | [ |
脂肪酸及其 衍生物 | 大肠 杆菌 | 葡萄糖 | 无机盐基础培养基,多种氨基酸 | 脂肪酸异丙酯(203.4 mg/L) | 脂肪酸合成途径,逆β氧化途径 | ①表达酰基CoA-酰基转移酶(ATOB),乙酰乙酰 CoA转移酶(ATOAD),乙酰乙酸脱羧酶(ADC),乙醇脱氢酶(ADH) ②硫酯酶(TESA),脂酰辅酶A合成酶(FADD),酰基转移酶(DGAT) | 逆β氧化途径和脂肪酸合成途径共同作用 | [ |
脂肪酸及其 衍生物 | 大肠 杆菌 | 甘油 | 无机盐基础培养基,酵母提取物 | 脂肪酸短链酯(1 g/L) | 酮酸途径,脂肪酸合成途径 | ①表达酮酸脱羧酶(ARO10),乙醇脱氢酶(ADH),酰基转酯酶(DGAT) ②表达硫酯酶(TESA),脂酰辅酶A合成酶(FADD) ③ΔfadE | ①酮酸途径合成短链醇,脂肪酸合成途径提供乙酰CoA,随后酯化形成终产物 ②6 L生物反应器 | [ |
脂肪酸及其 衍生物 | 大肠 杆菌 | 甘油 | 无机盐基础培养基,酵母提取物,胰蛋白胨 | 脂肪酸乙酯(813 mg/L) | 脂肪酸合成途径 | 表达酰基辅酶A:二酰基甘油酰基转移酶(ATFA) | 5 L生物反应器 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | YP | C12~C18脂肪醇(6 g/L) | 脂肪酸合成途径 | ①Δhfd1,Δadh6,Δgdh1,Δdga1 ②表达脂肪酸还原酶(FAR,Mus musculus),乙酰CoA羧化酶(ACC1),脂肪酸合成酶(FAS),脂肪酸去饱和酶(OLE1) | ①木质纤维素作为原材料 ②产率:葡萄糖最大理论转化率的20% ③2 L生物反应器 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖,半乳糖 | 无机盐基础培养基 | 超长链脂肪酸(83.5 mg/L) | 脂肪酸合成途径 | ①表达脂肪酸合成酶(FAS) ②ΔElo3,Δgal1 ③表达脂肪酸还原酶(FAR) ④表达乙酰CoA羧化酶(ACC1),延长酶(ELO1,ELO2) | ①C22脂肪酸及脂肪醇为主 ②不同链长脂肪酸表达不同的延长酶 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基 | 脂肪酸(33.4 g/L) | 脂肪酸合成途径 | ①增强乙酰CoA供应:表达丙酮酸羧化酶(PYC1),乙酰CoA羧化酶(ACC1),线粒体丙酮酸载体(MPC),柠檬酸合成酶(CIT1),柠檬酸裂解酶(ACL),胞质异柠檬酸脱氢酶(IDP2),柠檬酸穿梭蛋白(YHM2) ②加强PPP途径,降低葡萄糖磷酸异构酶(PGI1) ③降低异柠檬酸脱氢酶1(IDH1) ④Δpdc(丙酮酸脱羧酶) ⑤pyk突变 | ①葡萄糖生产脂肪酸的最高产量 ②1 L生物反应器 ③挖掘进化的关键基因并通过反向工程验证,阐明高产油机制 | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基 | 中链脂肪酸[C6~C12:(1.39±0.05) g/L] | 脂肪酸合成途径 | ①工程化改造脂肪酸合成酶(FAS) ②Δhfd1 ③膜转运蛋白(TOP1)易错PCR结合进化筛选 ④菌株进化结合代谢重塑 | ①摇瓶发酵 ②产率:18.9%±0.6% | [ |
脂肪酸及其 衍生物 | 酿酒 酵母 | 葡萄糖 | 无机盐基础培养基 | 脂肪酸(20 g/L) | 脂肪酸合成途径 | ①Δpgi,Δpdc1,Δpdc5,Δpdc6 ②增加胞质 NADH:表达谷氨酸脱氢酶(GDH1, GDH2) ③表达琥珀酸生成途径:延胡索酸酶(FUM1),苹果酸酶(tMDH3),丙酮酸羧化酶(PYC2),富马酸还原酶(FDR1) ④下调PFK1,Δpfk2 ⑤捕获胞质NADH进入呼吸链:表达NADH脱氢酶(NDE1,NDE2) ⑥表达脂肪酸合成途径:脂肪酸合成酶(FAS),硫酯酶(TESA),乙酰CoA羧化酶(ACC1) ⑦Δfaa1,Δfaa4,Δpox1 ⑧过表达PPP途径:6-磷酸葡萄糖脱氢酶(ZWF1),磷酸葡糖酸脱氢酶(GND1),转酮酶(TKL1),转醛酶(TAL1) ⑨利用不同启动子表达不同来源的果糖1,6-二磷酸酶(FBP) ⑩表达NOG途径:磷酸转酮酶(XFPK),磷脂酰转移酶(PTA) | ①利用合成的能量系统代替TCA进行能量供应,用于脂肪酸生成 ②产率:0.134 g/g 葡萄糖,40%的产率为已知报道的最高 | [ |
脂肪酸及其 衍生物 | 斯达油脂酵母(Lipomyces starkeyi),解脂耶氏酵母(Yarrowialipolytica) | 葡萄糖,木糖 | 无机盐基础培养基 | 脂肪醇 | 脂肪酸合成途径 | 表达脂肪酰辅酶A还原酶(FAR,Marinobactor aquaeolei VT8) | ①十六烷醇(C16∶0)和十八醇(C18∶0)占主导地位 ②不同底物所得的脂肪醇产量不同 | [ |
| 生物燃料 | 宿主 | 主要底物 | 发酵培养基 | 产物和产量 | 主要途径 | 改造策略及相关基因 | 备注 | 参考文献 |
|---|---|---|---|---|---|---|---|---|
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇 | 木糖,核糖 | 乙醇(4.6 g/L);1-丁醇(2 g/L) | RuMP,逆β氧化途径 | ①表达RuMP相关酶 ②ΔAdhE(甲醛脱氢酶),Δald(乙醛脱氢酶),ΔrpiAB(核糖磷酸异构酶) ③表达腺苷酸环化酶 ④表达丙酮酸脱羧酶(PDC),乙醛脱氢酶(ADH) ⑤表达丁醇合成途径 | ①构建甲醇依赖型木糖菌株 ②甲醇与木糖摩尔利用率为1∶1 ③RuMP和逆β氧化途径共同作用 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇,甲醛 | 葡萄糖,硫酸素焦磷酸盐 | 1,3-丙二醇[(508.3±9.1) mg/L] | 一磷酸核酮糖途径(RuMP),酮酸途径 | ①表达甲醇脱氢酶 ②ΔfrmA(甲醛脱氢酶) ③表达羟丁酸醛缩酶,酮酸脱羧酶,1,3-丁二酸氧化还原酶 | ①首次实现利用甲醇和丙酮酸合成1,3-丙二醇 ②缩短途径,并有效提高1,3-丙二醇产量 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇 | 葡萄糖,酵母提取物 | 丙酮(13 mmol/L) | RuMP,酮酸途径 | ①Δpgi(6-磷酸葡萄糖异构酶),Δedd(磷酸葡萄糖酸脱氢酶),ΔrpiAB(核糖磷酸异构酶),ΔfrmA(甲醛脱氢酶) ②表达RuMP相关酶 ③表达丙酮生成途径(Clostridium acetobutylicum) | ①显著提升甲醇向丙酮的转化 ②构建了甲醇依赖的菌株底盘 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇 | 葡萄糖,酵母提取物 | 丙酮[(45.0±8.7)mmol/L] | RuMP,酮酸途径 | ①Δpgi(6-磷酸葡萄糖异构酶)、ΔfrmA(甲醛脱氢酶) ②表达RuMP途径相关酶 ③表达磷酸核糖异构酶(RPE),转酮酶(TKT) ④表达二磷酸果糖醛缩酶(FBA),景天庚糖双磷酸酶(GLPX),磷酸果糖激酶(PFK) ⑤表达丙酮生成途径(C.acetobutylicum):硫解酶(THL),辅酶A转移酶(CTFAB),乙酰乙酸脱羧酶(ADC) | ①两种策略共同提高甲醇利用率 ②大肠杆菌利用甲醇合成丙酮 | [ |
| 生物高级醇、酮,短链酸类物质 | 扭脱甲基杆菌AM1(Methylobact-erium extorquens) | 甲醇 | 无机盐基础培养基 | 异丁醇(19 mg/L) | 丝氨酸循环,EMC途径,酮酸途径 | ①ΔldhA | 摇瓶培养 | [ |
| ②表达2-酮异戊酸脱羧酶(Lactococcus lactis),醇脱氢酶(Lactococcus lactis),乙酰乳酸合酶(Bacillus subtilis) | ||||||||
| 生物高级醇、酮,短链酸类物质 | 扭脱甲基杆菌AM1(Methylobact-erium extorquens) | 甲醇 | 无机盐基础培养基 | 1-丁醇(25.5 mg/L) | 丝氨酸循环,EMC途径,酮酸途径 | 表达烯酰辅酶A还原酶(Treponema denticola),乙醇脱氢酶(Clostridium acetobutylicum),巴豆酸酶(Methylobacterium extorquens AM1) | ①适应性进化筛选突变株耐受丁醇达到0.5% | [ |
| ②摇瓶培养 | ||||||||
| 生物高级醇、酮,短链酸类物质 | 扭脱甲基杆菌AM1(Methylobact-erium extorquens) | 甲醇 | 无机盐基础培养基 | 3-羟基丙酸(0.857 g/L) | RuMP途径,EMC途径 | ①ΔhprA ②表达己糖磷酸合成酶(Bacillus methanolicus),磷酸己糖异构酶(Bacillus methanolicus),磷酸果糖激酶(Bacillus methanolicus),6-磷酸葡萄糖脱氢酶(Bacillus methanolicus);③丙二酰辅酶A还原酶(Chlorofexus aurantiacus) | 5 L生物反应器 | [ |
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌H16 | CO2 | 果糖,无机盐基础培养基 | 高级醇混合物(140 mg/L) | 酮酸途径 | ①表达α-乙酰乳酸合酶(ALSS,Bacillus subtilis),酮醇酸还原异构酶(ILVC),二羟基酸脱氢酶(ILVD) ②敲除PHB合成基因:ΔphaB,ΔphaC | ①电催化产生甲酸,甲酸经由微生物转化为异丁醇或3-甲基-1-丁醇 ②以缬氨酸和亮氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌Re2133(Cupriavidus necator) | H2,O2,CO2,N2 | 果糖,无机盐基础培养基 | 异丙醇(3.5 g/L) | 酮酸途径 | 异丙醇产生菌株 | ①多气体供给的加压生物反应器 ②首次报道工程化自养菌利用CO2产生克级别的化合物 ③70%~80%的CO2被回收 | [ |
| 生物高级醇、酮,短链酸类物质 | 杨氏梭菌(Clostridium ljungdahlii) | CO2,H2 | — | 丁醇(109 mg/L);己醇(393 mg/L) | 还原型乙酰CoA途径,酮酸途径 | ①表达硫解酶(THLA),羟基丁基CoA脱氢酶(HBD),巴豆酸酶(CRT),丁基CoA脱氢酶(BCD), ②表达电子转移蛋白(ETF),醛醇脱氢酶(ADHE) | ①微生物可以利用CO和CO2作为碳源 ②CO2和H2作为碳源 ③2 L生物反应器 | [ |
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌(Ralstonia eutropha) | CO2,H2O | 无机盐基础培养基 | 异丙醇(600 mg/L);异丁醇+3-甲基-1-丁醇(220 mg/L) | 酮酸途径 | 异丙醇产生菌株 | ①CO2在水电混合反应装置中转化为H2,微生物利用H2产生异丁醇等生物燃料 ②CO2还原效率达到10%,超过自然光合效率 | [ |
| 脂肪酸及其衍生物 | 酿酒酵母 | CO2 | 无机盐基础培养基,乙酸 | 脂肪酸(500 mg/L) | 脂肪酸合成途径 | ①Δfaa1,Δfaa4,Δpox1 | 电催化和生物系统结合:CO2经电催化合成乙酸,酿酒酵母利用乙酸合成长链化合物 | [ |
| ②表达硫酯酶(TESA),酰基CoA羧化酶(ACC1),脂肪酸合成酶(FAS) | ||||||||
| 脂肪酸及其衍生物 | 毕赤酵母 | 甲醇 | 无机盐培养基 | 脂肪酸(23.4g/L);脂肪醇(2.0 g/L) | 脂肪酸合成途径 | ①Δfaa1,Δfaa4,Δpox1 ②加强甲醇利用途径:过表达二酰丙酮磷酸合酶(DAS) ③增加乙酰CoA前体供应:过表达酰基磷酸转移酶(PTA),磷酸转酮酶(XFPK) ④加强NADPH再生 | ①成功利用甲醇作为唯一碳源合成脂肪酸 ②1 L生物反应器 | [ |
| 脂肪酸及其衍生物 | 富养罗尔斯通氏菌(Ralstonia eutroph) | H2,CO2,O2 | 果糖,无机盐基础培养基 | 脂肪酸(124.48 mg/g 果糖)(60.64 mg/g CO2) | 脂肪酸合成途径 | ①ΔphaC ②表达脂肪酸合成酶(FAS),硫酯酶(TESA),乙酰CoA羧化酶(ACC1) ③ACP合成酶(ACPS) | 结合多气体生物反应器,自养菌利用CO2生成脂肪酸 | [ |
| 脂肪酸及其衍生物 | 汉逊酵母(Ogataeapolym orpha) | 甲醇 | 无机盐培养基 | 脂肪酸(15.9 g/L) | 脂肪酸合成途径 | ①Δfaa1 ②加强前体供应及辅因子供应:过表达果糖-1,6-二磷酸酶(FBP),磷酸核糖异构酶(RPE),柠檬酸裂解酶(ACL),异柠檬酸脱氢酶(ICL1),果糖6-磷酸脱氢酶(ZWF1) | ①适应性进化使得敲除细胞生长恢复,并解析机制是由于LPL1和IZH3缺失引起 ②1 L生物反应器 | [ |
| 脂肪酸及其衍生物 | 蓝藻(Synechocystis sp. PCC 6803) | CO2 | — | 脂肪酸甲酯(120 mg/L) | — | ①Δaas ②过表达硫酯酶(UcFatB1),O-甲基转移酶(DmJHAMT) ③引入S-腺苷甲硫氨酸(SAM)循环供应甲基 | ①不利用甲醇作为甲基供体,利用SAM合成酶供应甲基 ②可能产生的脂肪酸甲酯类物质:C12∶0,C14∶0,C16∶0 | [ |
| 脂肪酸及其衍生物 | 解脂耶氏酵母(Yarrowia lipolytica) | CO2 | 酵母合成培养基,酵母提取物 | 脂肪酸(10.7 g/L) | — | ①增强脂肪酸合成:过表达生物素羧化酶(BC) ②引入CO2利用途径:过表达碳酸酐酶(CA) | ①循环利用CO2生产脂肪酸 ②250 mL摇瓶发酵 | [ |
| 萜类物质 | 扭脱甲基杆菌AM1(Methylobact-eriumextorquens) | 甲醇 | 无机盐基础培养基 | 甲羟戊酸(2.22 g/L) | 丝氨酸循环,EMC途径,类异戊二烯途径 | 表达HMG-CoA合成酶(Enterococcus faecalistiters),HMG-CoA还原酶(Enterococcus faecalistiters),乙酰乙酰CoA硫解酶(Ralstonia eutropha) | ①产率:28.4 mg/g甲醇 ②5 L生物反应器 | [ |
| 萜类物质 | 类黄色噬氢菌DSM1084(Hydrogenop-haga pseudoflava) | CO2,合成气 | 醋酸盐,果糖,蔗糖等,无机盐基础培养基 | α-红没药烯[(59.0±7.9) μg] | 卡尔文循环,Wood-Ljungdahl途径(WL),类异戊二烯途径 | 表达没药烯合成酶(TPS,Abies grandis) | ①自养和异养条件皆可生长:自养条件下利用合成气作为碳源,异养条件下可以利用果糖、蔗糖等作为碳源 | [ |
| ②自养条件利用卡尔文循环和WL途径,异养条件利用MEP途径 | ||||||||
| 萜类物质 | Cupriavidus necator | CO2,H2,O2 | 果糖,无机盐基础培养基 | α-蛇麻烯[(10.8±2.5)mg/g DCW 或17 mg/g DCW] | 类异戊二烯途径 | 表达MVA途径:焦磷酸法呢合成酶(ERG20),IPP异构酶,α-蛇麻烯合酶(ZSSI) | 化学自养和电自养均可:化能自养主要利用CO2等,电自养需要电极和水辅助 | [ |
| 萜类物质 | 嗜甲烷菌20Z(Methylomicr-obium alcaliphilum) | 50%甲烷 | 硝酸矿物盐培养基(NMS) | α-蛇麻烯(0.75 mg/g DCW,835 μg/L) | 类异戊二烯途径 | ①表达α-蛇麻烯合成酶(ZSS1) ②表达1-脱氧木酮糖-5-磷酸合酶(DXS),HMBPP合成酶(ISPG),FPP合成酶(ISPA) ③Δpgi ④提高NADPH:表达转氢酶(PNTAB),葡萄糖6-磷酸脱氢酶(ZWF),磷酸葡糖酸脱氢酶(PGD) | 优化MEP途径 | [ |
Table 2 Engineered microbial chassis to synthetic advanced biofuels derived from C1 substrates
| 生物燃料 | 宿主 | 主要底物 | 发酵培养基 | 产物和产量 | 主要途径 | 改造策略及相关基因 | 备注 | 参考文献 |
|---|---|---|---|---|---|---|---|---|
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇 | 木糖,核糖 | 乙醇(4.6 g/L);1-丁醇(2 g/L) | RuMP,逆β氧化途径 | ①表达RuMP相关酶 ②ΔAdhE(甲醛脱氢酶),Δald(乙醛脱氢酶),ΔrpiAB(核糖磷酸异构酶) ③表达腺苷酸环化酶 ④表达丙酮酸脱羧酶(PDC),乙醛脱氢酶(ADH) ⑤表达丁醇合成途径 | ①构建甲醇依赖型木糖菌株 ②甲醇与木糖摩尔利用率为1∶1 ③RuMP和逆β氧化途径共同作用 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇,甲醛 | 葡萄糖,硫酸素焦磷酸盐 | 1,3-丙二醇[(508.3±9.1) mg/L] | 一磷酸核酮糖途径(RuMP),酮酸途径 | ①表达甲醇脱氢酶 ②ΔfrmA(甲醛脱氢酶) ③表达羟丁酸醛缩酶,酮酸脱羧酶,1,3-丁二酸氧化还原酶 | ①首次实现利用甲醇和丙酮酸合成1,3-丙二醇 ②缩短途径,并有效提高1,3-丙二醇产量 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇 | 葡萄糖,酵母提取物 | 丙酮(13 mmol/L) | RuMP,酮酸途径 | ①Δpgi(6-磷酸葡萄糖异构酶),Δedd(磷酸葡萄糖酸脱氢酶),ΔrpiAB(核糖磷酸异构酶),ΔfrmA(甲醛脱氢酶) ②表达RuMP相关酶 ③表达丙酮生成途径(Clostridium acetobutylicum) | ①显著提升甲醇向丙酮的转化 ②构建了甲醇依赖的菌株底盘 | [ |
| 生物高级醇、酮,短链酸类物质 | 大肠杆菌 | 甲醇 | 葡萄糖,酵母提取物 | 丙酮[(45.0±8.7)mmol/L] | RuMP,酮酸途径 | ①Δpgi(6-磷酸葡萄糖异构酶)、ΔfrmA(甲醛脱氢酶) ②表达RuMP途径相关酶 ③表达磷酸核糖异构酶(RPE),转酮酶(TKT) ④表达二磷酸果糖醛缩酶(FBA),景天庚糖双磷酸酶(GLPX),磷酸果糖激酶(PFK) ⑤表达丙酮生成途径(C.acetobutylicum):硫解酶(THL),辅酶A转移酶(CTFAB),乙酰乙酸脱羧酶(ADC) | ①两种策略共同提高甲醇利用率 ②大肠杆菌利用甲醇合成丙酮 | [ |
| 生物高级醇、酮,短链酸类物质 | 扭脱甲基杆菌AM1(Methylobact-erium extorquens) | 甲醇 | 无机盐基础培养基 | 异丁醇(19 mg/L) | 丝氨酸循环,EMC途径,酮酸途径 | ①ΔldhA | 摇瓶培养 | [ |
| ②表达2-酮异戊酸脱羧酶(Lactococcus lactis),醇脱氢酶(Lactococcus lactis),乙酰乳酸合酶(Bacillus subtilis) | ||||||||
| 生物高级醇、酮,短链酸类物质 | 扭脱甲基杆菌AM1(Methylobact-erium extorquens) | 甲醇 | 无机盐基础培养基 | 1-丁醇(25.5 mg/L) | 丝氨酸循环,EMC途径,酮酸途径 | 表达烯酰辅酶A还原酶(Treponema denticola),乙醇脱氢酶(Clostridium acetobutylicum),巴豆酸酶(Methylobacterium extorquens AM1) | ①适应性进化筛选突变株耐受丁醇达到0.5% | [ |
| ②摇瓶培养 | ||||||||
| 生物高级醇、酮,短链酸类物质 | 扭脱甲基杆菌AM1(Methylobact-erium extorquens) | 甲醇 | 无机盐基础培养基 | 3-羟基丙酸(0.857 g/L) | RuMP途径,EMC途径 | ①ΔhprA ②表达己糖磷酸合成酶(Bacillus methanolicus),磷酸己糖异构酶(Bacillus methanolicus),磷酸果糖激酶(Bacillus methanolicus),6-磷酸葡萄糖脱氢酶(Bacillus methanolicus);③丙二酰辅酶A还原酶(Chlorofexus aurantiacus) | 5 L生物反应器 | [ |
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌H16 | CO2 | 果糖,无机盐基础培养基 | 高级醇混合物(140 mg/L) | 酮酸途径 | ①表达α-乙酰乳酸合酶(ALSS,Bacillus subtilis),酮醇酸还原异构酶(ILVC),二羟基酸脱氢酶(ILVD) ②敲除PHB合成基因:ΔphaB,ΔphaC | ①电催化产生甲酸,甲酸经由微生物转化为异丁醇或3-甲基-1-丁醇 ②以缬氨酸和亮氨酸合成途径为基础 | [ |
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌Re2133(Cupriavidus necator) | H2,O2,CO2,N2 | 果糖,无机盐基础培养基 | 异丙醇(3.5 g/L) | 酮酸途径 | 异丙醇产生菌株 | ①多气体供给的加压生物反应器 ②首次报道工程化自养菌利用CO2产生克级别的化合物 ③70%~80%的CO2被回收 | [ |
| 生物高级醇、酮,短链酸类物质 | 杨氏梭菌(Clostridium ljungdahlii) | CO2,H2 | — | 丁醇(109 mg/L);己醇(393 mg/L) | 还原型乙酰CoA途径,酮酸途径 | ①表达硫解酶(THLA),羟基丁基CoA脱氢酶(HBD),巴豆酸酶(CRT),丁基CoA脱氢酶(BCD), ②表达电子转移蛋白(ETF),醛醇脱氢酶(ADHE) | ①微生物可以利用CO和CO2作为碳源 ②CO2和H2作为碳源 ③2 L生物反应器 | [ |
| 生物高级醇、酮,短链酸类物质 | 富养罗尔斯通氏菌(Ralstonia eutropha) | CO2,H2O | 无机盐基础培养基 | 异丙醇(600 mg/L);异丁醇+3-甲基-1-丁醇(220 mg/L) | 酮酸途径 | 异丙醇产生菌株 | ①CO2在水电混合反应装置中转化为H2,微生物利用H2产生异丁醇等生物燃料 ②CO2还原效率达到10%,超过自然光合效率 | [ |
| 脂肪酸及其衍生物 | 酿酒酵母 | CO2 | 无机盐基础培养基,乙酸 | 脂肪酸(500 mg/L) | 脂肪酸合成途径 | ①Δfaa1,Δfaa4,Δpox1 | 电催化和生物系统结合:CO2经电催化合成乙酸,酿酒酵母利用乙酸合成长链化合物 | [ |
| ②表达硫酯酶(TESA),酰基CoA羧化酶(ACC1),脂肪酸合成酶(FAS) | ||||||||
| 脂肪酸及其衍生物 | 毕赤酵母 | 甲醇 | 无机盐培养基 | 脂肪酸(23.4g/L);脂肪醇(2.0 g/L) | 脂肪酸合成途径 | ①Δfaa1,Δfaa4,Δpox1 ②加强甲醇利用途径:过表达二酰丙酮磷酸合酶(DAS) ③增加乙酰CoA前体供应:过表达酰基磷酸转移酶(PTA),磷酸转酮酶(XFPK) ④加强NADPH再生 | ①成功利用甲醇作为唯一碳源合成脂肪酸 ②1 L生物反应器 | [ |
| 脂肪酸及其衍生物 | 富养罗尔斯通氏菌(Ralstonia eutroph) | H2,CO2,O2 | 果糖,无机盐基础培养基 | 脂肪酸(124.48 mg/g 果糖)(60.64 mg/g CO2) | 脂肪酸合成途径 | ①ΔphaC ②表达脂肪酸合成酶(FAS),硫酯酶(TESA),乙酰CoA羧化酶(ACC1) ③ACP合成酶(ACPS) | 结合多气体生物反应器,自养菌利用CO2生成脂肪酸 | [ |
| 脂肪酸及其衍生物 | 汉逊酵母(Ogataeapolym orpha) | 甲醇 | 无机盐培养基 | 脂肪酸(15.9 g/L) | 脂肪酸合成途径 | ①Δfaa1 ②加强前体供应及辅因子供应:过表达果糖-1,6-二磷酸酶(FBP),磷酸核糖异构酶(RPE),柠檬酸裂解酶(ACL),异柠檬酸脱氢酶(ICL1),果糖6-磷酸脱氢酶(ZWF1) | ①适应性进化使得敲除细胞生长恢复,并解析机制是由于LPL1和IZH3缺失引起 ②1 L生物反应器 | [ |
| 脂肪酸及其衍生物 | 蓝藻(Synechocystis sp. PCC 6803) | CO2 | — | 脂肪酸甲酯(120 mg/L) | — | ①Δaas ②过表达硫酯酶(UcFatB1),O-甲基转移酶(DmJHAMT) ③引入S-腺苷甲硫氨酸(SAM)循环供应甲基 | ①不利用甲醇作为甲基供体,利用SAM合成酶供应甲基 ②可能产生的脂肪酸甲酯类物质:C12∶0,C14∶0,C16∶0 | [ |
| 脂肪酸及其衍生物 | 解脂耶氏酵母(Yarrowia lipolytica) | CO2 | 酵母合成培养基,酵母提取物 | 脂肪酸(10.7 g/L) | — | ①增强脂肪酸合成:过表达生物素羧化酶(BC) ②引入CO2利用途径:过表达碳酸酐酶(CA) | ①循环利用CO2生产脂肪酸 ②250 mL摇瓶发酵 | [ |
| 萜类物质 | 扭脱甲基杆菌AM1(Methylobact-eriumextorquens) | 甲醇 | 无机盐基础培养基 | 甲羟戊酸(2.22 g/L) | 丝氨酸循环,EMC途径,类异戊二烯途径 | 表达HMG-CoA合成酶(Enterococcus faecalistiters),HMG-CoA还原酶(Enterococcus faecalistiters),乙酰乙酰CoA硫解酶(Ralstonia eutropha) | ①产率:28.4 mg/g甲醇 ②5 L生物反应器 | [ |
| 萜类物质 | 类黄色噬氢菌DSM1084(Hydrogenop-haga pseudoflava) | CO2,合成气 | 醋酸盐,果糖,蔗糖等,无机盐基础培养基 | α-红没药烯[(59.0±7.9) μg] | 卡尔文循环,Wood-Ljungdahl途径(WL),类异戊二烯途径 | 表达没药烯合成酶(TPS,Abies grandis) | ①自养和异养条件皆可生长:自养条件下利用合成气作为碳源,异养条件下可以利用果糖、蔗糖等作为碳源 | [ |
| ②自养条件利用卡尔文循环和WL途径,异养条件利用MEP途径 | ||||||||
| 萜类物质 | Cupriavidus necator | CO2,H2,O2 | 果糖,无机盐基础培养基 | α-蛇麻烯[(10.8±2.5)mg/g DCW 或17 mg/g DCW] | 类异戊二烯途径 | 表达MVA途径:焦磷酸法呢合成酶(ERG20),IPP异构酶,α-蛇麻烯合酶(ZSSI) | 化学自养和电自养均可:化能自养主要利用CO2等,电自养需要电极和水辅助 | [ |
| 萜类物质 | 嗜甲烷菌20Z(Methylomicr-obium alcaliphilum) | 50%甲烷 | 硝酸矿物盐培养基(NMS) | α-蛇麻烯(0.75 mg/g DCW,835 μg/L) | 类异戊二烯途径 | ①表达α-蛇麻烯合成酶(ZSS1) ②表达1-脱氧木酮糖-5-磷酸合酶(DXS),HMBPP合成酶(ISPG),FPP合成酶(ISPA) ③Δpgi ④提高NADPH:表达转氢酶(PNTAB),葡萄糖6-磷酸脱氢酶(ZWF),磷酸葡糖酸脱氢酶(PGD) | 优化MEP途径 | [ |
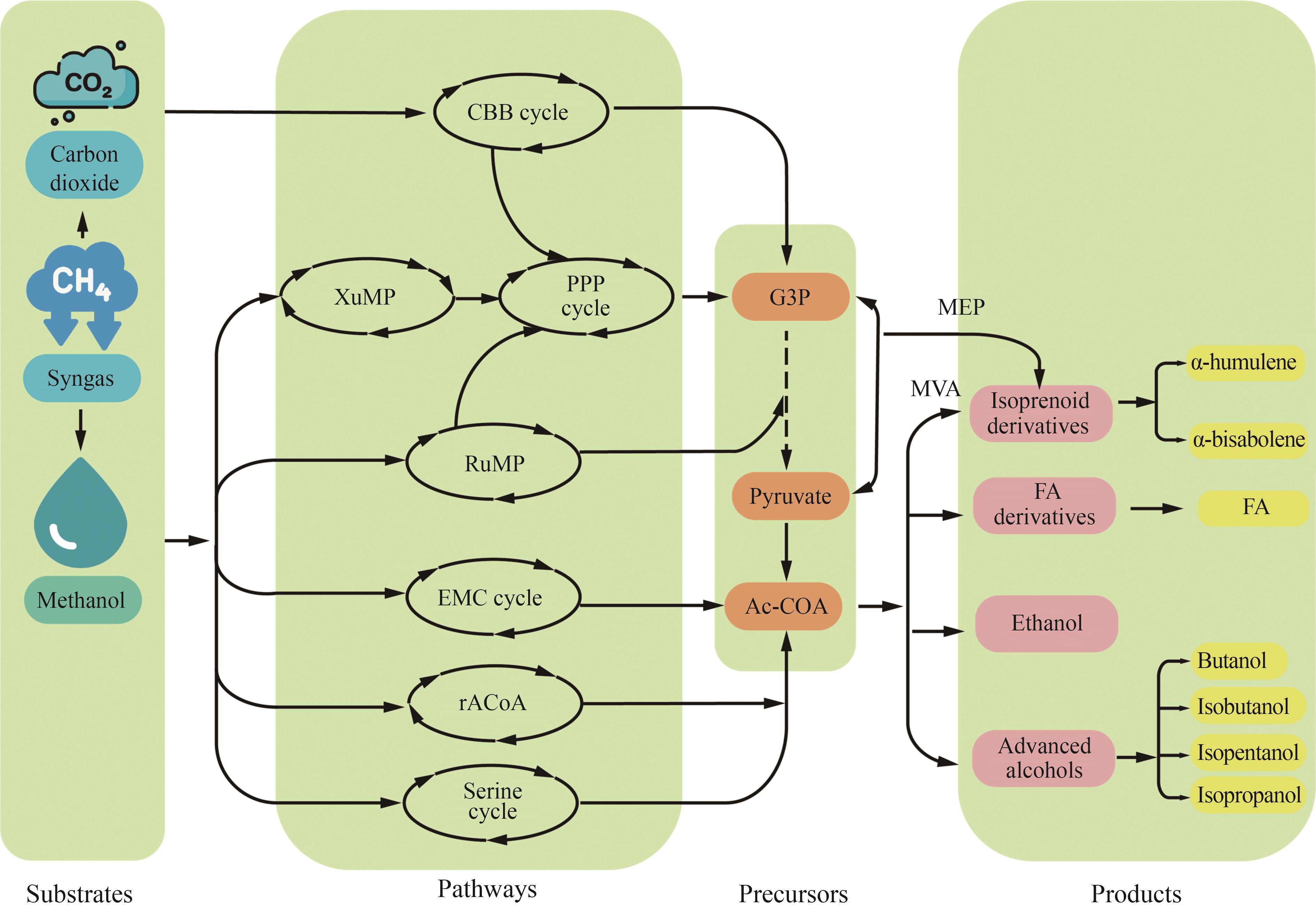
Fig. 1 Synthesis of advanced biofuels based on one-carbon substances(Pathway: CBB cycle—Calvin-Benson-Bassham cycle; XuMP—xylulose monophosphate pathway; PPP—pentose phosphate pathway; RuMP—ribulose monophosphate pathway; EMC—ethylmalonyl-CoA; rACoAP—reductive acetyl-CoA pathway, also known as Wood-Ljungdahl pathway; MVA—mevalonate; MEP—methylerythritol‑4‑phosphate. Metabolites: G3P—glyceraldehyde-3-phosphate; Ac-CoA—acetyl-CoA; FA—fatty acid)
| 途径 | 前体物质 | 主要中间物 | 关键酶 | 合成产物 | 主要应用 | 备注 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 酮酸途径 | 丙酮酸 | 2-酮酸 | 2-酮酸脱羧酶,醇脱氢酶 | 1-丙醇,异丁醇,1-丁醇,2-甲基-1-丁醇,3-甲基-1-丁醇,2-苯乙醇 | 生物汽油 | ①丙酮酸经过氨基酸生物合成途径转化为不同长度碳链的2-酮酸 | [ |
| ②2-酮酸经过延伸酶、脱羧酶、水解酶形成终产物 | |||||||
| 类异戊二烯途径 | 乙酰CoA,3-磷酸甘油醛(G3P),丙酮酸 | 异戊烯焦磷酸(IPP),二甲基丙烯焦磷酸酯(DMAPP),香叶基焦磷酸酯(GPP),法呢基焦磷酸酯(FPP) | 萜烯合成酶 | 异戊醇,异戊烯醇,3-甲基-2-丁烯醇,松萜,柠烯,红没药烯,法呢烯 | 生物汽油,航空用油,发动机燃料 | ①乙酰CoA经MVA途径合成IPP和DMAPP | [ |
| ②G3P和丙酮酸经MEP途径合成IPP和DMAPP | |||||||
| 逆β氧化途径 | 乙酰CoA,CoA | 酰基-CoA | 酰基转移酶 | 1-丁醇,丁酸,异丙醇,1-乙醇,1-辛醇 | 生物汽油,生物柴油,航空燃料 | ①CoA直接作为供体 | [ |
| ②不同类型的硫酯酶可形成不同类型的化合物 | |||||||
| 脂肪酸生物合成途径 | 乙酰CoA,酰基载体蛋白(ACP) | 酰基-ACP | 脂肪酸合成酶,硫酯酶,还原酶 | 脂肪酸,脂肪醇,脂肪酸甲酯,脂肪酸乙酯,烷烃 | 生物汽油,生物柴油 | CoA供体为ACP | [ |
| 聚酮生物合成途径 | 乙酰CoA,酰基载体蛋白(ACP) | β-酮酰基-ACP | 聚酮合成酶 | 1-丁烯,1-己烯,1-己醇,乙基酮,甲基酮,支链醇 | 生物汽油,航空用油,发动机燃料 | CoA供体为ACP | [ |
Table 3 Comparison of synthetic pathways for the production of advanced biofuels
| 途径 | 前体物质 | 主要中间物 | 关键酶 | 合成产物 | 主要应用 | 备注 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 酮酸途径 | 丙酮酸 | 2-酮酸 | 2-酮酸脱羧酶,醇脱氢酶 | 1-丙醇,异丁醇,1-丁醇,2-甲基-1-丁醇,3-甲基-1-丁醇,2-苯乙醇 | 生物汽油 | ①丙酮酸经过氨基酸生物合成途径转化为不同长度碳链的2-酮酸 | [ |
| ②2-酮酸经过延伸酶、脱羧酶、水解酶形成终产物 | |||||||
| 类异戊二烯途径 | 乙酰CoA,3-磷酸甘油醛(G3P),丙酮酸 | 异戊烯焦磷酸(IPP),二甲基丙烯焦磷酸酯(DMAPP),香叶基焦磷酸酯(GPP),法呢基焦磷酸酯(FPP) | 萜烯合成酶 | 异戊醇,异戊烯醇,3-甲基-2-丁烯醇,松萜,柠烯,红没药烯,法呢烯 | 生物汽油,航空用油,发动机燃料 | ①乙酰CoA经MVA途径合成IPP和DMAPP | [ |
| ②G3P和丙酮酸经MEP途径合成IPP和DMAPP | |||||||
| 逆β氧化途径 | 乙酰CoA,CoA | 酰基-CoA | 酰基转移酶 | 1-丁醇,丁酸,异丙醇,1-乙醇,1-辛醇 | 生物汽油,生物柴油,航空燃料 | ①CoA直接作为供体 | [ |
| ②不同类型的硫酯酶可形成不同类型的化合物 | |||||||
| 脂肪酸生物合成途径 | 乙酰CoA,酰基载体蛋白(ACP) | 酰基-ACP | 脂肪酸合成酶,硫酯酶,还原酶 | 脂肪酸,脂肪醇,脂肪酸甲酯,脂肪酸乙酯,烷烃 | 生物汽油,生物柴油 | CoA供体为ACP | [ |
| 聚酮生物合成途径 | 乙酰CoA,酰基载体蛋白(ACP) | β-酮酰基-ACP | 聚酮合成酶 | 1-丁烯,1-己烯,1-己醇,乙基酮,甲基酮,支链醇 | 生物汽油,航空用油,发动机燃料 | CoA供体为ACP | [ |
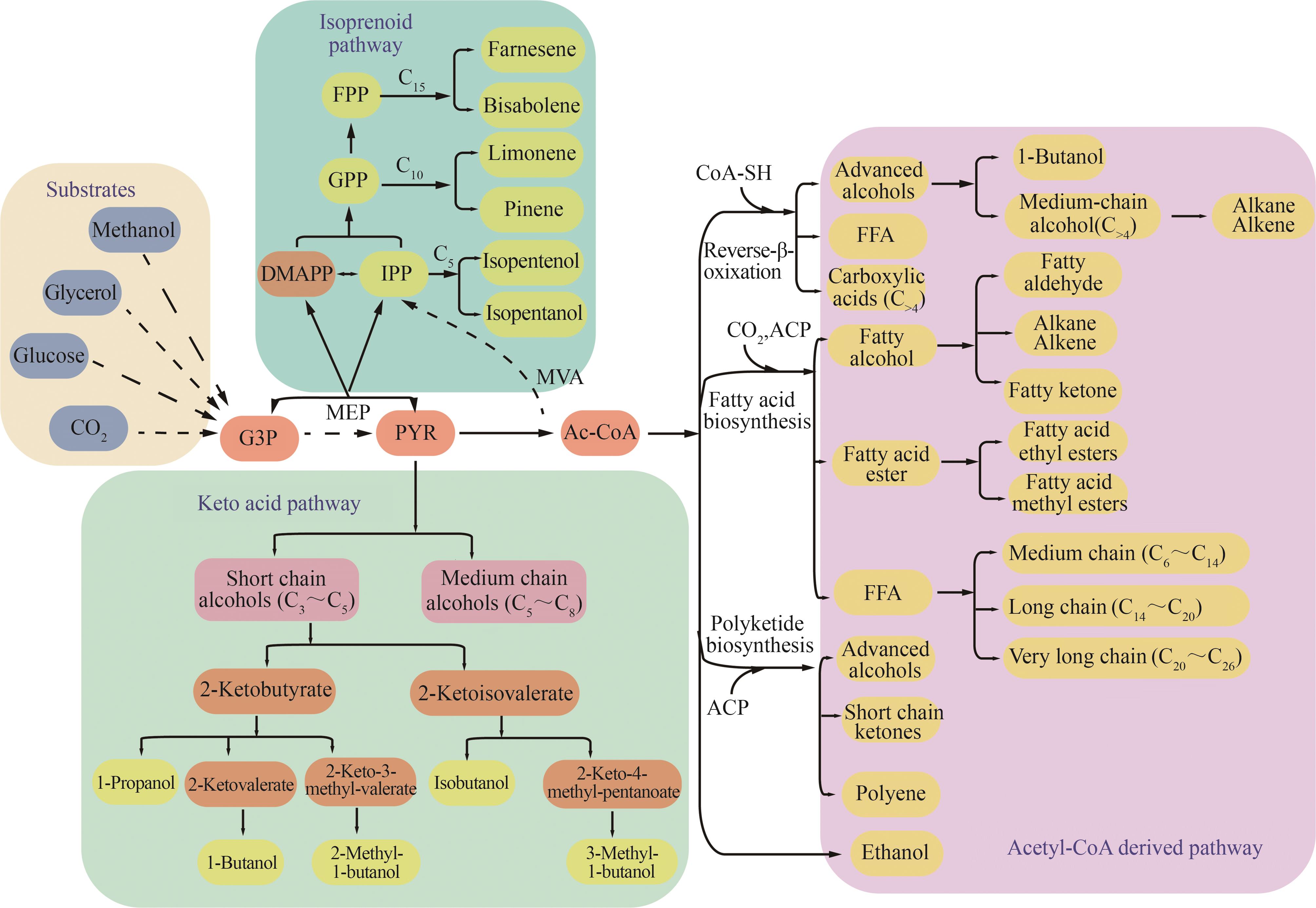
Fig. 2 Metabolic pathways for producing advanced biofuels(Pathway: MVA—mevalonate; MEP—methylerythritol‑4‑phosphate. Metabolites: G3P—glyceraldehyde-3-phosphate; PYR—pyruvate; Ac-CoA—acetyl-CoA; FFA—free fatty acid; IPP—isopentenyl pyrophosphate; DMAPP—dimethylallyl pyrophosphate; GPP—geranyl pyrophosphate; FPP—farnesyl pyrophosphate)
| 1 | IEA. World Energy Outlook 2022[R/OL]. [2023-04-01]. . |
| 2 | LIAO J C, MI L, PONTRELLI S, et al. Fuelling the future: microbial engineering for the production of sustainable biofuels[J]. Nature Reviews Microbiology, 2016, 14(5): 288-304. |
| 3 | AMJITH L, BAVANISH B. A review on biomass and wind as renewable energy for sustainable environment[J]. Chemosphere, 2022, 293: 133579. |
| 4 | INGANÄS O, SUNDSTRÖM V. Solar energy for electricity and fuels[J]. Ambio, 2016, 45(): S15-S23. |
| 5 | CHOWDHURY F A. Recent advances and demonstrated potentials for clean hydrogen via overall solar water splitting[J]. MRS Advances, 2019, 4(51): 2771-2785. |
| 6 | LYU S L, YOUNIS M A, LIU Z B, et al. Rational design on photoelectrodes and devices to boost photoelectrochemical performance of solar-driven water splitting: a mini review[J]. Frontiers of Chemical Science and Engineering, 2022, 16(6): 777-798. |
| 7 | DAS J, RAVISHANKAR H, LENS P N L. Biological biogas purification: recent developments, challenges and future prospects[J]. Journal of Environmental Management, 2022, 304: 114198. |
| 8 | DE TISSERA S, KÖPKE M, SIMPSON S D, et al. Syngas biorefinery and syngas utilization[J]. Advances in Biochemical Engineering/Biotechnology, 2019, 166: 247-280. |
| 9 | LIU Y Z, CRUZ-MORALES P, ZARGAR A, et al. Biofuels for a sustainable future[J]. Cell, 2021, 184(6): 1636-1647. |
| 10 | 王冉, 常家辉, 白子龙. 国外生物能源研究进展及应用概述[J]. 国防制造技术, 2021(4): 4-7. |
| WANG R, CHANG J H, BAI Z L. Overview of research progress and application of bioenergy abroad[J]. Defense Manufacturing Technology, 2021(4): 4-7. | |
| 11 | CAVELIUS P, ENGELHART-STRAUB S, MEHLMER N, et al. The potential of biofuels from first to fourth generation[J]. PLoS Biology, 2023, 21(3): e3002063. |
| 12 | MUSSATTO S I, DRAGONE G, GUIMARÃES P M R, et al. Technological trends, global market, and challenges of bio-ethanol production[J]. Biotechnology Advances, 2010, 28(6): 817-830. |
| 13 | 徐雪雯, 李鹏辉, 童国林. 第二代生物乙醇制备研究进展[J]. 中国造纸, 2023, 42(2): 94-101. |
| XU X W, LI P H, TONG G L. Research progress of the preparation of second-generation bioethanol[J]. China Pulp & Paper, 2023, 42(2): 94-101. | |
| 14 | BRODA M, YELLE D J, SERWAŃSKA K. Bioethanol production from lignocellulosic biomass—challenges and solutions[J]. Molecules, 2022, 27(24): 8717. |
| 15 | ISIKGOR F H, BECER C R. Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers[J]. Polymer Chemistry, 2015, 6(25): 4497-4559. |
| 16 | SCHWARZ W H. The cellulosome and cellulose degradation by anaerobic bacteria[J]. Applied Microbiology and Biotechnology, 2001, 56(5-6): 634-649. |
| 17 | 方志锋, 刘昆仑, 陈复生, 等. 木质纤维素类生物质制备生物乙醇研究进展[J]. 现代化工, 2013, 33(1): 30-33. |
| FANG Z F, LIU K L, CHEN F S, et al. Research development of bioethanol preparation from lignocellulosic biomass[J]. Modern Chemical Industry, 2013, 33(1): 30-33. | |
| 18 | SARAVANAN A, SENTHIL KUMAR P, JEEVANANTHAM S, et al. Recent advances and sustainable development of biofuels production from lignocellulosic biomass[J]. Bioresource Technology, 2022, 344: 126203. |
| 19 | OJEDA K, SÁNCHEZ E, EL-HALWAGI M, et al. Exergy analysis and process integration of bioethanol production from acid pre-treated biomass: comparison of SHF, SSF and SSCF pathways[J]. Chemical Engineering Journal, 2011, 176-177: 195-201. |
| 20 | MOHD AZHAR S H, ABDULLA R, JAMBO S A, et al. Yeasts in sustainable bioethanol production: a review[J]. Biochemistry and Biophysics Reports, 2017, 10: 52-61. |
| 21 | JOSHI S, MISHRA S. Recent advances in biofuel production through metabolic engineering[J]. Bioresource Technology, 2022, 352: 127037. |
| 22 | KAZEMI SHARIAT PANAHI H, DEHHAGHI M, DEHHAGHI S, et al. Engineered bacteria for valorizing lignocellulosic biomass into bioethanol[J]. Bioresource Technology, 2022, 344: 126212. |
| 23 | SIMAS-RODRIGUES C, VILLELA H D M, MARTINS A P, et al. Microalgae for economic applications: advantages and perspectives for bioethanol[J]. Journal of Experimental Botany, 2015, 66(14): 4097-4108. |
| 24 | MEDIPALLY S R, YUSOFF F M, BANERJEE S, et al. Microalgae as sustainable renewable energy feedstock for biofuel production[J]. BioMed Research International, 2015, 2015: 519513. |
| 25 | 刘雪艳, 苏忠亮. 微藻生物燃料的研究进展[J]. 化学与生物工程, 2017, 34(3): 11-14. |
| LIU X Y, SU Z L. Research progress on microalgae biofuel[J]. Chemistry & Bioengineering, 2017, 34(3): 11-14. | |
| 26 | GILMOUR D J. Microalgae for biofuel production[J]. Advances in Applied Microbiology, 2019, 109: 1-30. |
| 27 | MAIA J L D, CARDOSO J S, MASTRANTONIO D J D S, et al. Microalgae starch: a promising raw material for the bioethanol production[J]. International Journal of Biological Macromolecules, 2020, 165(Pt B): 2739-2749. |
| 28 | LAKATOS G E, RANGLOVÁ K, MANOEL J C, et al. Bioethanol production from microalgae polysaccharides[J]. Folia Microbiologica, 2019, 64(5): 627-644. |
| 29 | GRAMA S B, LIU Z Y, LI J. Emerging trends in genetic engineering of microalgae for commercial applications[J]. Marine Drugs, 2022, 20(5): 285. |
| 30 | JAISWAL A, BABU V, BAISHYA B, et al. Biochemical pathways regulated by algae to mitigate global carbon emissions: a review[J]. Journal of Environmental Pathology, Toxicology and Oncology, 2020, 39(4): 317-334. |
| 31 | LIU H, SUN J L, CHANG J S, et al. Engineering microbes for direct fermentation of cellulose to bioethanol[J]. Critical Reviews in Biotechnology, 2018, 38(7): 1089-1105. |
| 32 | SINGH A, SINGHANIA R R, SOAM S, et al. Production of bioethanol from food waste: status and perspectives[J]. Bioresource Technology, 2022, 360: 127651. |
| 33 | SHARMA J, KUMAR V, PRASAD R, et al. Engineering of Saccharomyces cerevisiae as a consolidated bioprocessing host to produce cellulosic ethanol: recent advancements and current challenges[J]. Biotechnology Advances, 2022, 56: 107925. |
| 34 | KUMAR K, DASGUPTA C N, NAYAK B, et al. Development of suitable photobioreactors for CO2 sequestration addressing global warming using green algae and cyanobacteria[J]. Bioresource Technology, 2011, 102(8): 4945-4953. |
| 35 | 林海龙, 林鑫, 岳国君. 我国生物燃料乙醇产业新进展[J]. 新能源进展, 2020, 8(3): 165-171. |
| LIN H L, LIN X, YUE G J. New advances in China’s biofuel ethanol industry[J]. Advances in New and Renewable Energy, 2020, 8(3): 165-171. | |
| 36 | KAMARAJ R, RAO Y K S S, BALAKRISHNA B. Biodiesel blends: a comprehensive systematic review on various constraints[J]. Environmental Science and Pollution Research International, 2022, 29(29): 43770-43785. |
| 37 | MATHEW G M, RAINA D, NARISETTY V, et al. Recent advances in biodiesel production: challenges and solutions[J]. Science of the Total Environment, 2021, 794: 148751. |
| 38 | 王欣竹. 从工程微藻中提取制备生物柴油的研究进展[J]. 当代化工研究, 2022(2): 162-164. |
| WANG X Z. Research progress of biodiesel extraction from engineering microalgae[J]. Modern Chemical Research, 2022(2): 162-164. | |
| 39 | SINGH A, NIGAM P S, MURPHY J D. Mechanism and challenges in commercialisation of algal biofuels[J]. Bioresource Technology, 2011, 102(1): 26-34. |
| 40 | BRAR A, KUMAR M, SONI T, et al. Insights into the genetic and metabolic engineering approaches to enhance the competence of microalgae as biofuel resource: a review[J]. Bioresource Technology, 2021, 339: 125597. |
| 41 | TAPARIA T, MVSS M, MEHROTRA R, et al. Developments and challenges in biodiesel production from microalgae: a review[J]. Biotechnology and Applied Biochemistry, 2016, 63(5): 715-726. |
| 42 | GONG Y, MIAO X L. Short chain fatty acid biosynthesis in microalgae Synechococcus sp. PCC 7942[J]. Marine Drugs, 2019, 17(5): 255. |
| 68 | SHI T Q, LI Y W, ZHU L, et al. Engineering the oleaginous yeast Yarrowia lipolytica for β-farnesene overproduction[J]. Biotechnology Journal, 2021, 16(7): e2100097. |
| 69 | LIU Y H, JIANG X, CUI Z Y, et al. Engineering the oleaginous yeast Yarrowia lipolytica for production of α-farnesene[J]. Biotechnology for Biofuels, 2019, 12: 296. |
| 70 | FANG L X, FAN J, LUO S L, et al. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids[J]. Nature Communications, 2021, 12(1): 4976. |
| 71 | PAN H, ZHANG L H, LI X, et al. Biosynthesis of the fatty acid isopropyl esters by engineered Escherichia coli [J]. Enzyme and Microbial Technology, 2017, 102: 49-52. |
| 72 | GUO D Y, ZHU J, DENG Z X, et al. Metabolic engineering of Escherichia coli for production of fatty acid short-chain esters through combination of the fatty acid and 2-keto acid pathways[J]. Metabolic Engineering, 2014, 22: 69-75. |
| 73 | YANG L, ZHU Z, WANG W H, et al. Microbial recycling of glycerol to biodiesel[J]. Bioresource Technology, 2013, 150: 1-8. |
| 74 | D’ESPAUX L, GHOSH A, RUNGUPHAN W, et al. Engineering high-level production of fatty alcohols by Saccharomyces cerevisiae from lignocellulosic feedstocks[J]. Metabolic Engineering, 2017, 42: 115-125. |
| 75 | YU T, ZHOU Y J, WENNING L, et al. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals[J]. Nature Communications, 2017, 8: 15587. |
| 76 | YU T, ZHOU Y J, HUANG M T, et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis[J]. Cell, 2018, 174(6): 1549-1558.e14. |
| 77 | ZHU Z W, HU Y T, TEIXEIRA P G, et al. Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids[J]. Nature Catalysis, 2020, 3: 64-74. |
| 78 | YU T, LIU Q L, WANG X, et al. Metabolic reconfiguration enables synthetic reductive metabolism in yeast[J]. Nature Metabolism, 2022, 4(11): 1551-1559. |
| 79 | WANG W, WEI H, KNOSHAUG E, et al. Fatty alcohol production in Lipomyces starkeyi and Yarrowia lipolytica [J]. Biotechnology for Biofuels, 2016, 9: 227. |
| 43 | SUN X M, REN L J, ZHAO Q Y, et al. Enhancement of lipid accumulation in microalgae by metabolic engineering[J]. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 2019, 1864(4): 552-566. |
| 44 | 董文婉. 航空生物燃料作为新型能源的发展与利弊[J]. 民航管理, 2013(11): 111-112. |
| DONG W W. The development of aviation biofuel and the pros and cons[J]. Civil Aviation Management, 2013(11): 111-112. | |
| 45 | MASCAL M, DUTTA S. Synthesis of highly-branched alkanes for renewable gasoline[J]. Fuel Processing Technology, 2020, 197: 106192. |
| 46 | EL-DALATONY M M, SAHA S, GOVINDWAR S P, et al. Biological conversion of amino acids to higher alcohols[J]. Trends in Biotechnology, 2019, 37(8): 855-869. |
| 47 | MEHRER C R, INCHA M R, POLITZ M C, et al. Anaerobic production of medium-chain fatty alcohols via a β-reduction pathway[J]. Metabolic Engineering, 2018, 48: 63-71. |
| 48 | DELLOMONACO C, CLOMBURG J M, MILLER E N, et al. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals[J]. Nature, 2011, 476(7360): 355-359. |
| 49 | GUO L, DIAO W W, GAO C, et al. Engineering Escherichia coli lifespan for enhancing chemical production[J]. Nature Catalysis, 2020, 3: 307-318. |
| 50 | SHEN C R, LAN E I, DEKISHIMA Y, et al. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli [J]. Applied and Environmental Microbiology, 2011, 77(9): 2905-2915. |
| 51 | ATSUMI S, HANAI T, LIAO J C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels[J]. Nature, 2008, 451(7174): 86-89. |
| 52 | PARK S H, HAHN J S. Development of an efficient cytosolic isobutanol production pathway in Saccharomyces cerevisiae by optimizing copy numbers and expression of the pathway genes based on the toxic effect of α-acetolactate[J]. Scientific Reports, 2019, 9(1): 3996. |
| 53 | AVALOS J L, FINK G R, STEPHANOPOULOS G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols[J]. Nature Biotechnology, 2013, 31(4): 335-341. |
| 54 | WESS J, BRINEK M, BOLES E. Improving isobutanol production with the yeast Saccharomyces cerevisiae by successively blocking competing metabolic pathways as well as ethanol and glycerol formation[J]. Biotechnology for Biofuels, 2019, 12: 173. |
| 55 | VOGT M, BRÜSSELER C, VAN OOYEN J, et al. Production of 2-methyl-1-butanol and 3-methyl-1-butanol in engineered Corynebacterium glutamicum [J]. Metabolic Engineering, 2016, 38: 436-445. |
| 56 | SIRIPONG W, ANGELA C, TANAPONGPIPAT S, et al. Metabolic engineering of Pichia pastoris for production of isopentanol (3-methyl-1-butanol)[J]. Enzyme and Microbial Technology, 2020, 138: 109557. |
| 57 | SIRIPONG W, WOLF P, KUSUMOPUTRI T P, et al. Metabolic engineering of Pichia pastoris for production of isobutanol and isobutyl acetate[J]. Biotechnology for Biofuels, 2018, 11: 1. |
| 58 | SU H F, LIN J F, WANG Y H, et al. Engineering Brevibacterium flavum for the production of renewable bioenergy: C4–C5 advanced alcohols[J]. Biotechnology and Bioengineering, 2017, 114(9): 1946-1958. |
| 59 | LI S S, WEN J P, JIA X Q. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression[J]. Applied Microbiology and Biotechnology, 2011, 91(3): 577-589. |
| 60 | SHI X Y, PARK H M, KIM M H, et al. Isopropanol biosynthesis from crude glycerol using fatty acid precursors via engineered oleaginous yeast Yarrowia lipolytica [J]. Microbial Cell Factories, 2022, 21(1): 168. |
| 61 | YUZAWA S, MIRSIAGHI M, JOCIC R, et al. Short-chain ketone production by engineered polyketide synthases in Streptomyces albus [J]. Nature Communications, 2018, 9(1): 4569. |
| 62 | GROUSSEAU E, LU J N, GORRET N, et al. Isopropanol production with engineered Cupriavidus necator as bioproduction platform[J]. Applied Microbiology and Biotechnology, 2014, 98(9): 4277-4290. |
| 63 | ZHANG Y, SONG X H, LAI Y M, et al. High-yielding terpene-based biofuel production in Rhodobacter capsulatus [J]. ACS Synthetic Biology, 2021, 10(6): 1545-1552. |
| 64 | PERALTA-YAHYA P P, OUELLET M, CHAN R, et al. Identification and microbial production of a terpene-based advanced biofuel[J]. Nature Communications, 2011, 2: 483. |
| 65 | LIU C L, BI H R, BAI Z H, et al. Engineering and manipulation of a mevalonate pathway in Escherichia coli for isoprene production[J]. Applied Microbiology and Biotechnology, 2019, 103(1): 239-250. |
| 80 | JANG Y S, LEE J Y, LEE J M, et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum [J]. mBio, 2012, 3(5): e00314-12. |
| 81 | LIU H H, SONG Y L, FAN X, et al. Yarrowia lipolytica as an oleaginous platform for the production of value-added fatty acid-based bioproducts[J]. Frontiers in Microbiology, 2021, 11: 608662. |
| 82 | ZHOU Y J, BUIJS N A, ZHU Z W, et al. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition[J]. Journal of the American Chemical Society, 2016, 138(47): 15368-15377. |
| 83 | ZHAO Y K, ZHU K, LI J, et al. High-efficiency production of bisabolene from waste cooking oil by metabolically engineered Yarrowia lipolytica [J]. Microbial Biotechnology, 2021, 14(6): 2497-2513. |
| 84 | LI H, OPGENORTH P H, WERNICK D G, et al. Integrated electromicrobial conversion of CO2 to higher alcohols[J]. Science, 2012, 335(6076): 1596. |
| 85 | LIU C, COLÓN B C, ZIESACK M, et al. Water splitting-biosynthetic system with CO₂ reduction efficiencies exceeding photosynthesis[J]. Science, 2016, 352(6290): 1210-1213. |
| 86 | HENSTRA A M, SIPMA J, RINZEMA A, et al. Microbiology of synthesis gas fermentation for biofuel production[J]. Current Opinion in Biotechnology, 2007, 18(3): 200-206. |
| 87 | WILLCOX D, CHAPPELL B G N, HOGG K F, et al. A general catalytic β-C-H carbonylation of aliphatic amines to β-lactams[J]. Science, 2016, 354(6314): 851-857. |
| 88 | KIRST H, GABILLY S T, NIYOGI K K, et al. Photosynthetic antenna engineering to improve crop yields[J]. Planta, 2017, 245(5): 1009-1020. |
| 89 | SOUTH P F, CAVANAGH A P, LIU H W, et al. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field[J]. Science, 2019, 363(6422): eaat9077. |
| 90 | WU I, ARNOLD F H. Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures[J]. Biotechnology and Bioengineering, 2013, 110(7): 1874-1883. |
| 91 | LI P H, REN J P, JIANG Z W, et al. Review on the preparation of fuels and chemicals based on lignin[J]. RSC Advances, 2022, 12(17): 10289-10305. |
| 92 | ZHANG Y H P, LYND L R. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(20): 7321-7325. |
| 93 | LEE J W, YOOK S D, KOH H G, et al. Engineering xylose metabolism in yeasts to produce biofuels and chemicals[J]. Current Opinion in Biotechnology, 2021, 67: 15-25. |
| 94 | RUCHALA J, SIBIRNY A A. Pentose metabolism and conversion to biofuels and high-value chemicals in yeasts[J]. FEMS Microbiology Reviews, 2021, 45(4): fuaa069. |
| 95 | GONG C J, CAO L P, FANG D L, et al. Genetic manipulation strategies for ethanol production from bioconversion of lignocellulose waste[J]. Bioresource Technology, 2022, 352: 127105. |
| 96 | KRICKA W, FITZPATRICK J, BOND U. Challenges for the production of bioethanol from biomass using recombinant yeasts[J]. Advances in Applied Microbiology, 2015, 92: 89-125. |
| 97 | KATAHIRA S, MIZUIKE A, FUKUDA H, et al. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain[J]. Applied Microbiology and Biotechnology, 2006, 72(6): 1136-1143. |
| 98 | STEEN E J, KANG Y S, BOKINSKY G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass[J]. Nature, 2010, 463(7280): 559-562. |
| 99 | XUE D S, YAO D H, SUKUMARAN R K, et al. Tandem integration of aerobic fungal cellulase production, lignocellulose substrate saccharification and anaerobic ethanol fermentation by a modified gas lift bioreactor[J]. Bioresource Technology, 2020, 302: 122902. |
| 100 | JIANG Y J, DONG W L, XIN F X, et al. Designing synthetic microbial consortia for biofuel production[J]. Trends in Biotechnology, 2020, 38(8): 828-831. |
| 101 | BROWN J L, PERISIN M A, SWIFT C L, et al. Co-cultivation of anaerobic fungi with Clostridium acetobutylicum bolsters butyrate and butanol production from cellulose and lignocellulose[J]. Journal of Industrial Microbiology & Biotechnology, 2023, 49(6): kuac024. |
| 102 | YOU S P, CHANG H X, ZHANG C Y, et al. Recycling strategy and repression elimination for lignocellulosic-based farnesene production with an engineered Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2019, 67(35): 9858-9867. |
| 103 | WANG S M, WANG Z B, WANG Y C, et al. Production of isoprene, one of the high-density fuel precursors, from peanut hull using the high-efficient lignin-removal pretreatment method[J]. Biotechnology for Biofuels, 2017, 10: 297. |
| 104 | CHEN C T, CHEN F Y H, BOGORAD I W, et al. Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production[J]. Metabolic Engineering, 2018, 49: 257-266. |
| 105 | WANG C, REN J, ZHOU L B, et al. An aldolase-catalyzed new metabolic pathway for the assimilation of formaldehyde and methanol to synthesize 2-keto-4-hydroxybutyrate and 1,3-propanediol in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8(11): 2483-2493. |
| 106 | BENNETT R K, DILLON M, GERALD HAR J R, et al. Engineering Escherichia coli for methanol-dependent growth on glucose for metabolite production[J]. Metabolic Engineering, 2020, 60: 45-55. |
| 107 | BENNETT R K, GONZALEZ J E, WHITAKER W B, et al. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph[J]. Metabolic Engineering, 2018, 45: 75-85. |
| 108 | MA Z X, ZHANG M, ZHANG C T, et al. Metabolomic analysis improves bioconversion of methanol to isobutanol in Methylorubrum extorquens AM1[J]. Biotechnology Journal, 2021, 16(6): e2000413. |
| 109 | HU B, YANG Y M, BECK D A, et al. Comprehensive molecular characterization of Methylobacterium extorquens AM1 adapted for 1-butanol tolerance[J]. Biotechnology for Biofuels, 2016, 9: 84. |
| 110 | YUAN X J, CHEN W J, MA Z X, et al. Rewiring the native methanol assimilation metabolism by incorporating the heterologous ribulose monophosphate cycle into Methylorubrum extorquens [J]. Metabolic Engineering, 2021, 64: 95-110. |
| 111 | DAVIES F K, WORK V H, BELIAEV A S, et al. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002[J]. Frontiers in Bioengineering and Biotechnology, 2014, 2: 21. |
| 112 | GARRIGUES L, MAIGNIEN L, LOMBARD E, et al. Isopropanol production from carbon dioxide in Cupriavidus necator in a pressurized bioreactor[J]. New Biotechnology, 2020, 56: 16-20. |
| 113 | LAUER I, PHILIPPS G, JENNEWEIN S. Metabolic engineering of Clostridium ljungdahlii for the production of hexanol and butanol from CO2 and H2 [J]. Microbial Cell Factories, 2022, 21(1): 85. |
| 114 | ZHENG T T, ZHANG M L, WU L H, et al. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering[J]. Nature Catalysis, 2022, 5: 388-396. |
| 115 | CAI P, WU X Y, DENG J, et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(29): e2201711119. |
| 66 | ALONSO-GUTIERREZ J, CHAN R, BATTH T S, et al. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production[J]. Metabolic Engineering, 2013, 19: 33-41. |
| 67 | MEADOWS A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production[J]. Nature, 2016, 537: 694-697. |
| 116 | LI Z K, XIONG B, LIU L, et al. Development of an autotrophic fermentation technique for the production of fatty acids using an engineered Ralstonia eutropha cell factory[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(6): 783-790. |
| 117 | GAO J Q, LI Y X, YU W, et al. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol[J]. Nature Metabolism, 2022, 4(7): 932-943. |
| 118 | YUNUS I S, PALMA A, TRUDEAU D L, et al. Methanol-free biosynthesis of fatty acid methyl ester (FAME) in Synechocystis sp. PCC 6803[J]. Metabolic Engineering, 2020, 57: 217-227. |
| 119 | YOU S K, PARK H M, LEE M E, et al. Non-photosynthetic CO2 utilization to increase fatty acid production in Yarrowia lipolytica [J]. Journal of Agricultural and Food Chemistry, 2021, 69(40): 11912-11918. |
| 120 | ZHU W L, CUI J Y, CUI L Y, et al. Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway[J]. Applied Microbiology and Biotechnology, 2016, 100(5): 2171-2182. |
| 121 | GRENZ S, BAUMANN P T, RÜCKERT C, et al. Exploiting Hydrogenophaga pseudoflava for aerobic syngas-based production of chemicals[J]. Metabolic Engineering, 2019, 55: 220-230. |
| 122 | KRIEG T, SYDOW A, FAUST S, et al. CO2 to terpenes: autotrophic and electroautotrophic α-humulene production with Cupriavidus necator [J]. Angewandte Chemie International Edition, 2018, 57(7): 1879-1882. |
| 123 | NGUYEN A D, KIM D H, LEE E Y. Unlocking the biosynthesis of sesquiterpenoids from methane via the methylerythritol phosphate pathway in methanotrophic bacteria, using α-humulene as a model compound[J]. Metabolic Engineering, 2020, 61: 69-78. |
| 124 | YUNUS I S, ANFELT J, SPORRE E, et al. Synthetic metabolic pathways for conversion of CO2 into secreted short-to medium-chain hydrocarbons using cyanobacteria[J]. Metabolic Engineering, 2022, 72: 14-23. |
| 125 | LI M, LONG B, DAI S Y, et al. Altered carbon partitioning enhances CO2 to terpene conversion in cyanobacteria[J]. Biodesign Research, 2022, 2022: 9897425. |
| 126 | ANTHONY J, RANGAMARAN V R, GOPAL D, et al. Ultraviolet and 5' fluorodeoxyuridine induced random mutagenesis in Chlorella vulgaris and its impact on fatty acid profile: a new insight on lipid-metabolizing genes and structural characterization of related proteins[J]. Marine Biotechnology, 2015, 17(1): 66-80. |
| 127 | BOYLE N R, PAGE M D, LIU B S, et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas [J]. The Journal of Biological Chemistry, 2012, 287(19): 15811-15825. |
| 128 | BECKMANN J, LEHR F, FINAZZI G, et al. Improvement of light to biomass conversion by de-regulation of light-harvesting protein translation in Chlamydomonas reinhardtii [J]. Journal of Biotechnology, 2009, 142(1): 70-77. |
| 129 | MELIS A. Solar energy conversion efficiencies in photosynthesis: minimizing the chlorophyll antennae to maximize efficiency[J]. Plant Science, 2009, 177(4): 272-280. |
| 130 | MUSSGNUG J H, THOMAS-HALL S, RUPPRECHT J, et al. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion[J]. Plant Biotechnology Journal, 2007, 5(6): 802-814. |
| 131 | LIANG F Y, ENGLUND E, LINDBERG P, et al. Engineered cyanobacteria with enhanced growth show increased ethanol production and higher biofuel to biomass ratio[J]. Metabolic Engineering, 2018, 46: 51-59. |
| 132 | LI-BEISSON Y, THELEN J J, FEDOSEJEVS E, et al. The lipid biochemistry of eukaryotic algae[J]. Progress in Lipid Research, 2019, 74: 31-68. |
| 133 | PANICH J, FONG B, SINGER S W. Metabolic engineering of Cupriavidus necator H16 for sustainable biofuels from CO2 [J]. Trends in Biotechnology, 2021, 39(4): 412-424. |
| 134 | KROG A, HEGGESET T M B, MÜLLER J E N, et al. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties[J]. PLoS One, 2013, 8(3): e59188. |
| 135 | KRACKE F, LAI B, YU S Q, et al. Balancing cellular redox metabolism in microbial electrosynthesis and electro fermentation — a chance for metabolic engineering[J]. Metabolic Engineering, 2018, 45: 109-120. |
| 136 | NEVIN K P, HENSLEY S A, FRANKS A E, et al. Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms[J]. Applied and Environmental Microbiology, 2011, 77(9): 2882-2886. |
| 137 | 郭姝媛, 吴良焕, 刘香健, 等. 微生物中一碳代谢网络构建的进展与挑战[J]. 合成生物学, 2022, 3(1): 116-137. |
| GUO S Y, WU L H, LIU X J, et al. Developing C1-based metabolic network in methylotrophy for biotransformation[J]. Synthetic Biology Journal, 2022, 3(1): 116-137. | |
| 138 | YU H, LIAO J C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9(1): 3992. |
| 139 | BOGORAD I W, CHEN C T, THEISEN M K, et al. Building carbon-carbon bonds using a biocatalytic methanol condensation cycle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(45): 15928-15933. |
| 140 | BAR-EVEN A. Formate assimilation: the metabolic architecture of natural and synthetic pathways[J]. Biochemistry, 2016, 55(28): 3851-3863. |
| 141 | YISHAI O, LINDNER S N, GONZALEZ DE LA CRUZ J, et al. The formate bio-economy[J]. Current Opinion in Chemical Biology, 2016, 35: 1-9. |
| 142 | HU G P, LI Z H, MA D L, et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals[J]. Nature Catalysis, 2021, 4: 395-406. |
| 143 | LING H, PRATOMO JUWONO N K, TEO W S, et al. Engineering transcription factors to improve tolerance against alkane biofuels in Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2015, 8: 231. |
| 144 | YAO P, YOU S P, QI W, et al. Investigation of fermentation conditions of biodiesel by-products for high production of β-farnesene by an engineered Escherichia coli [J]. Environmental Science and Pollution Research International, 2020, 27(18): 22758-22769. |
| 145 | ZHOU Y K, LI G, DONG J K, et al. MiYA, an efficient machine-learning workflow in conjunction with the YeastFab assembly strategy for combinatorial optimization of heterologous metabolic pathways in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2018, 47: 294-302. |
| 146 | LIU Z H, WANG K, CHEN Y, et al. Third-generation biorefineries as the means to produce fuels and chemicals from CO2 [J]. Nature Catalysis, 2020, 3: 274-288. |
| 147 | TAN X Y, NIELSEN J. The integration of bio-catalysis and electrocatalysis to produce fuels and chemicals from carbon dioxide[J]. Chemical Society Reviews, 2022, 51(11): 4763-4785. |
| 148 | HU Q S, HU H T, CUI L, et al. Ultrafast electron transfer in Au-cyanobacteria hybrid for solar to chemical production[J]. ACS Energy Letters, 2023, 8(1): 677-684. |
| [1] | Xue BAI, Nan LIANG, Xinqi LI, Zhipeng MO, Shuhuan TONG, Meiqi YUE, Xiaojing JIA, Kang REN, Xiaojie XI, Wei CHAO. Research on the equipment, process, and commercialization progress of syngas fermentation [J]. Synthetic Biology Journal, 2025, (): 1-22. |
| [2] | Ping ZHANG, Weijiao ZHANG, Ruirui XU, Jianghua LI, Jian CHEN, Zhen KANG. Research advances on Mycosporine-like amino acids biosynthesis [J]. Synthetic Biology Journal, 2024, (): 1-14. |
| [3] | Chuangen TANG, Jing WANG, Shuo ZHANG, Haoning ZHANG, Zhen KANG. Recent advances in synthesis and mining strategies of functional peptides [J]. Synthetic Biology Journal, 2024, (): 1-18. |
| [4] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [5] | BAI Zhonghu, REN He, NIE Jianqi, SUN Yang. The recent progresses and applications of in-parallel fermentation technology [J]. Synthetic Biology Journal, 2023, 4(5): 904-915. |
| [6] | Fei TAO, Tao SUN, Yu WANG, Ting WEI, Jun NI, Ping XU. Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality [J]. Synthetic Biology Journal, 2022, 3(5): 932-952. |
| [7] | Jinyu CUI, Aidi ZHANG, Guodong LUAN, Xuefeng LYU. Engineering microalgae for photosynthetic biosynthesis: progress and prospect [J]. Synthetic Biology Journal, 2022, 3(5): 884-900. |
| [8] | Chenkai CAO, Jialong LI, Kechun ZHANG. Progress in artificial metabolic pathways for biosynthesis of organic alcohols & acids [J]. Synthetic Biology Journal, 2021, 2(6): 902-919. |
| [9] | Xiaolong ZHANG, Chenyun WANG, Yanfeng LIU, Jianghua LI, Long LIU, Guocheng DU. Research progress of constructing efficient biomanufacturing system based on synthetic biotechnology [J]. Synthetic Biology Journal, 2021, 2(6): 863-875. |
| [10] | Zhengjie HOU, Huizhong SUN, Song BAI, Xinyue CHEN, Chunyang CAO, Jingsheng CHENG. Research progress of cyclic lipopeptide biosynthesis [J]. Synthetic Biology Journal, 2021, 2(4): 577-597. |
| [11] | Zhiqiang WEN, Xiaoman SUN, Qingzhuo WANG, Yanan LI, Wenzheng LIU, Yu JIANG, Sheng YANG. Recent advances in metabolic engineering of clostridia for n-butanol production [J]. Synthetic Biology Journal, 2021, 2(2): 194-221. |
| [12] | Hui ZHANG, Yaomeng YUAN, Chong ZHANG, Song YANG, Xinhui XING. Research progresses and future prospects of synthetic methylotrophic cell factory for methanol assimilation [J]. Synthetic Biology Journal, 2021, 2(2): 222-233. |
| [13] | Jiaoqi GAO, Yongjin ZHOU. Advances in methanol bio-transformation [J]. Synthetic Biology Journal, 2020, 1(2): 158-173. |
| [14] | Sen XIAO, Litao HU, Zhicheng SHI, Fayin WANG, Siting YU, Guocheng DU, Jian CHEN, Zhen KANG. Research advances in biosynthesis of hyaluronic acid with controllable molecular weights [J]. Synthetic Biology Journal, 2024, (): 1-16. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||