Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (2): 461-478.DOI: 10.12211/2096-8280.2024-067
• Invited Review • Previous Articles Next Articles
Advances in synthesis and mining strategies for functional peptides
TANG Chuan′gen1,2,3,4, WANG Jing1, ZHANG Shuo1, ZHANG Haoning1, KANG Zhen2,4
- 1.Nanjing Hanxin Pharmaceutical Technology Co. ,Ltd. ,Nanjing 210033,Jiangsu,China
2.Science Center for Future Foods,Jiangnan University,Wuxi 214122,Jiangsu,China
3.Key Laboratory of Industrial Biotechnology,Ministry of Education,School of Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
4.Key Laboratory of Carbohydrate Chemistry and Biotechnology,Ministry of Education,School of Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2024-08-28Revised:2024-11-11Online:2025-05-20Published:2025-04-30 -
Contact:TANG Chuan′gen
功能肽合成和挖掘策略研究进展
汤传根1,2,3,4, 王璟1, 张烁1, 张昊宁1, 康振2,4
- 1.南京汉欣医药科技有限公司,江苏 南京 210033
2.江南大学,未来食品科学中心,江苏 无锡 214122
3.江南大学生物工程学院,工业生物技术教育部重点实验室,江苏 无锡 214122
4.江南大学生物工程学院,糖化学与生物技术教育部重点实验室,江苏 无锡 214122
-
通讯作者:汤传根 -
作者简介:汤传根 (1985—),男,博士研究生,工程师。研究方向为重组多肽/蛋白药物、合成多肽药物、功能肽研究、合成生物学产业化应用。E-mail:rootyt@hanxinpharm.com
CLC Number:
Cite this article
TANG Chuan′gen, WANG Jing, ZHANG Shuo, ZHANG Haoning, KANG Zhen. Advances in synthesis and mining strategies for functional peptides[J]. Synthetic Biology Journal, 2025, 6(2): 461-478.
汤传根, 王璟, 张烁, 张昊宁, 康振. 功能肽合成和挖掘策略研究进展[J]. 合成生物学, 2025, 6(2): 461-478.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-067
| 分类 | 作用机制和典型氨基酸序列 | 特点和应用场景 | 参考 文献 |
|---|---|---|---|
| 抗菌肽 | ①破坏细胞膜结构:通过与细菌细胞膜中的磷脂双层结合,增加膜通透性,导致细胞内容物泄漏,从而杀死细菌 ②抑制细菌代谢途径:干扰细菌DNA、RNA和蛋白质合成,抑制细菌生长和繁殖 ③典型氨基酸序列:Phe-Trp-Lys-Phe-Lys | 广谱抗菌、抗感染,特别是对抗耐药性菌株感染有很好效果;促进伤口愈合;预防感染;治疗女性阴道念珠菌感染等 | [ |
| 抗病毒肽 | ①阻断病毒进入细胞:通过与病毒外壳蛋白或宿主细胞受体结合,阻止病毒附着和进入宿主细胞 ②抑制病毒复制:干扰病毒RNA或DNA合成,从而抑制病毒复制 ③典型氨基酸序列:Ala-Ala-Val-Ala-Leu-Leu-Pro-Ala-Val--Leu-Leu-Ala-Leu-Leu-Ala-Pro | 通过抑制病毒蛋白的功能,阻断病毒生命周期;可预防病毒感染,对于已感染病毒患者,则可阻止病毒在体内的进一步繁殖 | [ |
| 抗氧化肽 | ①清除自由基:通过捐献电子或氢原子,中和体内自由基,减少自由基对细胞损伤 ②提高抗氧化酶活性:通过提高超氧化物歧化酶(superoxide dismutase, SOD)和过氧化氢酶等抗氧化酶的活性,增强机体抗氧化能力 ③典型氨基酸序列:Leu-Ala-Asn-Ala-Lys | 安全性高,可以用在食品领域抗氧化上;也可开发相应抗氧化化妆品、保健食品或者药品 | [ |
| 抗紫外肽 | ①吸收紫外线:通过吸收紫外线光子,减少其对皮肤的穿透和损伤 ②修复紫外线损伤:通过促进DNA修复机制,修复紫外线引起的DNA损伤,减少皮肤癌的发生 ③典型氨基酸序列:Leu-Val-Asn-Glu-Leu-Thr-Glu-Phe-Ala-Gln | 增强皮肤对紫外线损伤的防御和修护能力以及防止光老化,在防晒护肤品中应用前景广阔 | [ |
| 抗癌肽 | ①诱导细胞凋亡:通过激活凋亡信号通路,如Caspase级联反应,诱导癌细胞凋亡 ②抑制血管生成:通过抑制血管内皮生长因子(vascular endothelial growth factor, VEGF)活性,减少肿瘤血管生成,从而抑制肿瘤生长 ③典型氨基酸序列:Xxx-Arg-Gly-Asp-Xxx | 肽易于人体吸收,可选择性地抑制肿瘤细胞生长,同时对正常细胞影响较小,亦可降低化疗药物毒副作用 | [ |
| 免疫调节肽 | ①增强免疫细胞功能:促进T细胞、B细胞和自然杀伤细胞(natural killer cell, NK cell)的增殖和活化,提高免疫系统整体功能 ②调节细胞因子分泌:通过调节细胞因子分泌,平衡促炎和抗炎反应,维持免疫稳态 ③典型氨基酸序列:Gly-Arg-Gly-(Asp)9 | 增强机体免疫反应或抑制过度免疫反应,达到免疫平衡效果,在调节免疫方面有良好的应用前景 | [ |
| 美白肽 | ①抑制酪氨酸酶活性:美白肽通过与酪氨酸酶结合,抑制其活性,从而减少黑色素的合成 ②干扰黑色素转运:某些美白肽可干扰黑色素小体从黑素细胞向角质形成细胞的转运,减少皮肤色素沉着 ③典型氨基酸序列: H-Met-Pro-D-Phe-Arg-D-Trp-Phe-Lys-Pro-Val-NH2 | 具有更好的安全性和美白活性,可在美白护肤品中广泛应用 | [ |
| 减肥肽 | ①促进胰岛素分泌:胰高血糖素样肽-1受体激动剂(glucagon-like peptide-1 receptor agonists, GLP-1RA)可增加胰岛素分泌,降低血糖水平 ②抑制胰高血糖素分泌:GLP-1RA可减少胰高血糖素的分泌,减缓肝脏释放葡萄糖 ③延缓胃排空:GLP-1RA能延缓胃排空速度,使饭后血糖上升更为温和 ④减少食欲:GLP-1RA可降低食欲 ⑤典型氨基酸序列:His-Aib-Glu-Gly-Thr-Phe-Thr-Ser-Asp-Val-Ser-Ser-Tyr-Leu-Glu-Gly-Gln-Ala-Ala-Lys-Glu-Phe-Ile-Ala-Trp-Leu-Val-Arg-Gly-Arg-Gly | GLP-1RA不仅能够良好安全地控制血糖水平,适用于2型糖尿病患者的血糖控制;而且能有效减少热量摄入,帮助控制体重,达到有效减肥效果 | [ |
Table 1 Classification of functional peptides and their action mechanism, characteristics and application scenarios
| 分类 | 作用机制和典型氨基酸序列 | 特点和应用场景 | 参考 文献 |
|---|---|---|---|
| 抗菌肽 | ①破坏细胞膜结构:通过与细菌细胞膜中的磷脂双层结合,增加膜通透性,导致细胞内容物泄漏,从而杀死细菌 ②抑制细菌代谢途径:干扰细菌DNA、RNA和蛋白质合成,抑制细菌生长和繁殖 ③典型氨基酸序列:Phe-Trp-Lys-Phe-Lys | 广谱抗菌、抗感染,特别是对抗耐药性菌株感染有很好效果;促进伤口愈合;预防感染;治疗女性阴道念珠菌感染等 | [ |
| 抗病毒肽 | ①阻断病毒进入细胞:通过与病毒外壳蛋白或宿主细胞受体结合,阻止病毒附着和进入宿主细胞 ②抑制病毒复制:干扰病毒RNA或DNA合成,从而抑制病毒复制 ③典型氨基酸序列:Ala-Ala-Val-Ala-Leu-Leu-Pro-Ala-Val--Leu-Leu-Ala-Leu-Leu-Ala-Pro | 通过抑制病毒蛋白的功能,阻断病毒生命周期;可预防病毒感染,对于已感染病毒患者,则可阻止病毒在体内的进一步繁殖 | [ |
| 抗氧化肽 | ①清除自由基:通过捐献电子或氢原子,中和体内自由基,减少自由基对细胞损伤 ②提高抗氧化酶活性:通过提高超氧化物歧化酶(superoxide dismutase, SOD)和过氧化氢酶等抗氧化酶的活性,增强机体抗氧化能力 ③典型氨基酸序列:Leu-Ala-Asn-Ala-Lys | 安全性高,可以用在食品领域抗氧化上;也可开发相应抗氧化化妆品、保健食品或者药品 | [ |
| 抗紫外肽 | ①吸收紫外线:通过吸收紫外线光子,减少其对皮肤的穿透和损伤 ②修复紫外线损伤:通过促进DNA修复机制,修复紫外线引起的DNA损伤,减少皮肤癌的发生 ③典型氨基酸序列:Leu-Val-Asn-Glu-Leu-Thr-Glu-Phe-Ala-Gln | 增强皮肤对紫外线损伤的防御和修护能力以及防止光老化,在防晒护肤品中应用前景广阔 | [ |
| 抗癌肽 | ①诱导细胞凋亡:通过激活凋亡信号通路,如Caspase级联反应,诱导癌细胞凋亡 ②抑制血管生成:通过抑制血管内皮生长因子(vascular endothelial growth factor, VEGF)活性,减少肿瘤血管生成,从而抑制肿瘤生长 ③典型氨基酸序列:Xxx-Arg-Gly-Asp-Xxx | 肽易于人体吸收,可选择性地抑制肿瘤细胞生长,同时对正常细胞影响较小,亦可降低化疗药物毒副作用 | [ |
| 免疫调节肽 | ①增强免疫细胞功能:促进T细胞、B细胞和自然杀伤细胞(natural killer cell, NK cell)的增殖和活化,提高免疫系统整体功能 ②调节细胞因子分泌:通过调节细胞因子分泌,平衡促炎和抗炎反应,维持免疫稳态 ③典型氨基酸序列:Gly-Arg-Gly-(Asp)9 | 增强机体免疫反应或抑制过度免疫反应,达到免疫平衡效果,在调节免疫方面有良好的应用前景 | [ |
| 美白肽 | ①抑制酪氨酸酶活性:美白肽通过与酪氨酸酶结合,抑制其活性,从而减少黑色素的合成 ②干扰黑色素转运:某些美白肽可干扰黑色素小体从黑素细胞向角质形成细胞的转运,减少皮肤色素沉着 ③典型氨基酸序列: H-Met-Pro-D-Phe-Arg-D-Trp-Phe-Lys-Pro-Val-NH2 | 具有更好的安全性和美白活性,可在美白护肤品中广泛应用 | [ |
| 减肥肽 | ①促进胰岛素分泌:胰高血糖素样肽-1受体激动剂(glucagon-like peptide-1 receptor agonists, GLP-1RA)可增加胰岛素分泌,降低血糖水平 ②抑制胰高血糖素分泌:GLP-1RA可减少胰高血糖素的分泌,减缓肝脏释放葡萄糖 ③延缓胃排空:GLP-1RA能延缓胃排空速度,使饭后血糖上升更为温和 ④减少食欲:GLP-1RA可降低食欲 ⑤典型氨基酸序列:His-Aib-Glu-Gly-Thr-Phe-Thr-Ser-Asp-Val-Ser-Ser-Tyr-Leu-Glu-Gly-Gln-Ala-Ala-Lys-Glu-Phe-Ile-Ala-Trp-Leu-Val-Arg-Gly-Arg-Gly | GLP-1RA不仅能够良好安全地控制血糖水平,适用于2型糖尿病患者的血糖控制;而且能有效减少热量摄入,帮助控制体重,达到有效减肥效果 | [ |
| 对比维度 | 固相合成法(SPPS) | 液相合成法(LPPS) |
|---|---|---|
| 原理与过程 | 固相载体上逐步缩合氨基酸 | 溶液中氨基酸/肽片段进行耦合 |
| 自动化程度 | 高,流程化的操作可以通过自动化的多肽合成设备实现自动化生产 | 较低,操作过程较为复杂,目前难以实现大规模自动化生产 |
| 适用范围 | 中长链多肽(通常40个氨基酸以内) | 短肽及特定结构要求的多肽(通常10个氨基酸以内) |
| 产率 | 较高,随着肽链延长收率下降 | 适中,随着肽链延长收率迅速下降 |
| 纯度 | 较高,随着肽链延长纯度下降 | 适中,随着肽链延长纯度迅速下降 |
| 耗时 | 较长,每个氨基酸通常需2~3 h | 适中,合成简洁迅速 |
| 成本 | 较高,树脂和大量溶剂 | 相对较低 |
Table 2 Advantages/disadvantages of the solid/liquid phase synthesis for the production of functional peptides
| 对比维度 | 固相合成法(SPPS) | 液相合成法(LPPS) |
|---|---|---|
| 原理与过程 | 固相载体上逐步缩合氨基酸 | 溶液中氨基酸/肽片段进行耦合 |
| 自动化程度 | 高,流程化的操作可以通过自动化的多肽合成设备实现自动化生产 | 较低,操作过程较为复杂,目前难以实现大规模自动化生产 |
| 适用范围 | 中长链多肽(通常40个氨基酸以内) | 短肽及特定结构要求的多肽(通常10个氨基酸以内) |
| 产率 | 较高,随着肽链延长收率下降 | 适中,随着肽链延长收率迅速下降 |
| 纯度 | 较高,随着肽链延长纯度下降 | 适中,随着肽链延长纯度迅速下降 |
| 耗时 | 较长,每个氨基酸通常需2~3 h | 适中,合成简洁迅速 |
| 成本 | 较高,树脂和大量溶剂 | 相对较低 |
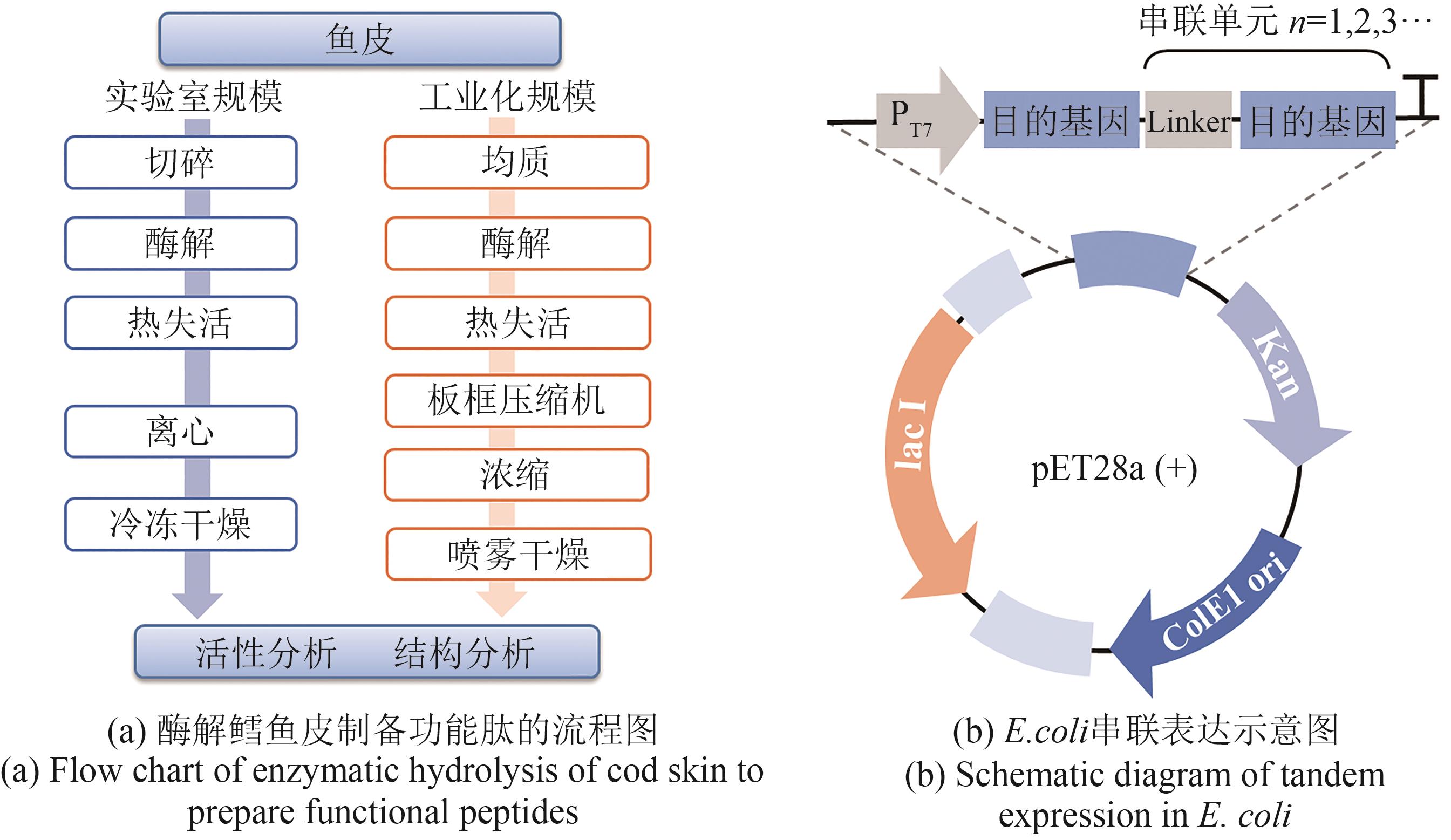
| 名称 | 原料 | 蛋白酶种类 | 酶解条件 | 分离纯化 | 功能描述 | 参考文献 |
|---|---|---|---|---|---|---|
| 降压肽 | 大豆 蛋白 | 嗜热菌蛋白酶、 胃蛋白酶、胰蛋白酶 | 50 g/L,55 °C,pH 8, 3 h;37 °C,pH 8, 3 h; 37 °C,pH 7.6,3 h | 离心、LC-MS | ACE抑制活性 | [ |
| 降压肽 | 诃子 果实 | 胃蛋白酶 | 1∶25(质量比),37°C,12 h | 离心、RP-HPLC | ACE抑制活性 | [ |
| 降压肽 | 米糠 | 胰蛋白酶 | 50 mg/mL,1500 U/mg,37 °C,pH 8,2 h | 离心 | ACE抑制活性、抗氧化 | [ |
| 降压肽 | 鸡皮 | 碱性蛋白酶 | 10 g/L,60 °C,pH 9.5,4 h | 离心、超滤 | ACE抑制活性 | [ |
| 降压肽 | 短鳍鱼 | 碱性蛋白酶 | 10 g/L | 离心、过滤 | ACE抑制活性 | [ |
| 抗氧化肽 | 牛毛 | 碱性蛋白酶 | 5%,55 °C,8 h | 离心、凝胶过滤、分子排阻色谱 | 抗氧化 | [ |
| 二肽酶抑制肽 | 三文鱼鱼白 | 蜂蜜曲霉蛋白酶 | 5种蛋白酶筛选,50 °C, >5 h,17.5 g酶/7.2 kg原料 | 硅藻土过滤、 超滤、浓缩 | 控制血糖 | [ |
| 血糖控制 以及抗炎肽 | 去骨 三文鱼 | 胃蛋白酶、胰蛋白酶、胰凝乳蛋白酶 | NA | 过滤、超滤 | 控制血糖、抗炎 | [ |
| 抗氧化肽 | 黄鳍 金枪鱼 | 碱性蛋白酶 | 酶量3000 U/g,50 °C,5 h,100 r/min | 离心、超滤Sephadex G-15 | 抗氧化 | [ |
| 抗氧化肽 | 鱿鱼 | 碱性蛋白酶 | 酶量5~30 U/g,5~180 min | 离心 | 抗氧化 | [ |
| 抗癌肽 | 虾壳 | Cryotin酶 | pH 8,50 °C,45 min | 离心、超滤 | 抗恶性细胞增殖 | [ |
Table 3 Summary of functional peptides produced by enzymatic hydrolysis
| 名称 | 原料 | 蛋白酶种类 | 酶解条件 | 分离纯化 | 功能描述 | 参考文献 |
|---|---|---|---|---|---|---|
| 降压肽 | 大豆 蛋白 | 嗜热菌蛋白酶、 胃蛋白酶、胰蛋白酶 | 50 g/L,55 °C,pH 8, 3 h;37 °C,pH 8, 3 h; 37 °C,pH 7.6,3 h | 离心、LC-MS | ACE抑制活性 | [ |
| 降压肽 | 诃子 果实 | 胃蛋白酶 | 1∶25(质量比),37°C,12 h | 离心、RP-HPLC | ACE抑制活性 | [ |
| 降压肽 | 米糠 | 胰蛋白酶 | 50 mg/mL,1500 U/mg,37 °C,pH 8,2 h | 离心 | ACE抑制活性、抗氧化 | [ |
| 降压肽 | 鸡皮 | 碱性蛋白酶 | 10 g/L,60 °C,pH 9.5,4 h | 离心、超滤 | ACE抑制活性 | [ |
| 降压肽 | 短鳍鱼 | 碱性蛋白酶 | 10 g/L | 离心、过滤 | ACE抑制活性 | [ |
| 抗氧化肽 | 牛毛 | 碱性蛋白酶 | 5%,55 °C,8 h | 离心、凝胶过滤、分子排阻色谱 | 抗氧化 | [ |
| 二肽酶抑制肽 | 三文鱼鱼白 | 蜂蜜曲霉蛋白酶 | 5种蛋白酶筛选,50 °C, >5 h,17.5 g酶/7.2 kg原料 | 硅藻土过滤、 超滤、浓缩 | 控制血糖 | [ |
| 血糖控制 以及抗炎肽 | 去骨 三文鱼 | 胃蛋白酶、胰蛋白酶、胰凝乳蛋白酶 | NA | 过滤、超滤 | 控制血糖、抗炎 | [ |
| 抗氧化肽 | 黄鳍 金枪鱼 | 碱性蛋白酶 | 酶量3000 U/g,50 °C,5 h,100 r/min | 离心、超滤Sephadex G-15 | 抗氧化 | [ |
| 抗氧化肽 | 鱿鱼 | 碱性蛋白酶 | 酶量5~30 U/g,5~180 min | 离心 | 抗氧化 | [ |
| 抗癌肽 | 虾壳 | Cryotin酶 | pH 8,50 °C,45 min | 离心、超滤 | 抗恶性细胞增殖 | [ |
| 表达系统 | 优势 | 不足 | 应用范围 | 产业化现状 |
|---|---|---|---|---|
| 大肠杆菌 | 繁殖速度快、培养周期短、遗传背景清晰、工艺放大技术成熟、成本低 | 易形成包涵体、存在内毒素风险 | 抗菌肽、抗氧化肽、减肥肽、化妆品肽等 | 技术成熟,可实现产业化放大 |
| 枯草芽 孢杆菌 | 遗传背景清晰、安全性强、可分泌表达至培养基中、纯化简单 | 表达量偏低、重组质粒易丢失 | 淀粉酶、蛋白酶、维生素等 | 技术成熟,产业化程度较低 |
| 酵母菌 | 一定的翻译后修饰、耐受能力强 | 发酵周期长、表达量不高 | 抗菌肽、免疫调节肽等 | 技术成熟,可实现产业化放大 |
Table 4 Applications of different expression systems in the preparation of functional peptides
| 表达系统 | 优势 | 不足 | 应用范围 | 产业化现状 |
|---|---|---|---|---|
| 大肠杆菌 | 繁殖速度快、培养周期短、遗传背景清晰、工艺放大技术成熟、成本低 | 易形成包涵体、存在内毒素风险 | 抗菌肽、抗氧化肽、减肥肽、化妆品肽等 | 技术成熟,可实现产业化放大 |
| 枯草芽 孢杆菌 | 遗传背景清晰、安全性强、可分泌表达至培养基中、纯化简单 | 表达量偏低、重组质粒易丢失 | 淀粉酶、蛋白酶、维生素等 | 技术成熟,产业化程度较低 |
| 酵母菌 | 一定的翻译后修饰、耐受能力强 | 发酵周期长、表达量不高 | 抗菌肽、免疫调节肽等 | 技术成熟,可实现产业化放大 |
| 68 | 陈清, 曾鑫, 彭永亮, 等. 重组串联融合蛋白制备目标多肽的方法: ZL201910563692.3[P]. 2022-05-13. |
| CHEN Q, ZENG X, PENG Y L,et al. Method for preparing target polypeptide by recombinant tandem fusion protein ZL201910563692.3[P]. 2022-05-13. | |
| 69 | ZHANG L C, WEI D D, ZHAN N, et al. Heterologous expression of the novel α-helical hybrid peptide PR-FO in Bacillus subtilis [J]. Bioprocess and Biosystems Engineering, 2020, 43(9): 1619-1627. |
| 70 | CHEN M L, LIN N F, LIU X D, et al. A novel antimicrobial peptide screened by a Bacillus subtilis expression system, derived from Larimichthys crocea Ferritin H, exerting bactericidal and parasiticidal activities[J]. Frontiers in Immunology, 2023, 14: 1168517. |
| 71 | SUN W F, WU Y M, DING W W, et al. An auto-inducible expression and high cell density fermentation of Beefy Meaty Peptide with Bacillus subtilis [J]. Bioprocess and Biosystems Engineering, 2020, 43(4): 701-710. |
| 72 | FU G, YUE J, LI D D, et al. An operator-based expression toolkit for Bacillus subtilis enables fine-tuning of gene expression and biosynthetic pathway regulation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(11): e2119980119. |
| 73 | LU Z H, YANG S H, YUAN X, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis [J]. Nucleic Acids Research, 2019, 47(7): e40. |
| 74 | ZHU X Y, LUO H, YU X R, et al. Genome-wide CRISPRi screening of key genes for recombinant protein expression in Bacillus subtilis [J]. Advanced Science, 2024, 11(33): 2404313. |
| 75 | ILGEN C, LIN‐CEREGHINO J, CREGG J M. Pichia pastoris[M/OL]//GELLISSEN G. Production of recombinant proteins. 1st ed. New York: Wiley, 2004: 143-162 [2024-08-08]. . |
| 76 | KARBALAEI M, REZAEE S A, FARSIANI H. Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins[J]. Journal of Cellular Physiology, 2020, 235(9): 5867-5881. |
| 77 | CAO X T, ZHANG Y, MAO R Y, et al. Design and recombination expression of a novel plectasin-derived peptide MP1106 and its properties against Staphylococcus aureus [J]. Applied Microbiology and Biotechnology, 2015, 99(6): 2649-2662. |
| 78 | ZHANG K, YANG N, TENG D, et al. Expression and characterization of the new antimicrobial peptide AP138L-arg26 anti Staphylococcus aureus [J]. Applied Microbiology and Biotechnology, 2024, 108(1): 111. |
| 79 | CAO J C, DE LA FUENTE-NUNEZ C, OU R W, et al. Yeast-based synthetic biology platform for antimicrobial peptide production[J]. ACS Synthetic Biology, 2018, 7(3): 896-902. |
| 80 | LI X H, FAN Y, LIN Q, et al. Expression of chromogranin A-derived antifungal peptide CGA-N12 in Pichia pastoris [J]. Bioengineered, 2020, 11(1): 318-327. |
| 81 | RENYE J A JR, SOMKUTI G A. Nisin-induced expression of a recombinant antihypertensive peptide in dairy lactic acid bacteria[J]. Biotechnology Letters, 2015, 37(7): 1447-1454. |
| 82 | HUYNH E, LI J L. Generation of Lactococcus lactis capable of coexpressing epidermal growth factor and trefoil factor to enhance in vitro wound healing[J]. Applied Microbiology and Biotechnology, 2015, 99(11): 4667-4677. |
| 83 | NONGONIERMA A B, O’KEEFFE M B, FITZGERALD R J. Milk protein hydrolysates and bioactive peptides[M/OL]//MCSWEENEY P L H, O’MAHONY J A. Advanced dairy chemistry: volume 1B: proteins: applied aspects. New York, NY: Springer New York, 2016: 417-482 [2024-08-10]. . |
| 84 | LIAO W, JAHANDIDEH F, FAN H B, et al. Egg protein-derived bioactive peptides: preparation, efficacy, and absorption[M/OL]//Advances in food and nutrition research. New York: Elsevier, 2018, 85: 1-58 [2024-08-10]. . |
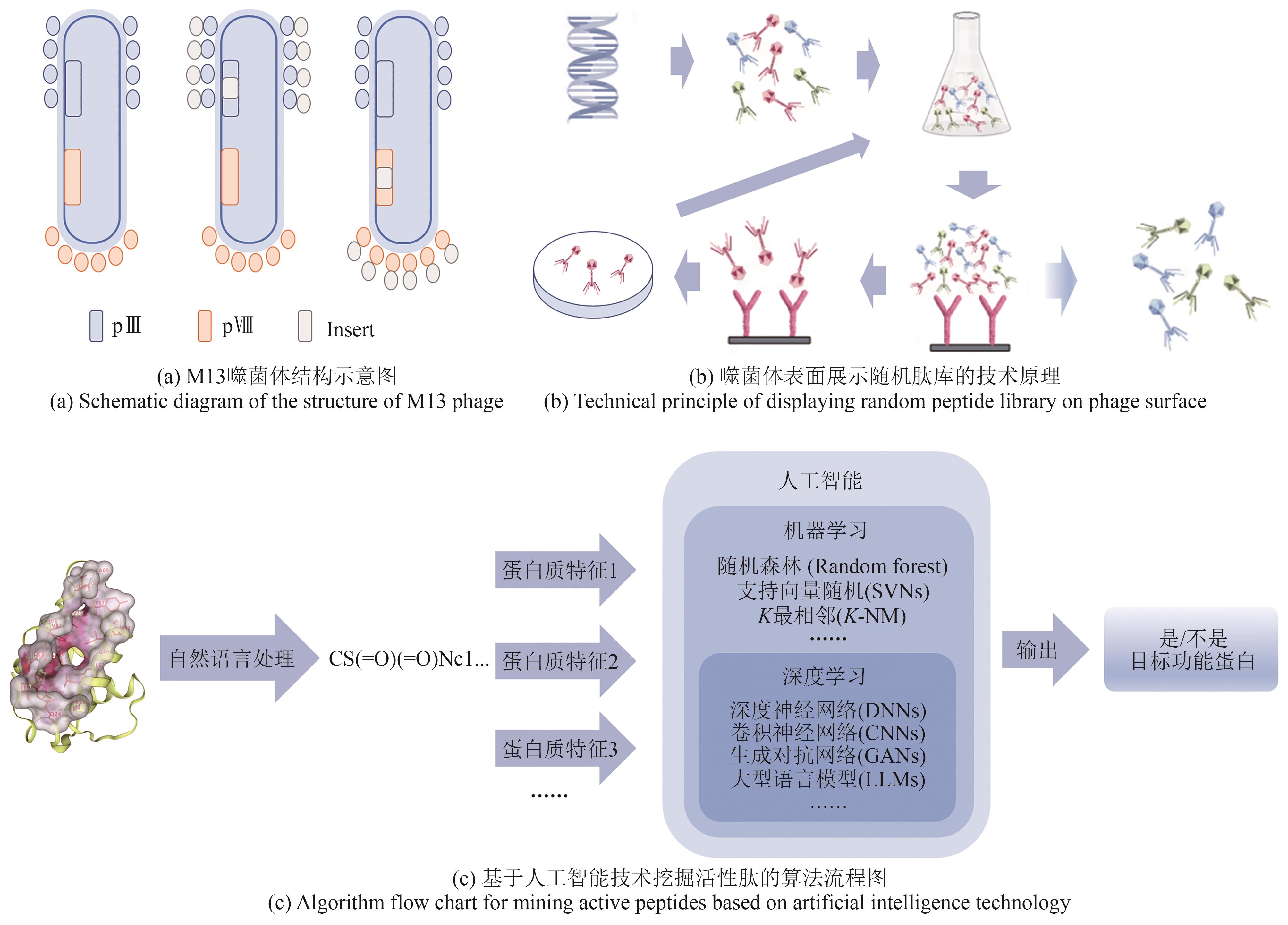
| 85 | SMITH G P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface[J]. Science, 1985, 228(4705): 1315-1317. |
| 86 | FLING M, HOROWITZ N H, HEINEMANN S F. The isolation and properties of crystalline tyrosinase from Neurospora [J]. Journal of Biological Chemistry, 1963, 238(6): 2045-2053. |
| 87 | SUN Y J, SHUKLA G S, WEAVER D, et al. Phage-display selection on tumor histological specimens with laser capture microdissection[J]. Journal of Immunological Methods, 2009, 347(1-2): 46-53. |
| 88 | YANG M, LIU C W, NIU M C, et al. Phage-display library biopanning and bioinformatic analysis yielded a high-affinity peptide to inflamed vascular endothelium both in vitro and in vivo [J]. Journal of Controlled Release, 2014, 174: 72-80. |
| 89 | GUAN M Q, WANG J, YANG L B, et al. Targeting osteosarcoma vasculature with peptide obtained by phage display[J]. Contemporary Oncology/ Współczesna Onkologia, 2014, 18(3): 165-170. |
| 90 | ZHOU C, KANG J L, WANG X X, et al. Phage display screening identifies a novel peptide to suppress ovarian cancer cells in vitro and in vivo in mouse models[J]. BMC Cancer, 2015, 15: 889. |
| 91 | WENG X J, LIAO Q D, LI K H, et al. Screening serum biomarker of knee osteoarthritis using a phage display technique[J]. Clinical Biochemistry, 2012, 45(4-5): 303-308. |
| 92 | YIN L, LUO Y Z, LIANG B, et al. Specific ligands for classical swine fever virus screened from landscape phage display library[J]. Antiviral Research, 2014, 109: 68-71. |
| 93 | LIU Z P, LIU J F, WANG K, et al. Selection of phage-displayed peptides for the detection of imidacloprid in water and soil[J]. Analytical Biochemistry, 2015, 485: 28-33. |
| 94 | CHEN Y P, SHEN Y Y, GUO X, et al. Transdermal protein delivery by a coadministered peptide identified via phage display[J]. Nature Biotechnology, 2006, 24(4): 455-460. |
| 95 | DUERR D M, WHITE S J, SCHLUESENER H J. Identification of peptide sequences that induce the transport of phage across the gastrointestinal mucosal barrier[J]. Journal of Virological Methods, 2004, 116(2): 177-180. |
| 96 | PASQUALINI R, RUOSLAHTI E. Organ targeting in vivo using phage display peptide libraries[J]. Nature, 1996, 380(6572): 364-366. |
| 97 | ZHANG X C, ZHANG X Y, GAO H L, et al. Phage display derived peptides for Alzheimer’s disease therapy and diagnosis[J]. Theranostics, 2022, 12(5): 2041-2062. |
| 98 | MANAVALAN B, BASITH S, SHIN T H, et al. MLACP: machine-learning-based prediction of anticancer peptides[J]. Oncotarget, 2017, 8(44): 77121-77136. |
| 99 | GUPTA S, SHARMA A K, SHASTRI V, et al. Prediction of anti-inflammatory proteins/peptides: an in silico approach[J]. Journal of Translational Medicine, 2017, 15(1): 7. |
| 100 | ILYAS S, LEE J, HWANG Y, et al. Deciphering Cathepsin K inhibitors: a combined QSAR, docking and MD simulation based machine learning approaches for drug design[J]. SAR and QSAR in Environmental Research, 2024, 35(9): 771-793. |
| 101 | GULL S, SHAMIM N, MINHAS F. AMAP: hierarchical multi-label prediction of biologically active and antimicrobial peptides[J]. Computers in Biology and Medicine, 2019, 107: 172-181. |
| 102 | CHO C Y, LEE S S, BANG D M, et al. ChemAP: predicting drug approval with chemical structures before clinical trial phase by leveraging multi-modal embedding space and knowledge distillation[J]. Scientific Reports, 2024, 14(1): 23010. |
| 1 | HARTMANN R, MEISEL H. Food-derived peptides with biological activity: from research to food applications[J]. Current Opinion in Biotechnology, 2007, 18(2): 163-169. |
| 2 | KITTS D D, WEILER K. Bioactive proteins and peptides from food sources. Applications of bioprocesses used in isolation and recovery[J]. Current Pharmaceutical Design, 2003, 9(16): 1309-1323. |
| 3 | HANCOCK R E W, SAHL H G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies[J]. Nature Biotechnology, 2006, 24(12): 1551-1557. |
| 4 | WANG G S, LI X, WANG Z. APD3: the antimicrobial peptide database as a tool for research and education[J]. Nucleic Acids Research, 2016, 44(D1): D1087-D1093. |
| 5 | MENDIS E, RAJAPAKSE N, BYUN H G, et al. Investigation of jumbo squid (Dosidicus gigas) skin gelatin peptides for their in vitro antioxidant effects[J]. Life Sciences, 2005, 77(17): 2166-2178. |
| 6 | SUETSUNA K. Antioxidant peptides from the protease digest of prawn (Penaeus japonicus) muscle[J]. Marine Biotechnology, 2000, 2(1): 5-10. |
| 7 | SARMADI B H, ISMAIL A. Antioxidative peptides from food proteins: a review[J]. Peptides, 2010, 31(10): 1949-1956. |
| 8 | PIHLANTO A. Antioxidative peptides derived from milk proteins[J]. International Dairy Journal, 2006, 16(11): 1306-1314. |
| 9 | FREITAS A C, ANDRADE J C, SILVA F M, et al. Antioxidative peptides: trends and perspectives for future research[J]. Current Medicinal Chemistry, 2013, 20(36): 4575-4594. |
| 10 | ZHANG X L, ZHUANG H, WU S J, et al. Marine bioactive peptides: anti-photoaging mechanisms and potential skin protective effects[J]. Current Issues in Molecular Biology, 2024, 46(2): 990-1009. |
| 11 | DOUNGAPAI C, SIRIWOHARN T, MALILA Y, et al. UV-B protective and antioxidant activities of protein hydrolysate from sea cucumber (Holothuria scabra) using enzymatic hydrolysis[J]. Frontiers in Marine Science, 2022, 9: 892255. |
| 12 | ZHANG Y H, WANG C, ZHANG W H, et al. Bioactive peptides for anticancer therapies[J]. Biomaterials Translational, 2023, 4(1): 5-17. |
| 13 | WANG L H, DONG C, LI X, et al. Anticancer potential of bioactive peptides from animal sources (Review)[J]. Oncology Reports, 2017, 38(2): 637-651. |
| 103 | GIGUÈRE S, LAVIOLETTE F, MARCHAND M, et al. Machine learning assisted design of highly active peptides for drug discovery[J]. PLoS Computational Biology, 2015, 11(4): e1004074. |
| 104 | JOO S H, PEI D H. Synthesis and screening of support-bound combinatorial peptide libraries with free C-termini: determination of the sequence specificity of PDZ domains[J]. Biochemistry, 2008, 47(9): 3061-3072. |
| 105 | FANG C, MORIWAKI Y, LI C H, et al. Prediction of antifungal peptides by deep learning with character embedding[J]. IPSJ Transactions on Bioinformatics, 2019, 12: 21-29. |
| 106 | AGRAWAL P, BHALLA S, CHAUDHARY K, et al. In silico approach for prediction of antifungal peptides[J]. Frontiers in Microbiology, 2018, 9: 323. |
| 107 | BJERRUM E J, SATTAROV B. Improving chemical autoencoder latent space and molecular de novo generation diversity with heteroencoders[J]. Biomolecules, 2018, 8(4): 131. |
| 108 | CAO D S, LIU S, XU Q S, et al. Large-scale prediction of drug-target interactions using protein sequences and drug topological structures[J]. Analytica Chimica Acta, 2012, 752: 1-10. |
| 109 | KAO P Y, YANG Y C, CHIANG W Y, et al. Exploring the advantages of quantum generative adversarial networks in generative chemistry[J]. Journal of Chemical Information and Modeling, 2023, 63(11): 3307-3318. |
| 110 | MÜLLER A T, HISS J A, SCHNEIDER G. Recurrent neural network model for constructive peptide design[J]. Journal of Chemical Information and Modeling, 2018, 58(2): 472-479. |
| 111 | MA Y, GUO Z Y, XIA B B, et al. Identification of antimicrobial peptides from the human gut microbiome using deep learning[J]. Nature Biotechnology, 2022, 40(6): 921-931. |
| 112 | KHABBAZ H, KARIMI-JAFARI M H, SABOURY A A, et al. Prediction of antimicrobial peptides toxicity based on their physico-chemical properties using machine learning techniques[J]. BMC Bioinformatics, 2021, 22(1): 549. |
| 113 | XIAO X, WANG P, LIN W Z, et al. iAMP-2L: a two-level multi-label classifier for identifying antimicrobial peptides and their functional types[J]. Analytical Biochemistry, 2013, 436(2): 168-177. |
| 114 | WEI L Y, ZHOU C, CHEN H R, et al. ACPred-FL: a sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides[J]. Bioinformatics, 2018, 34(23): 4007-4016. |
| 14 | SOON T N, CHIA A Y Y, YAP W H, et al. Anticancer mechanisms of bioactive peptides[J]. Protein & Peptide Letters, 2020, 27(9): 823-830. |
| 15 | PAVLICEVIC M, MARMIROLI N, MAESTRI E. Immunomodulatory peptides: a promising source for novel functional food production and drug discovery[J]. Peptides, 2022, 148: 170696. |
| 16 | CLEMENTE A. Enzymatic protein hydrolysates in human nutrition[J]. Trends in Food Science & Technology, 2000, 11(7): 254-262. |
| 17 | KORHONEN H, PIHLANTO A. Bioactive peptides: production and functionality[J]. International Dairy Journal, 2006, 16(9): 945-960. |
| 18 | UDENIGWE C C, ALUKO R E. Food protein-derived bioactive peptides: production, processing, and potential health benefits[J]. Journal of Food Science, 2012, 77(1): R11-R24. |
| 19 | ALTMANN S E, JONES J C, SCHULTZ-CHERRY S, et al. Inhibition of Vaccinia virus entry by a broad spectrum antiviral peptide[J]. Virology, 2009, 388(2): 248-259. |
| 20 | WANG Y G, WANG X, ZHANG Y F, et al. RGD-modified polymeric micelles as potential carriers for targeted delivery to integrin-overexpressing tumor vasculature and tumor cells[J]. Journal of Drug Targeting, 2009, 17(6): 459-467. |
| 21 | NGOC L T N, MOON J Y, LEE Y C. Insights into bioactive peptides in cosmetics[J]. Cosmetics, 2023, 10(4): 111. |
| 22 | ZHAO W, YANG A Q, WANG J, et al. Potential application of natural bioactive compounds as skin-whitening agents: a review[J]. Journal of Cosmetic Dermatology, 2022, 21(12): 6669-6687. |
| 23 | SURYANINGTYAS I T, JE J Y. Bioactive peptides from food proteins as potential anti-obesity agents: mechanisms of action and future perspectives[J]. Trends in Food Science & Technology, 2023, 138: 141-152. |
| 24 | KUMAR M S. Peptides and peptidomimetics as potential antiobesity agents: overview of current status[J]. Frontiers in Nutrition, 2019, 6: 11. |
| 25 | MUTTENTHALER M, KING G F, ADAMS D J, et al. Trends in peptide drug discovery[J]. Nature Reviews Drug Discovery, 2021, 20(4): 309-325. |
| 115 | 丁明珠, 李炳志, 王颖, 等. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28. |
| DING M Z, LI B Z, WANG Y, et al. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28. | |
| 116 | KOWALCZYK R, HARRIS P W R, WILLIAMS G M, et al. Peptide lipidation-a synthetic strategy to afford peptide based therapeutics[M/OL]. Advances in experimental medicine and biology: peptides and peptide-based biomaterials and their biomedical applications. Cham: Springer, 2017, 1030: 185-227. (2017-10-29)[2024-08-01]. . |
| 117 | GU S P. Applying machine learning algorithms for the analysis of biological sequences and medical records[D/OL]. Electronic Theses and Dissertations. Brookings: South Dakota State University, 2019, 3666[2024-08-01]. . |
| 118 | TRIPATHI N M, BANDYOPADHYAY A. High throughput virtual screening (HTVS) of peptide library: technological advancement in ligand discovery[J]. European Journal of Medicinal Chemistry, 2022, 243: 114766. |
| 119 | DANG Y, GUAN J J. Nanoparticle-based drug delivery systems for cancer therapy[J]. Smart Materials in Medicine, 2020, 1: 10-19. |
| 26 | MERRIFIELD R B. Solid phase peptide synthesis. Ⅰ. The synthesis of a tetrapeptide[J]. Journal of the American Chemical Society, 1963, 85(14): 2149-2154. |
| 27 | ABDILDINOVA D A, KURTH P M J, GONG P Y. Solid-phase synthesis of peptidomimetics with peptide backbone modifications[J]. Asian Journal of Organic Chemistry, 2021, 10(9): 2300-2317. |
| 28 | ZHANG R F, YAN H, WANG X J, et al. Screening of a short chain antimicrobial peptide - FWKFK and its application in wound healing[J]. Biomaterials Science, 2023, 11(5): 1867-1875. |
| 29 | SABANA I, NAUFAL M, WIANI I, et al. Synthesis of antioxidant peptide SCAP1 (Leu-Ala-Asn-Ala-Lys)[J]. Egyptian Journal of Chemistry, 2020, 63(3): 921-926. |
| 30 | BAHARLOUI M, MIRSHOKRAEE S A, MONFARED A, et al. Design and synthesis of novel triazole-based peptide analogues as anticancer agents[J]. Iranian Journal of Pharmaceutical Research, 2019, 18(3): 1299-1308. |
| 31 | WALEWSKA A, KOSIKOWSKA-ADAMUS P, TOMCZYKOWSKA M, et al. Improving fmoc solid phase synthesis of human beta defensin 3[J]. International Journal of Molecular Sciences, 2022, 23(20): 12562. |
| 32 | COLLINS J M, SINGH S K, WHITE T A, et al. Total wash elimination for solid phase peptide synthesis[J]. Nature Communications, 2023, 14(1): 8168. |
| 33 | BARREDO-VACCHELLI G R, RODRÍGUEZ J A, ELOY J A, et al. A novel method for liraglutide synthesis and purification[J]. Peptide Science, 2024, 116(5): e24351. |
| 34 | CHRISTOU G A, KATSIKI N, BLUNDELL J, et al. Semaglutide as a promising antiobesity drug[J]. Obesity Reviews, 2019, 20(6): 805-815. |
| 35 | PENG D Z, LI Y, SI L L, et al. A two-step method preparation of semaglutide through solid-phase synthesis and inclusion body expression[J]. Protein Expression and Purification, 2024, 219: 106477. |
| 36 | ANDRAOS J, MUHAR H, SMITH S R. Beyond glycemia: comparing tirzepatide to GLP-1 analogues[J]. Reviews in Endocrine & Metabolic Disorders, 2023, 24(6): 1089-1101. |
| 37 | FREDERICK M O, BOYSE R A, BRADEN T M, et al. Kilogram-scale GMP manufacture of tirzepatide using a hybrid SPPS/LPPS approach with continuous manufacturing[J]. Organic Process Research & Development, 2021, 25(7): 1628-1636. |
| 38 | ROSENSTOCK J, FRIAS J, JASTREBOFF A M, et al. Retatrutide, a GIP, GLP-1 and glucagon receptor agonist, for people with type 2 diabetes: a randomised, double-blind, placebo and active-controlled, parallel-group, phase 2 trial conducted in the USA[J]. The Lancet, 2023, 402(10401): 529-544. |
| 39 | KENT S B H. Chemical synthesis of peptides and proteins[J]. Annual Review of Biochemistry, 1988, 57: 957-989. |
| 40 | DAWSON P E, MUIR T W, CLARK-LEWIS I, et al. Synthesis of proteins by native chemical ligation[J]. Science, 1994, 266(5186): 776-779. |
| 41 | TYMECKA D, MISICKA A. Solution phase peptide synthesis: the case of biphalin[M/OL]//HUSSEIN W M, SKWARCZYNSKI M, TOTH I. Peptide synthesis: peptide synthesis. New York, NY: Springer US, 2020: 1-11 [2024-10-15]. . |
| 42 | GU X T, CHEN W J, GUO T, et al. A novel synthetic method for backbone-cyclized polypeptide POL7080 with the help of hydrophobic-support materials[J]. Organic & Biomolecular Chemistry, 2024, 22(1): 85-89. |
| 43 | LI H D, WANG L J, ZHANG L Y, et al. Scalable preparation of green C-terminal amidation peptide-synthesis TAGs and the optimized TAG-assisted liquid-phase synthesis of eptifibatide[J]. Sustainable Chemistry and Pharmacy, 2024, 41: 101684. |
| 44 | 蔡木易. 食源性低聚肽[M]. 北京: 中国轻工业出版社, 2021. |
| CAI M Y. Food-derived oligopeptides[M]. Beijing: China Light Industry Press, 2021. | |
| 45 | GU Y C, WU J P. LC-MS/MS coupled with QSAR modeling in characterising of angiotensin Ⅰ-converting enzyme inhibitory peptides from soybean proteins[J]. Food Chemistry, 2013, 141(3): 2682-2690. |
| 46 | SORNWATANA T, BANGPHOOMI K, ROYTRAKUL S, et al. Chebulin: Terminalia chebula Retz. fruit-derived peptide with angiotensin-Ⅰ-converting enzyme inhibitory activity[J]. Biotechnology and Applied Biochemistry, 2015, 62(6): 746-753. |
| 47 | WANG X M, CHEN H X, FU X G, et al. A novel antioxidant and ACE inhibitory peptide from rice bran protein: Biochemical characterization and molecular docking study[J]. LWT, 2017, 75: 93-99. |
| 48 | BABJI A S, DAUD N A, HUSAIN S G. Effect of molecular weight reduction of polypeptides on angiotensin converting enzyme (ACE) inhibitory activity in chicken skin hydrolysate (collagen)[J]. Journal of Nutritional Therapeutics, 2014, 3(2): 81-86. |
| 49 | RASLI H I, SARBON N M. Optimization of enzymatic hydrolysis conditions and characterization of Shortfin scad (Decapterus Macrosoma) skin gelatin hydrolysate sate using response surface methodology[J]. International Food Research Journal, 2018, 25(4): 1541-1549. |
| 50 | ZENG W C, ZHANG W H, HE Q, et al. Purification and characterization of a novel antioxidant peptide from bovine hair hydrolysates[J]. Process Biochemistry, 2015, 50(6): 948-954. |
| 51 | TAKAHASHI Y, KAMATA A, KONISHI T. Dipeptidyl peptidase-Ⅳ inhibitory peptides derived from salmon milt and their effects on postprandial blood glucose level[J]. Fisheries Science, 2021, 87(4): 619-626. |
| 52 | HENAUX L, PEREIRA K D, THIBODEAU J, et al. Glucoregulatory and anti-inflammatory activities of peptide fractions separated by electrodialysis with ultrafiltration membranes from salmon protein hydrolysate and identification of four novel glucoregulatory peptides[J]. Membranes, 2021, 11(7): 528. |
| 53 | CAI B N, WAN P, CHEN H, et al. Purification and identification of novel myeloperoxidase inhibitory antioxidant peptides from tuna (Thunnas albacares) protein hydrolysates[J]. Molecules, 2022, 27(9): 2681. |
| 54 | SINGH A, UTOMO PUTRI G A, MITTAL A, et al. Protein hydrolysate from splendid squid (Loligo formosana) fins: antioxidant, functional properties, and flavoring profile[J]. Turkish Journal of Fisheries and Aquatic Sciences, 2022, 22(6): TRJFAS21005. |
| 55 | KANNAN A, HETTIARACHCHY N S, MARSHALL M, et al. Shrimp shell peptide hydrolysates inhibit human cancer cell proliferation[J]. Journal of the Science of Food and Agriculture, 2011, 91(10): 1920-1924. |
| 56 | DE LA FUENTE B, PALLARÉS N, BERRADA H, et al. Salmon (Salmo salar) side streams as a bioresource to obtain potential antioxidant peptides after applying pressurized liquid extraction (PLE)[J]. Marine Drugs, 2021, 19(6): 323. |
| 57 | WANG B, LI L, CHI C F, et al. Purification and characterisation of a novel antioxidant peptide derived from blue mussel (Mytilus edulis) protein hydrolysate[J]. Food Chemistry, 2013, 138(2-3): 1713-1719. |
| 58 | KIM E K, KIM Y S, HWANG J W, et al. Purification of a novel nitric oxide inhibitory peptide derived from enzymatic hydrolysates of Mytilus coruscus [J]. Fish & Shellfish Immunology, 2013, 34(6): 1416-1420. |
| 59 | CHEN X L, PENG M, LI J, et al. Preparation and functional evaluation of collagen oligopeptide-rich hydrolysate from fish skin with the serine collagenolytic protease from Pseudoalteromonas sp. SM9913[J]. Scientific Reports, 2017, 7(1): 15716. |
| 60 | HERBEL V, SCHÄFER H, WINK M. Recombinant production of snakin-2 (an antimicrobial peptide from tomato) in E. coli and analysis of its bioactivity[J]. Molecules, 2015, 20(8): 14889-14901. |
| 61 | WU Y Y, MA Y K, LI L H, et al. Preparation and antioxidant activities in vitro of a designed antioxidant peptide from Pinctada fucata by recombinant Escherichia coli [J]. Journal of Microbiology and Biotechnology, 2018, 28(1): 1-11. |
| 62 | MOMEN A H, HARZANDI N, HADDADI A, et al. Implementation of a novel self-induced promoter for the expression of pharmaceutical peptides in Escherichia coli: YY(3-36) peptide[J]. Hormone Molecular Biology and Clinical Investigation, 2020, 41(1): 20180056. |
| 63 | RAUNIYAR K, AKHONDZADEH S, GĄCIARZ A, et al. Bioactive VEGF-C from E. coli [J]. Scientific Reports, 2022, 12: 18157. |
| 64 | XIE H L, LI J, LI L, et al. Enhanced proliferation and differentiation of neural stem cells grown on PHA films coated with recombinant fusion proteins[J]. Acta Biomaterialia, 2013, 9(8): 7845-7854. |
| 65 | 曹艳萍, 单安山, 马清泉, 等. 多拷贝策略在小肽表达中的应用[J]. 生物工程学报, 2011, 27(5): 684-689. |
| CAO Y P, SHAN A S, MA Q Q, et al. Application of multi-copies in expression of smaller peptides: a review[J]. Chinese Journal of Biotechnology, 2011, 27(5): 684-689. | |
| 66 | 黄欣媛, 邹礼平, 范红波. 豆类活性肽PA1b在大肠杆菌中的多拷贝串联表达[J]. 江苏农业科学, 2022, 50(15): 57-62. |
| HUANG X Y, ZOU L P, FAN H B. Prokaryotic expression of multicopies of legume peptide PA1b in Escherichia coli [J]. Jiangsu Agricultural Sciences, 2022, 50(15): 57-62. | |
| 67 | ZHANG J X, MOVAHEDI A, WEI Z H, et al. High-level SUMO-mediated fusion expression of ABP-dHC-cecropin A from multiple joined genes in Escherichia coli [J]. Analytical Biochemistry, 2016, 509: 15-23. |
| [1] | WU Ke, LUO Jiahao, LI Feiran. Applications of machine learning in the reconstruction and curation of genome-scale metabolic models [J]. Synthetic Biology Journal, 2025, 6(3): 566-584. |
| [2] | TIAN Xiao-jun, ZHANG Rixin. “Economics Paradox” with cells in synthetic gene circuits [J]. Synthetic Biology Journal, 2025, 6(3): 532-546. |
| [3] | ZHANG Yiqing, LIU Gaowen. Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(3): 685-700. |
| [4] | YANG Ying, LI Xia, LIU Lizhong. Applications of synthetic biology to stem-cell-derived modeling of early embryonic development [J]. Synthetic Biology Journal, 2025, 6(3): 669-684. |
| [5] | HUANG Yi, SI Tong, LU Anjing. Standardization for biomanufacturing: global landscape, critical challenges, and pathways forward [J]. Synthetic Biology Journal, 2025, 6(3): 701-714. |
| [6] | SONG Chengzhi, LIN Yihan. AI-enabled directed evolution for protein engineering and optimization [J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [7] | SHENG Zhouhuang, CHEN Zhixian, ZHANG Yan. Research progress of yeast mannoproteins [J]. Synthetic Biology Journal, 2025, 6(2): 408-421. |
| [8] | ZHANG Mengyao, CAI Peng, ZHOU Yongjin. Synthetic biology drives the sustainable production of terpenoid fragrances and flavors [J]. Synthetic Biology Journal, 2025, 6(2): 334-356. |
| [9] | ZHANG Lu’ou, XU Li, HU Xiaoxu, YANG Ying. Synthetic biology ushers cosmetic industry into the “bio-cosmetics” era [J]. Synthetic Biology Journal, 2025, 6(2): 479-491. |
| [10] | LU Jinchang, WU Yaokang, LV Xueqin, LIU Long, CHEN Jian, LIU Yanfeng. Green biomanufacturing of ceramide sphingolipids [J]. Synthetic Biology Journal, 2025, 6(2): 422-444. |
| [11] | YI Jinhang, TANG Yulin, LI Chunyu, WU Heyun, MA Qian, XIE Xixian. Applications and advances in the research of biosynthesis of amino acid derivatives as key ingredients in cosmetics [J]. Synthetic Biology Journal, 2025, 6(2): 254-289. |
| [12] | WEI Lingzhen, WANG Jia, SUN Xinxiao, YUAN Qipeng, SHEN Xiaolin. Biosynthesis of flavonoids and their applications in cosmetics [J]. Synthetic Biology Journal, 2025, 6(2): 373-390. |
| [13] | XIAO Sen, HU Litao, SHI Zhicheng, WANG Fayin, YU Siting, DU Guocheng, CHEN Jian, KANG Zhen. Research advances in biosynthesis of hyaluronic acid with controlled molecular weights [J]. Synthetic Biology Journal, 2025, 6(2): 445-460. |
| [14] | WANG Qian, GUO Shiting, XIN Bo, ZHONG Cheng, WANG Yu. Advances in biosynthesis of L-arginine using engineered microorganisms [J]. Synthetic Biology Journal, 2025, 6(2): 290-305. |
| [15] | ZUO Yimeng, ZHANG Jiaojiao, LIAN Jiazhang. Enabling technology for the biosynthesis of cosmetic raw materials with Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2025, 6(2): 233-253. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||