Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (2): 254-289.DOI: 10.12211/2096-8280.2024-060
• Invited Review • Previous Articles Next Articles
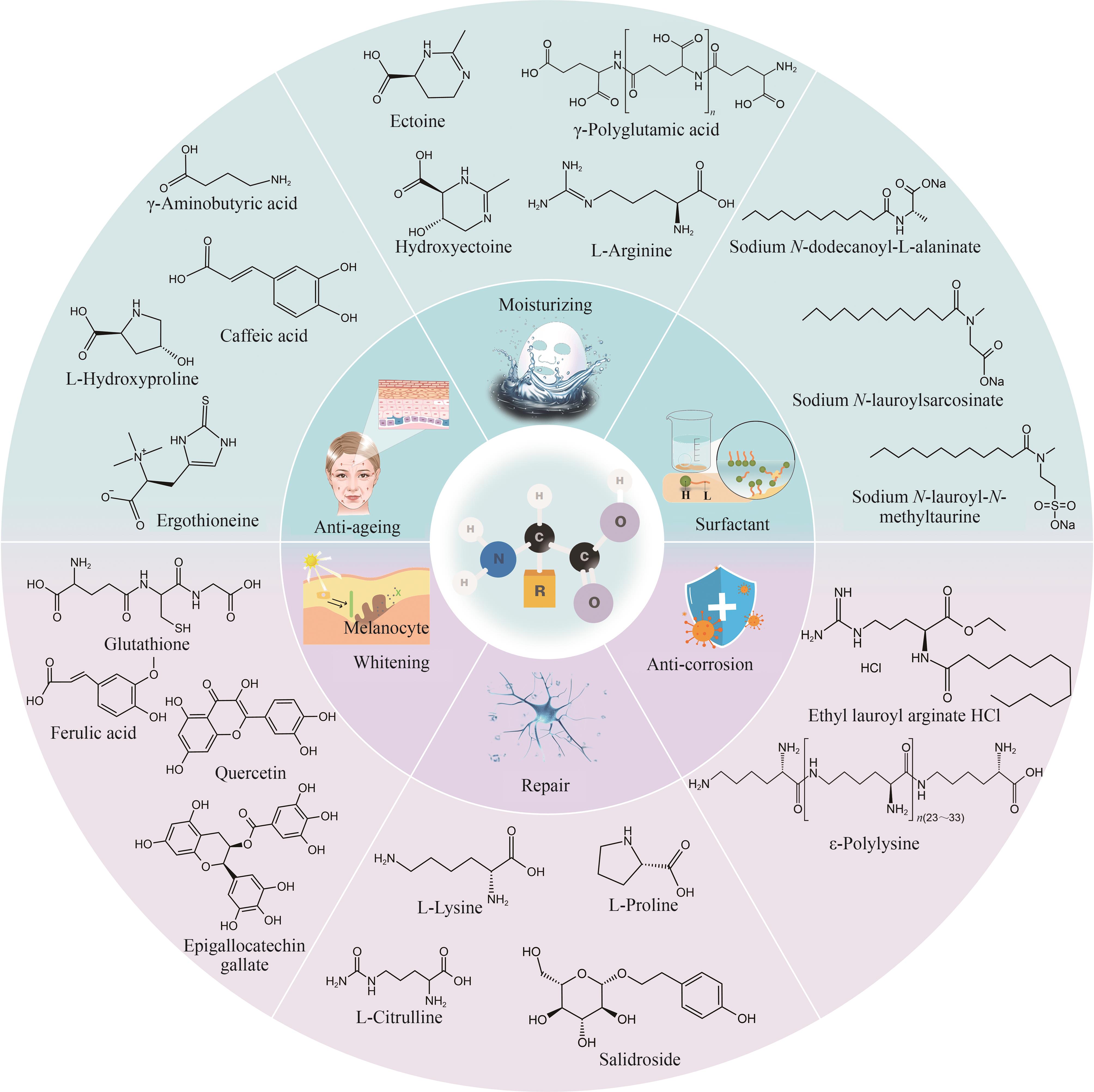
Applications and advances in the research of biosynthesis of amino acid derivatives as key ingredients in cosmetics
YI Jinhang1, TANG Yulin1, LI Chunyu1, WU Heyun1,2, MA Qian1,2, XIE Xixian1,2
- 1.College of Biotechnology,Tianjin University of Science and Technology,Tianjin 300457,China
2.Key Laboratory of Industrial Fermentation Microbiology,Ministry of Education,Tianjin University of Science and Technology,Tianjin 300457,China
-
Received:2024-08-01Revised:2024-10-11Online:2025-05-20Published:2025-04-30 -
Contact:MA Qian, XIE Xixian
氨基酸衍生物在化妆品中的应用及其生物合成研究进展
伊进行1, 唐宇琳1, 李春雨1, 吴鹤云1,2, 马倩1,2, 谢希贤1,2
- 1.天津科技大学生物工程学院,天津 300457
2.天津科技大学工业发酵微生物教育部重点实验室,天津 300457
-
通讯作者:马倩,谢希贤 -
作者简介:伊进行 (1998—),男,博士。研究方向为发酵工程、代谢工程和系统生物学。E-mail:yijin_hang@163.com马倩 (1987—),女,博士,副教授。研究方向为代谢工程、系统生物学。E-mail:qianma1987@tust.edu.cn谢希贤 (1976—),男,博士,教授,天津科技大学学术委员会委员。研究方向为合成生物学、代谢工程。E-mail:xixianxie@tust.edu.cn -
基金资助:国家重点研发计划(2022YFA0911800);国家自然科学基金(22378315);山东省重点研发计划(2022CXGC010506)
CLC Number:
Cite this article
YI Jinhang, TANG Yulin, LI Chunyu, WU Heyun, MA Qian, XIE Xixian. Applications and advances in the research of biosynthesis of amino acid derivatives as key ingredients in cosmetics[J]. Synthetic Biology Journal, 2025, 6(2): 254-289.
伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-060
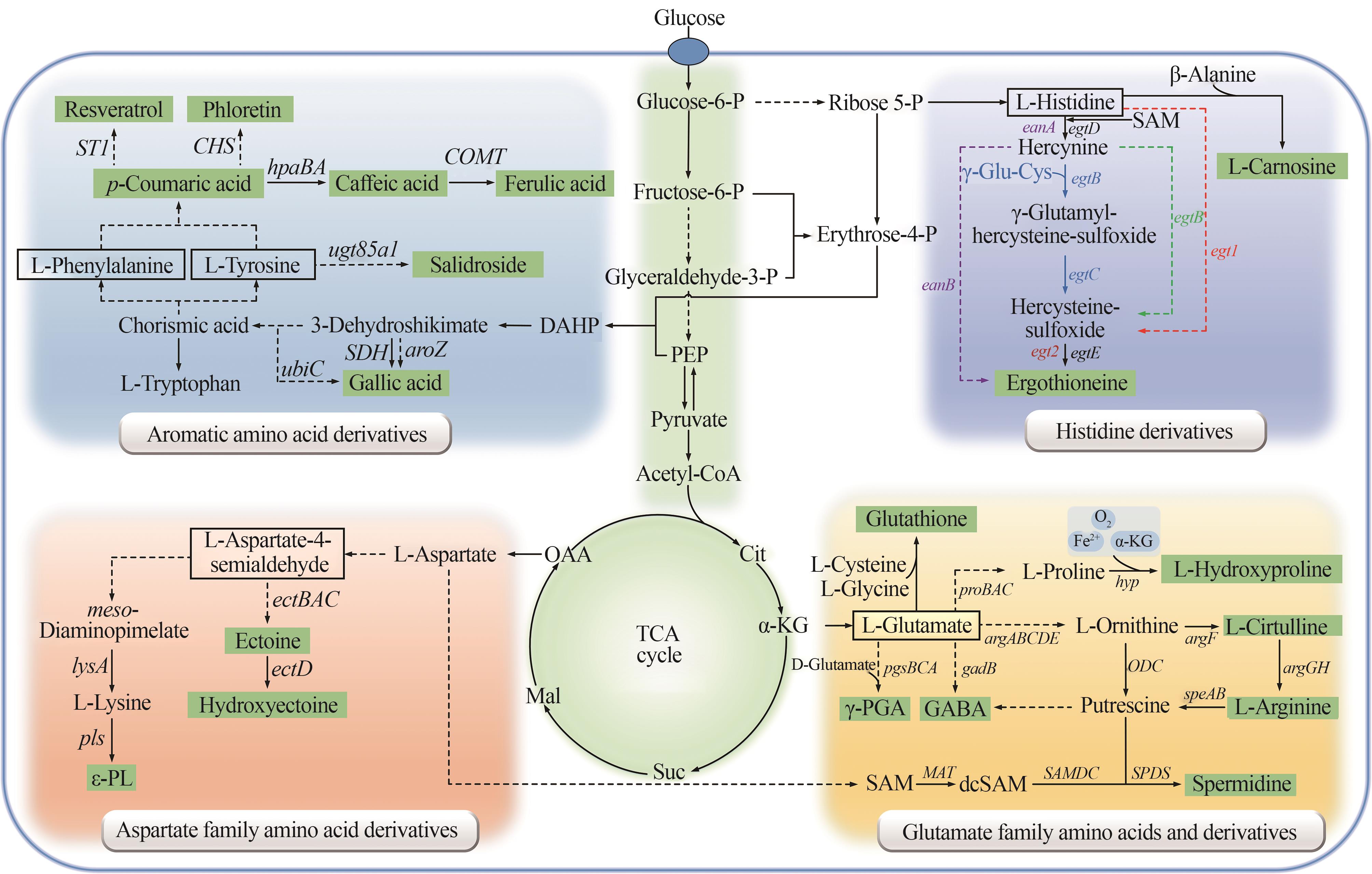
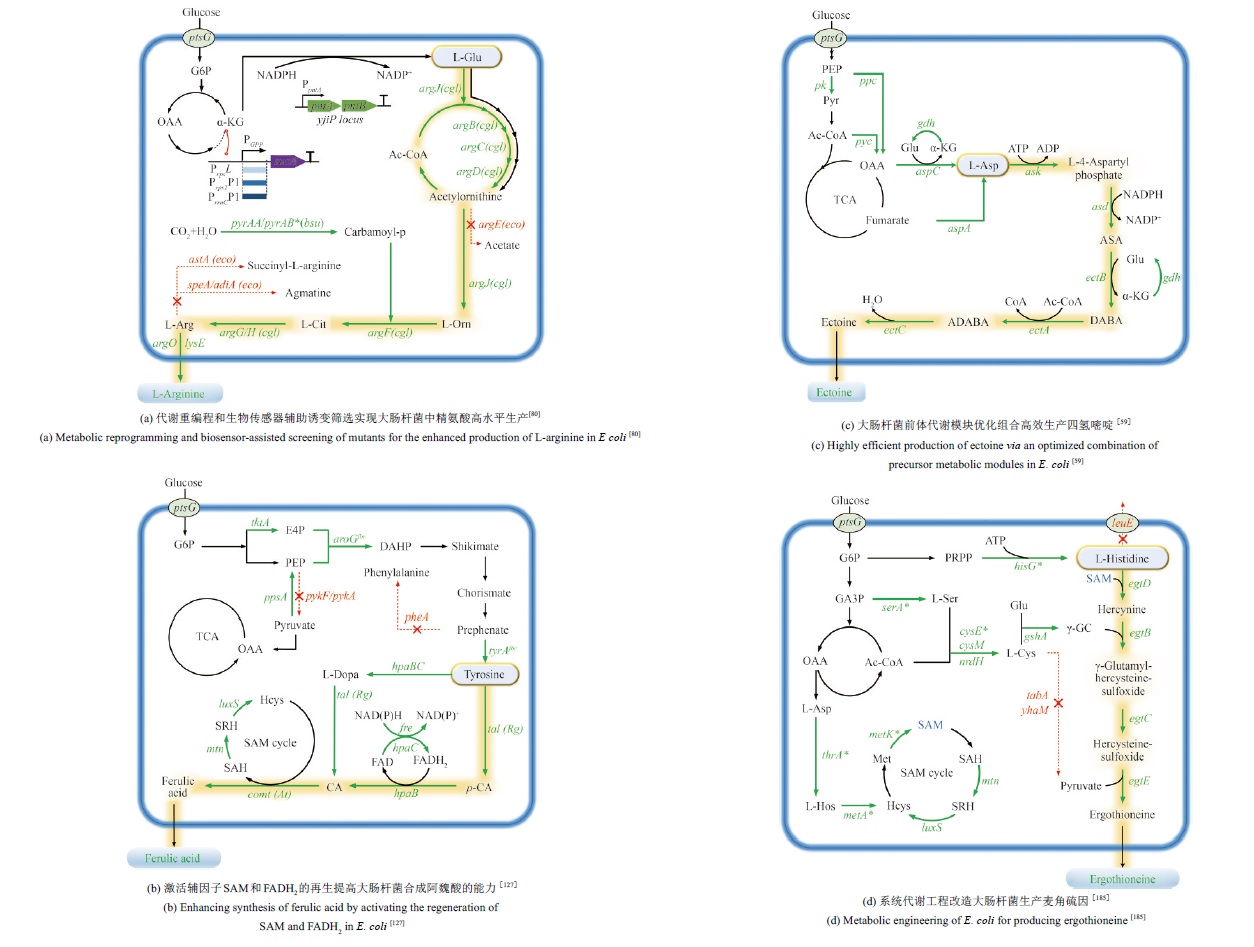
Fig. 2 Microbial synthesis pathways of amino acid derivatives as cosmetic raw materials(The biosynthetic pathway of ergothioneine: purple represents the anaerobic bacteria pathway; blue represents actinomycete pathway; green represents the bacterial pathway such as methylobacterium; red represents the fungal pathway. PEP—Phosphoenolpyruvate; Cit—Citrate; α-KG—α-Ketoglutarate; Suc—Succinate; Mal-Malate; OAA—Oxaloacetic acid; SAM—S-Adenosylmethionine; dcSAM—Decarboxylated S-adenosylmethionine; ε-PL—ε-poly-L-lysine)
| 氨基酸及衍生物 | 底盘菌株 | 生产 方法 | 主要策略 | 发酵规模 | 产量 | 生产强度 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 精氨酸 | 钝齿棒 杆菌 | 微生物发酵 | argB定向突变,解除精氨酸抑制 | 5 L发酵罐 | 45.6 g/L | 0.475 g/(L·h) | [ |
| 谷氨酸棒杆菌 | 微生物发酵 | 解除精氨酸反馈抑制;增加胞内NADPH水平;优化精氨酸代谢通量 | 5 L发酵罐 | 92.5 g/L | 1.29 g/(L·h) | [ | |
大肠 杆菌 | 微生物发酵 | 多层次合理代谢工程改造;构建生物传感器辅助的高通量筛选平台BHTS;全基因组测序和逆向工程鉴定和优化有益的突变基因 | 5 L发酵罐 | 132 g/L | 2.75 g/(L·h) | [ | |
| 瓜氨酸 | 粪链 球菌 | 全细胞催化 | 优化ADI固定化条件和催化反应条件 | 改进型填充床 反应器 | — | 95.6 g/(L·d) | [ |
大肠 杆菌 | 全细胞催化 | 大肠杆菌中表达乳酸乳球菌来源的ADI并通过易错PCR对酶进行突变;反应条件优化 | 30 L生物反应器 | 176.9 g/L | 22.1 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 阻断瓜氨酸降解;质粒过表达argJ基因,提高瓜氨酸的代谢通量 | 摇瓶 | 8.51 g/L | 0.12 g/(L·h) | [ | |
大肠 杆菌 | 微生物发酵 | 系统代谢工程对合成途径多模块耦合;Esa QS系统动态控制argG基因的表达 | 5 L发酵罐 | 82.1 g/L | 1.71 g/(L·h) | [ | |
| γ-聚谷氨酸 | 地衣芽孢杆菌 | 微生物发酵 | 60Co-γ射线辐照和ARTP诱变协同复合诱变技术;发酵培养基组分及条件优化 | 摇瓶 | 32.53 g/L | 0.45 g/(L·h) | [ |
| 地衣芽孢杆菌 | 微生物发酵 | 代谢工程改善ATP供应 | 1 L发酵罐 | 43.81 g/L | 1.37 g/(L·h) | [ | |
| 特基拉芽孢杆菌 | 微生物发酵 | 过表达外源ppc、aceE、pyk、icdh、gltA和gdhA基因;对关键酶Ppc、Pyk和AceE进行组装;低成本糖蜜作为发酵碳源 | 5 L发酵罐 | 25.73 g/L | 0.48 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 异源pgsBCA基因表达强度组合;优化发酵溶氧水平 | 5 L发酵罐 | 50.2 g/L | 1.05 g/(L·h) | [ | |
| γ-氨基丁酸 | 大肠杆菌 | 全细胞催化 | 过表达乳球菌来源gadB基因;敲除gabA和gabB基因阻断竞争通路;发酵条件优化 | 200 L生物反应器 | 614.15 g/L | 40.94 g/(L·h) | [ |
| 大肠杆菌 | 全细胞催化 | 定向进化和高通量筛选;过表达GadE;建立PLP自供系统 | 5 L生物反应器 | 307.5 g/L | 61.49 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 胞外分泌表达大肠杆菌来源突变体GadBmut;阻断GABA降解 | 3 L发酵罐 | 77.6 g/L | 1.21 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 强化甘油利用途径;敲除GABA降解途径并引入外源GABA合成途径;构建GABS动态调控GABA合成途径的基因表达 | 7.5 L发酵罐 | 45.6 g/L | 0.63 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 敲除ldhA、pqo和ack基因;过表达ppc、gltA、acn、icd、gdh和pdxST基因;PCP_2836odhA | 5 L发酵罐 | 81.31 g/L | 1.36 g/(L·h) | [ | |
| 短乳杆菌 | 全细胞催化 | pH自动维持系统 | 10 L发酵罐 | 321.9 g/L | 6.71 g/(L·h) | [ | |
| 反式-4-羟基-L-脯氨酸 | 大肠杆菌 | 微生物发酵 | 将地中海交替单胞菌来源PHP引入脯氨酸途径 | 5 L发酵罐 | 45.83 g/L | 1.27 g/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 建立木糖诱导表达体系;强化脯氨酸合成途径;引入小单孢菌属来源P4H | 5 L发酵罐 | 48.6 g/L | 1.22 g/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | 增加前体物脯氨酸合成;引入指孢囊菌来源P4H;引入NOG途径;发酵工艺优化 | 5 L发酵罐 | 89.4 g/L | 2.03 g/(L·h) | [ | |
| 亚精胺 | 解淀粉芽孢杆菌 | 微生物发酵 | 同源重组共表达异源speD和speE基因;发酵介质优化 | 摇瓶 | 227.4 mg/L | 3 mg/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 优化前体物供应;解除反馈抑制;强化转运途径 | 孔板 | 2.3 g/L | 20 mg/(L·h) | [ | |
| 大肠杆菌 | 全细胞催化 | 高亚精胺合成酶双重突变 | 摇瓶 | 933.5 mg/L | 155.6 mg/(L·h) | [ | |
| 大肠杆菌 | 全细胞催化 | 双酶级联催化系统;优化酶表达条件和反应条件 | 摇瓶 | 3.7 g/L | 463 mg/(L·h) | [ |
Table 1 Progress in the biosynthesis of glutamate family amino acids and derivatives
| 氨基酸及衍生物 | 底盘菌株 | 生产 方法 | 主要策略 | 发酵规模 | 产量 | 生产强度 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 精氨酸 | 钝齿棒 杆菌 | 微生物发酵 | argB定向突变,解除精氨酸抑制 | 5 L发酵罐 | 45.6 g/L | 0.475 g/(L·h) | [ |
| 谷氨酸棒杆菌 | 微生物发酵 | 解除精氨酸反馈抑制;增加胞内NADPH水平;优化精氨酸代谢通量 | 5 L发酵罐 | 92.5 g/L | 1.29 g/(L·h) | [ | |
大肠 杆菌 | 微生物发酵 | 多层次合理代谢工程改造;构建生物传感器辅助的高通量筛选平台BHTS;全基因组测序和逆向工程鉴定和优化有益的突变基因 | 5 L发酵罐 | 132 g/L | 2.75 g/(L·h) | [ | |
| 瓜氨酸 | 粪链 球菌 | 全细胞催化 | 优化ADI固定化条件和催化反应条件 | 改进型填充床 反应器 | — | 95.6 g/(L·d) | [ |
大肠 杆菌 | 全细胞催化 | 大肠杆菌中表达乳酸乳球菌来源的ADI并通过易错PCR对酶进行突变;反应条件优化 | 30 L生物反应器 | 176.9 g/L | 22.1 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 阻断瓜氨酸降解;质粒过表达argJ基因,提高瓜氨酸的代谢通量 | 摇瓶 | 8.51 g/L | 0.12 g/(L·h) | [ | |
大肠 杆菌 | 微生物发酵 | 系统代谢工程对合成途径多模块耦合;Esa QS系统动态控制argG基因的表达 | 5 L发酵罐 | 82.1 g/L | 1.71 g/(L·h) | [ | |
| γ-聚谷氨酸 | 地衣芽孢杆菌 | 微生物发酵 | 60Co-γ射线辐照和ARTP诱变协同复合诱变技术;发酵培养基组分及条件优化 | 摇瓶 | 32.53 g/L | 0.45 g/(L·h) | [ |
| 地衣芽孢杆菌 | 微生物发酵 | 代谢工程改善ATP供应 | 1 L发酵罐 | 43.81 g/L | 1.37 g/(L·h) | [ | |
| 特基拉芽孢杆菌 | 微生物发酵 | 过表达外源ppc、aceE、pyk、icdh、gltA和gdhA基因;对关键酶Ppc、Pyk和AceE进行组装;低成本糖蜜作为发酵碳源 | 5 L发酵罐 | 25.73 g/L | 0.48 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 异源pgsBCA基因表达强度组合;优化发酵溶氧水平 | 5 L发酵罐 | 50.2 g/L | 1.05 g/(L·h) | [ | |
| γ-氨基丁酸 | 大肠杆菌 | 全细胞催化 | 过表达乳球菌来源gadB基因;敲除gabA和gabB基因阻断竞争通路;发酵条件优化 | 200 L生物反应器 | 614.15 g/L | 40.94 g/(L·h) | [ |
| 大肠杆菌 | 全细胞催化 | 定向进化和高通量筛选;过表达GadE;建立PLP自供系统 | 5 L生物反应器 | 307.5 g/L | 61.49 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 胞外分泌表达大肠杆菌来源突变体GadBmut;阻断GABA降解 | 3 L发酵罐 | 77.6 g/L | 1.21 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 强化甘油利用途径;敲除GABA降解途径并引入外源GABA合成途径;构建GABS动态调控GABA合成途径的基因表达 | 7.5 L发酵罐 | 45.6 g/L | 0.63 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 敲除ldhA、pqo和ack基因;过表达ppc、gltA、acn、icd、gdh和pdxST基因;PCP_2836odhA | 5 L发酵罐 | 81.31 g/L | 1.36 g/(L·h) | [ | |
| 短乳杆菌 | 全细胞催化 | pH自动维持系统 | 10 L发酵罐 | 321.9 g/L | 6.71 g/(L·h) | [ | |
| 反式-4-羟基-L-脯氨酸 | 大肠杆菌 | 微生物发酵 | 将地中海交替单胞菌来源PHP引入脯氨酸途径 | 5 L发酵罐 | 45.83 g/L | 1.27 g/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 建立木糖诱导表达体系;强化脯氨酸合成途径;引入小单孢菌属来源P4H | 5 L发酵罐 | 48.6 g/L | 1.22 g/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | 增加前体物脯氨酸合成;引入指孢囊菌来源P4H;引入NOG途径;发酵工艺优化 | 5 L发酵罐 | 89.4 g/L | 2.03 g/(L·h) | [ | |
| 亚精胺 | 解淀粉芽孢杆菌 | 微生物发酵 | 同源重组共表达异源speD和speE基因;发酵介质优化 | 摇瓶 | 227.4 mg/L | 3 mg/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 优化前体物供应;解除反馈抑制;强化转运途径 | 孔板 | 2.3 g/L | 20 mg/(L·h) | [ | |
| 大肠杆菌 | 全细胞催化 | 高亚精胺合成酶双重突变 | 摇瓶 | 933.5 mg/L | 155.6 mg/(L·h) | [ | |
| 大肠杆菌 | 全细胞催化 | 双酶级联催化系统;优化酶表达条件和反应条件 | 摇瓶 | 3.7 g/L | 463 mg/(L·h) | [ |
| 氨基酸衍生物 | 底盘菌株 | 生产方法 | 主要策略 | 发酵规模 | 产量 | 生产强度 | 参考文献 |
|---|---|---|---|---|---|---|---|
对香 豆酸 | 大肠杆菌 | 微生物发酵 | 筛选p-CA合成基因;优化蛋白活性;增加辅因子利用率;优化发酵工艺 | 5 L发酵罐 | 3.09 g/L | 49.05 mg/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 筛选p-CA合成基因;增加前体物供应;阻断竞争途径;平衡PEP与E4P碳通量 | 1 L发酵罐 | 12.50 g/L | 130 mg/(L·h) | [ | |
| 解脂耶氏酵母 | 微生物发酵 | 增加TAL基因拷贝数;强化莽草酸途径通量;阻断苯丙氨酸的竞争途径 | 摇瓶 | 1.04 g/L | 8.63 mg/(L·h) | [ | |
白藜 芦醇 | 大肠杆菌 | 微生物发酵 | 引入异源丙二酸同化途径,增加关键前体丙二酰辅酶A的供应;CRISPRi技术下调脂肪酸合成途径基因,阻断丙二酰辅酶A消耗途径;引入并优化异源TAL途径 | 摇瓶 | 304.5 mg/L | 6.344 mg/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 混菌发酵;优化发酵条件(接种比例、碳源比例) | 摇瓶 | 204.8 mg/L | 2.44 mg/(L·h) | [ | |
| 解脂耶氏酵母 | 微生物发酵 | 引入白藜芦醇合成途径相关酶并采用刚性连接肽EAAAK连接;增加前体物供应;优化发酵条件(控制pH以维持酵母正常形态) | 5 L发酵罐 | 22.5 g/L | 0.16 g/(L·h) | [ | |
| — | 酶催化 | 虎杖苷-β-D-葡萄糖苷酶催化虎杖苷酶;反应条件优化 | 摇瓶 | 22.5 g/L | 5.63 g/(L·h) | [ | |
红景 天苷 | 大肠杆菌 | 微生物发酵 | 混菌发酵;优化发酵条件以平衡菌株生长(碳源比例、接种比例) | 5 L发酵罐 | 6.03 g/L | 0.05 g/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 引入红景天苷合成途径;增加前体物供应;敲除竞争途径 | 5 L发酵罐 | 26.55 g/L | 0.16 g/(L·h) | [ | |
| 咖啡酸 | 大肠杆菌 | 微生物发酵 | 引入p-CA合成途径;解除反馈抑制;阻断竞争途径;增加辅因子FAD供应;强化CA转运蛋白表达 | 5 L发酵罐 | 7.92 g/L | 0.12 g/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 引入p-CA合成途径;阻断竞争途径;增加前体物酪氨酸供应;增加辅因子FADH2供应 | 5 L发酵罐 | 6.17 g/L | 0.07 g/(L·h) | [ | |
| 酿酒酵母 | 微生物发酵 | 阻断苯丙氨酸和色氨酸合成,增加前体供应 | 5 L发酵罐 | 9.3 g/L | 0.09 g/(L·h) | [ | |
| 阿魏酸 | 大肠杆菌 | 微生物发酵 | 引入FA合成酶增加S-腺苷甲硫氨酸供应;强化合成途径;增加前体物供应;减少PEP向丙酮酸转化;阻断竞争途径;增加辅因子FADH2供应 | 3 L发酵罐 | 5.09 g/L | 0.07 g/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 引入FA合成途径;增加前体物p-CA供应;增加辅因子FADH2供应;增加辅因子NADPH供应;增加S-腺苷甲硫氨酸供应;回补菌株(HIS3, URA3) | 1.2 L发酵罐 | 3.80 g/L | 0.03 g/(L·h) | [ | |
没食 子酸 | 大肠杆菌 | 微生物发酵 | 引入GA合成所需酶、增加前体物供应 | 摇瓶 | 1266.39 mg/L | 35.18 mg/(L·h) | [ |
| 根皮素 | 酿酒酵母 | 微生物发酵 | 引入根皮素合成途径;增加丙二酰辅酶A供应;优化发酵条件 | 5 L发酵罐 | 619.50 mg/L | 7.74 mg/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 引入根皮素合成基因并对CHS酶进行诱变 | 摇瓶 | 1.85 mg/L | — | [ |
Table 2 Progress in the biosynthesis of aromatic amino acid derivatives
| 氨基酸衍生物 | 底盘菌株 | 生产方法 | 主要策略 | 发酵规模 | 产量 | 生产强度 | 参考文献 |
|---|---|---|---|---|---|---|---|
对香 豆酸 | 大肠杆菌 | 微生物发酵 | 筛选p-CA合成基因;优化蛋白活性;增加辅因子利用率;优化发酵工艺 | 5 L发酵罐 | 3.09 g/L | 49.05 mg/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 筛选p-CA合成基因;增加前体物供应;阻断竞争途径;平衡PEP与E4P碳通量 | 1 L发酵罐 | 12.50 g/L | 130 mg/(L·h) | [ | |
| 解脂耶氏酵母 | 微生物发酵 | 增加TAL基因拷贝数;强化莽草酸途径通量;阻断苯丙氨酸的竞争途径 | 摇瓶 | 1.04 g/L | 8.63 mg/(L·h) | [ | |
白藜 芦醇 | 大肠杆菌 | 微生物发酵 | 引入异源丙二酸同化途径,增加关键前体丙二酰辅酶A的供应;CRISPRi技术下调脂肪酸合成途径基因,阻断丙二酰辅酶A消耗途径;引入并优化异源TAL途径 | 摇瓶 | 304.5 mg/L | 6.344 mg/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 混菌发酵;优化发酵条件(接种比例、碳源比例) | 摇瓶 | 204.8 mg/L | 2.44 mg/(L·h) | [ | |
| 解脂耶氏酵母 | 微生物发酵 | 引入白藜芦醇合成途径相关酶并采用刚性连接肽EAAAK连接;增加前体物供应;优化发酵条件(控制pH以维持酵母正常形态) | 5 L发酵罐 | 22.5 g/L | 0.16 g/(L·h) | [ | |
| — | 酶催化 | 虎杖苷-β-D-葡萄糖苷酶催化虎杖苷酶;反应条件优化 | 摇瓶 | 22.5 g/L | 5.63 g/(L·h) | [ | |
红景 天苷 | 大肠杆菌 | 微生物发酵 | 混菌发酵;优化发酵条件以平衡菌株生长(碳源比例、接种比例) | 5 L发酵罐 | 6.03 g/L | 0.05 g/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 引入红景天苷合成途径;增加前体物供应;敲除竞争途径 | 5 L发酵罐 | 26.55 g/L | 0.16 g/(L·h) | [ | |
| 咖啡酸 | 大肠杆菌 | 微生物发酵 | 引入p-CA合成途径;解除反馈抑制;阻断竞争途径;增加辅因子FAD供应;强化CA转运蛋白表达 | 5 L发酵罐 | 7.92 g/L | 0.12 g/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 引入p-CA合成途径;阻断竞争途径;增加前体物酪氨酸供应;增加辅因子FADH2供应 | 5 L发酵罐 | 6.17 g/L | 0.07 g/(L·h) | [ | |
| 酿酒酵母 | 微生物发酵 | 阻断苯丙氨酸和色氨酸合成,增加前体供应 | 5 L发酵罐 | 9.3 g/L | 0.09 g/(L·h) | [ | |
| 阿魏酸 | 大肠杆菌 | 微生物发酵 | 引入FA合成酶增加S-腺苷甲硫氨酸供应;强化合成途径;增加前体物供应;减少PEP向丙酮酸转化;阻断竞争途径;增加辅因子FADH2供应 | 3 L发酵罐 | 5.09 g/L | 0.07 g/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 引入FA合成途径;增加前体物p-CA供应;增加辅因子FADH2供应;增加辅因子NADPH供应;增加S-腺苷甲硫氨酸供应;回补菌株(HIS3, URA3) | 1.2 L发酵罐 | 3.80 g/L | 0.03 g/(L·h) | [ | |
没食 子酸 | 大肠杆菌 | 微生物发酵 | 引入GA合成所需酶、增加前体物供应 | 摇瓶 | 1266.39 mg/L | 35.18 mg/(L·h) | [ |
| 根皮素 | 酿酒酵母 | 微生物发酵 | 引入根皮素合成途径;增加丙二酰辅酶A供应;优化发酵条件 | 5 L发酵罐 | 619.50 mg/L | 7.74 mg/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 引入根皮素合成基因并对CHS酶进行诱变 | 摇瓶 | 1.85 mg/L | — | [ |
| 氨基酸衍生物 | 底盘菌株 | 生产方法 | 主要策略 | 发酵规模 | 产量 | 生产强度 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 四氢嘧啶 | 大肠杆菌 | 微生物发酵 | 引入四氢嘧啶合成途径;增加前体物供应;优化补糖速率 | 15 L发酵罐 | 131.80 g/L | 1.37 g/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 增加前体物供应;优化培养基(碳氮比例) | 2.4 L发酵罐 | 34.27 g/L | 0.57 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 采用转录平衡技术设计启动子表达文库对菌株进行优化 | 1 L发酵罐 | 65 g/L | 1.16 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 引入四氢嘧啶合成途径;避免副产物积累;减少反馈抑制 | 5 L发酵罐 | 115.87 g/L | 1.49 g/(L·h) | [ | |
| 羟基四氢嘧啶 | 大肠杆菌 | 微生物发酵 | 引入羟基四氢嘧啶合成途径并进行优化;引入esaI/esaR群体感应系统控制sucA表达 | 摇瓶 | 14.93 g/L | 0.42 g/(L·h) | [ |
| 谷氨酸棒杆菌 | 微生物发酵 | 双菌株两步发酵 | 1 L发酵罐 | 74 g/L | 1.37 g/(L·h) | [ | |
ε-聚赖 氨酸 | 小白链霉菌 | 微生物发酵 | 增强ε-PL合成酶基因转录;赖氨酸合成过程中关键酶活性增强;优化发酵工艺(酸性pH冲击工艺) | 5 L发酵罐 | 70.3 g/L | 0.37 g/(L·h) | [ |
| 小白链霉菌 | 全细胞催化 | 表达异源lysp基因提升赖氨酸利用能力及底物转化效率;对培养基和培养条件进行优化 | 摇瓶 | 17.21 g/L | 0.18 g/(L·h) | [ | |
| 麦角硫因 | 大肠杆菌 | 微生物发酵 | 半理性设计和随机突变EgtD和TNcEgt1;流加前体氨基酸 | 5 L发酵罐 | 5.4 g/L | 56.3 mg/(L·h) | [ |
| 大肠杆菌 | 全细胞催化 | 构建EGT菌株高密度发酵方法;发酵工艺优化;流加前体氨基酸 | 2 L发酵罐 | 7 g/L | 90.9 mg/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | EGT合成模块、前体物组氨酸、半胱氨酸和腺苷蛋氨酸合成模块进行系统的代谢工程改造;发酵工艺优化 | 2 L发酵罐 | 7.2 g/L | 120 mg/(L·h) | [ | |
| 裂殖酵母 | 微生物发酵 | 紫外照射和氯化锂突变;流加前体氨基酸 | 5 L发酵罐 | 12.5 g/L | 84.5 mg/(L·h) | [ | |
| 肌肽 | — | 酶催化 | 定点饱和突变来改善酯酰基转移酶的底物特异性 | 摇瓶 | 105 mmol/L | — | [ |
| — | 酶催化 | 筛选来自黏质沙雷氏菌新型二肽酶SmPepD;反应条件优化;纳滤膜分离 | 5 L超滤膜 反应器 | 7.23 g/L | — | [ | |
| — | 酶催化 | 酶挖掘方法鉴定出来自巨大芽孢杆菌BmPepD并进行定向饱和诱变;反应条件优化 | 10 mL 反应体系 | 31.3 mmol/L | — | [ | |
| 大肠杆菌 | 全细胞催化 | 在大肠杆菌中表达SmPepD构建细胞工厂;对SmPepD理性设计获得更高活性双突变体Thr168Ser/Gly148Asp;敲除组氨酸输出蛋白yeaS基因 | 5 L生物 反应器 | 133.2 mmol/L | — | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 增加前体组氨酸和β-丙氨酸积累;引入来自哺乳动物的CARNS1基因;发酵优化;肌肽活性验证 | 2 L发酵罐 | 323.26 mg/L | 6.73 mg/(L·h) | [ | |
| 谷胱甘肽 | 酿酒酵母 | 微生物发酵 | 适应性进化;使用丙烯醛作为选择剂 | 发酵罐(1.2 L工作体积) | 320 mg/L | 8.28 mg/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 基于氧化应激和能量代谢的逐步控制策略 | 10 L发酵罐 | 5.76 g/L | 53 mg/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | 异源表达来自嗜热链球菌gshF基因;流加前体氨基酸 | 5 L发酵罐 | 15.21 g/L | 0.82 g/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | 代谢工程手段促进GSH生物合成;代谢组学分析 | 5 L发酵罐 | 22 g/(L·h) | 0.407 g/(L·h) | [ |
Table 3 Progress in the biosynthesis of aspartate family amino acid derivatives, ergothioneine and peptides
| 氨基酸衍生物 | 底盘菌株 | 生产方法 | 主要策略 | 发酵规模 | 产量 | 生产强度 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 四氢嘧啶 | 大肠杆菌 | 微生物发酵 | 引入四氢嘧啶合成途径;增加前体物供应;优化补糖速率 | 15 L发酵罐 | 131.80 g/L | 1.37 g/(L·h) | [ |
| 大肠杆菌 | 微生物发酵 | 增加前体物供应;优化培养基(碳氮比例) | 2.4 L发酵罐 | 34.27 g/L | 0.57 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 采用转录平衡技术设计启动子表达文库对菌株进行优化 | 1 L发酵罐 | 65 g/L | 1.16 g/(L·h) | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 引入四氢嘧啶合成途径;避免副产物积累;减少反馈抑制 | 5 L发酵罐 | 115.87 g/L | 1.49 g/(L·h) | [ | |
| 羟基四氢嘧啶 | 大肠杆菌 | 微生物发酵 | 引入羟基四氢嘧啶合成途径并进行优化;引入esaI/esaR群体感应系统控制sucA表达 | 摇瓶 | 14.93 g/L | 0.42 g/(L·h) | [ |
| 谷氨酸棒杆菌 | 微生物发酵 | 双菌株两步发酵 | 1 L发酵罐 | 74 g/L | 1.37 g/(L·h) | [ | |
ε-聚赖 氨酸 | 小白链霉菌 | 微生物发酵 | 增强ε-PL合成酶基因转录;赖氨酸合成过程中关键酶活性增强;优化发酵工艺(酸性pH冲击工艺) | 5 L发酵罐 | 70.3 g/L | 0.37 g/(L·h) | [ |
| 小白链霉菌 | 全细胞催化 | 表达异源lysp基因提升赖氨酸利用能力及底物转化效率;对培养基和培养条件进行优化 | 摇瓶 | 17.21 g/L | 0.18 g/(L·h) | [ | |
| 麦角硫因 | 大肠杆菌 | 微生物发酵 | 半理性设计和随机突变EgtD和TNcEgt1;流加前体氨基酸 | 5 L发酵罐 | 5.4 g/L | 56.3 mg/(L·h) | [ |
| 大肠杆菌 | 全细胞催化 | 构建EGT菌株高密度发酵方法;发酵工艺优化;流加前体氨基酸 | 2 L发酵罐 | 7 g/L | 90.9 mg/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | EGT合成模块、前体物组氨酸、半胱氨酸和腺苷蛋氨酸合成模块进行系统的代谢工程改造;发酵工艺优化 | 2 L发酵罐 | 7.2 g/L | 120 mg/(L·h) | [ | |
| 裂殖酵母 | 微生物发酵 | 紫外照射和氯化锂突变;流加前体氨基酸 | 5 L发酵罐 | 12.5 g/L | 84.5 mg/(L·h) | [ | |
| 肌肽 | — | 酶催化 | 定点饱和突变来改善酯酰基转移酶的底物特异性 | 摇瓶 | 105 mmol/L | — | [ |
| — | 酶催化 | 筛选来自黏质沙雷氏菌新型二肽酶SmPepD;反应条件优化;纳滤膜分离 | 5 L超滤膜 反应器 | 7.23 g/L | — | [ | |
| — | 酶催化 | 酶挖掘方法鉴定出来自巨大芽孢杆菌BmPepD并进行定向饱和诱变;反应条件优化 | 10 mL 反应体系 | 31.3 mmol/L | — | [ | |
| 大肠杆菌 | 全细胞催化 | 在大肠杆菌中表达SmPepD构建细胞工厂;对SmPepD理性设计获得更高活性双突变体Thr168Ser/Gly148Asp;敲除组氨酸输出蛋白yeaS基因 | 5 L生物 反应器 | 133.2 mmol/L | — | [ | |
| 谷氨酸棒杆菌 | 微生物发酵 | 增加前体组氨酸和β-丙氨酸积累;引入来自哺乳动物的CARNS1基因;发酵优化;肌肽活性验证 | 2 L发酵罐 | 323.26 mg/L | 6.73 mg/(L·h) | [ | |
| 谷胱甘肽 | 酿酒酵母 | 微生物发酵 | 适应性进化;使用丙烯醛作为选择剂 | 发酵罐(1.2 L工作体积) | 320 mg/L | 8.28 mg/(L·h) | [ |
| 酿酒酵母 | 微生物发酵 | 基于氧化应激和能量代谢的逐步控制策略 | 10 L发酵罐 | 5.76 g/L | 53 mg/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | 异源表达来自嗜热链球菌gshF基因;流加前体氨基酸 | 5 L发酵罐 | 15.21 g/L | 0.82 g/(L·h) | [ | |
| 大肠杆菌 | 微生物发酵 | 代谢工程手段促进GSH生物合成;代谢组学分析 | 5 L发酵罐 | 22 g/(L·h) | 0.407 g/(L·h) | [ |
| 111 | OHSAWA T, TSUKAHARA K, OGURA M. Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in γ-poly-glutamic acid synthesis[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(9): 2096-2102. |
| 112 | 周梦洁, 胡汶松, 胡刘秀, 等. 枯草芽孢杆菌聚谷氨酸合成途径相关基因功能研究[J]. 微生物学报, 2023, 63(1): 387-402. |
| ZHOU M J, HU W S, HU L X, et al. Functions of genes involved in polyglutamic acid synthesis in Bacillus subtilis [J]. Acta Microbiologica Sinica, 2023, 63(1): 387-402. | |
| 113 | GONG L C, REN C, XU Y. GlnR negatively regulates glutamate-dependent acid resistance in Lactobacillus brevis [J]. Applied and Environmental Microbiology, 2020, 86(7): e02615-19. |
| 114 | SHI X F, CHANG C Y, MA S X, et al. Efficient bioconversion of L-glutamate to γ-aminobutyric acid by Lactobacillus brevis resting cells[J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(4-5): 697-704. |
| 115 | PARK S H, SOHN Y J, PARK S J, et al. Effect of DR1558, a Deinococcus radiodurans response regulator, on the production of GABA in the recombinant Escherichia coli under low pH conditions[J]. Microbial Cell Factories, 2020, 19(1): 64. |
| 116 | SOMA Y, FUJIWARA Y, NAKAGAWA T, et al. Reconstruction of a metabolic regulatory network in Escherichia coli for purposeful switching from cell growth mode to production mode in direct GABA fermentation from glucose[J]. Metabolic Engineering, 2017, 43: 54-63. |
| 117 | SHIBASAKI T, MORI H, CHIBA S, et al. Microbial proline 4-hydroxylase screening and gene cloning[J]. Applied and Environmental Microbiology, 1999, 65(9): 4028-4031. |
| 118 | BALDWIN J E, FIELD R A, LAWRENCE C C, et al. Substrate specificity of proline-4-hydroxylase: chemical and enzymatic synthesis of 2S,3R,4S-epoxyproline[J]. Tetrahedron Letters, 1994, 35(26): 4649-4652. |
| 119 | LIU Q L, YU T, LI X W, et al. Rewiring carbon metabolism in yeast for high level production of aromatic chemicals[J]. Nature Communications, 2019, 10(1): 4976. |
| 120 | ZHU J R, YANG S, CAO Q, et al. Engineering Yarrowia lipolytica as a cellulolytic cell factory for production of p-coumaric acid from cellulose and hemicellulose[J]. Journal of Agricultural and Food Chemistry, 2024, 72(11): 5867-5877. |
| 121 | WU J J, ZHOU P, ZHANG X, et al. Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(7): 1083-1095. |
| 122 | LI J, QIU Z T, ZHAO G R. Modular engineering of E. coli coculture for efficient production of resveratrol from glucose and Arabinose mixture[J]. Synthetic and Systems Biotechnology, 2022, 7(2): 718-729. |
| 123 | CHEN M, LI D, GAO Z Q, et al. Enzymatic transformation of polydatin to resveratrol by piceid-β-D-glucosidase from Aspergillus oryzae [J]. Bioprocess and Biosystems Engineering, 2014, 37(7): 1411-1416. |
| 124 | LIU X, LI X B, JIANG J L, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides[J]. Metabolic Engineering, 2018, 47: 243-253. |
| 125 | SAKAE K, NONAKA D, KISHIDA M, et al. Caffeic acid production from glucose using metabolically engineered Escherichia coli [J]. Enzyme and Microbial Technology, 2023, 164: 110193. |
| 126 | 袁豆豆, 周秀琪, 庞雪晴, 等. 代谢工程改造酿酒酵母发酵生产咖啡酸[J]. 食品与发酵工业, 2024, 50(19): 17-24. |
| YUAN D D, ZHOU X Q, PANG X Q, et al. Metabolic engineering of Saccharomyces cerevisiae for biosynthesis of caffeic acid[J]. Food and Fermentation Industries, 2024, 50(19): 17-24. | |
| 127 | ZHOU Z, ZHANG X Y, WU J, et al. Targeting cofactors regeneration in methylation and hydroxylation for high level production of ferulic acid[J]. Metabolic Engineering, 2022, 73: 247-255. |
| 128 | CHEN Z Y, SHEN X L, WANG J, et al. Rational engineering of p-hydroxybenzoate hydroxylase to enable efficient gallic acid synthesis via a novel artificial biosynthetic pathway[J]. Biotechnology and Bioengineering, 2017, 114(11): 2571-2580. |
| 129 | JIANG C M, LIU X N, CHEN X Q, et al. Raising the production of phloretin by alleviation of by-product of Chalcone synthase in the engineered yeast[J]. Science China Life Sciences, 2020, 63(11): 1734-1743. |
| 130 | LIU X, LIU J C, LEI D W, et al. Modular metabolic engineering for production of phloretic acid, phloretin and phlorizin in Escherichia coli [J]. Chemical Engineering Science, 2022, 247: 116931. |
| 131 | BOO Y C. p-Coumaric acid as an active ingredient in cosmetics: a review focusing on its antimelanogenic effects[J]. Antioxidants, 2019, 8(8): 275. |
| 132 | JANG S, HA C W, KIM S H, et al. Dual suppressive effect of p-coumaric acid on pigmentation in B16F10 cells[J]. Molecular & Cellular Toxicology, 2024, 20(4): 1011-1023. |
| 1 | BENNER S A, SISMOUR A M. Synthetic biology[J]. Nature Reviews Genetics, 2005, 6: 533-543. |
| 2 | AUSLÄNDER D S, AUSLÄNDER D D, FUSSENEGGER P M. Synthetic biology: the synthesis of biology[J]. Angewandte Chemie International Edition, 2017, 56(23): 6396-6419. |
| 3 | RAMZI A B. Metabolic engineering and synthetic biology[M/OL]//AIZAT W, GOH H H, BAHARUM S. Omics applications for systems biology. Advances in experimental medicine and biology. Cham: Springer, 2018, 1102: 81-85. (2018-11-01)[2024-07-01]. . |
| 4 | SAKAMOTO K. Amino acids and derivatives[M]//Cosmeceuticals and active cosmetics. CRC Press, 2016: 163-175. |
| 5 | FARMER W R, LIAO J C. Progress in metabolic engineering[J]. Current Opinion in Biotechnology, 1996, 7(2): 198-204. |
| 6 | WU G Y. Amino acids: metabolism, functions, and nutrition[J]. Amino Acids, 2009, 37(1): 1-17. |
| 7 | KAMMEYER A, LUITEN R M. Oxidation events and skin aging[J]. Ageing Research Reviews, 2015, 21: 16-29. |
| 8 | GU Y P, HAN J X, JIANG C P, et al. Biomarkers, oxidative stress and autophagy in skin aging[J]. Ageing Research Reviews, 2020, 59: 101036. |
| 9 | AVILA RODRIGUEZ M I, RODRIGUEZ BARROSO L G, SANCHEZ M L. Collagen: a review on its sources and potential cosmetic applications[J]. Journal of Cosmetic Dermatology, 2018, 17(1): 20-26. |
| 10 | SHOULDERS M D, RAINES R T. Collagen structure and stability[J]. Annual Review of Biochemistry, 2009, 78: 929-958. |
| 11 | MADEO F, EISENBERG T, PIETROCOLA F, et al. Spermidine in health and disease[J]. Science, 2018, 359(6374): eaan2788. |
| 12 | VERDIER‐SEVRAIN S, BONTE F. Skin hydration: a review on its molecular mechanisms[J]. Journal of Cosmetic Dermatology, 2007, 6(2): 75-82. |
| 133 | ZHANG J Q, TANG H Z, YU X, et al. Co-production of ferulic acid and p-coumaric acid from distiller grain by a putative feruloyl esterase discovered in metagenome assembled genomes[J]. Journal of Cleaner Production, 2024, 439: 140814. |
| 134 | RODRIGUEZ A, KILDEGAARD K R, LI M J, et al. Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis[J]. Metabolic Engineering, 2015, 31: 181-188. |
| 135 | FAISAL Z, MAZHAR A, BATOOL S A, et al. Exploring the multimodal health-promoting properties of resveratrol: a comprehensive review[J]. Food Science & Nutrition, 2024, 12(4): 2240-2258. |
| 136 | BEJENARU L E, BIŢĂ A, BELU I, et al. Resveratrol: a review on the biological activity and applications[J]. Applied Sciences, 2024, 14(11): 4534. |
| 137 | MENG T T, XIAO D F, MUHAMMED A, et al. Anti-inflammatory action and mechanisms of resveratrol[J]. Molecules, 2021, 26(1): 229. |
| 138 | BEEKWILDER J, WOLSWINKEL R, JONKER H, et al. Production of resveratrol in recombinant microorganisms[J]. Applied and Environmental Microbiology, 2006, 72(8): 5670-5672. |
| 139 | WATTS K T, LEE P C, SCHMIDT-DANNERT C. Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli [J]. BMC Biotechnology, 2006, 6: 22. |
| 140 | AFONSO M S, FERREIRA S, DOMINGUES F C, et al. Resveratrol production in bioreactor: assessment of cell physiological states and plasmid segregational stability[J]. Biotechnology Reports, 2015, 5: 7-13. |
| 141 | LIM C G, FOWLER Z L, HUELLER T, et al. High-yield resveratrol production in engineered Escherichia coli [J]. Applied and Environmental Microbiology, 2011, 77(10): 3451-3460. |
| 142 | KATSUYAMA Y, FUNA N, MIYAHISA I, et al. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli [J]. Chemistry & Biology, 2007, 14(6): 613-621. |
| 143 | KATSUYAMA Y, FUNA N, HORINOUCHI S. Precursor-directed biosynthesis of stilbene methyl ethers in Escherichia coli [J]. Biotechnology Journal, 2007, 2(10): 1286-1293. |
| 144 | WU J J, LIU P R, FAN Y M, et al. Multivariate modular metabolic engineering of Escherichia coli to produce resveratrol from L-tyrosine[J]. Journal of Biotechnology, 2013, 167(4): 404-411. |
| 145 | IBRAHIM G G, PERERA M, ABDULMALEK S A, et al. De novo synthesis of resveratrol from sucrose by metabolically engineered Yarrowia lipolytica [J]. Biomolecules, 2024, 14(6): 712. |
| 146 | YANG Y P, LIN Y H, LI L Y, et al. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products[J]. Metabolic Engineering, 2015, 29: 217-226. |
| 13 | TFAYLI A, JAMAL D, VYUMVUHORE R, et al. Hydration effects on the barrier function of stratum corneum lipids: Raman analysis of ceramides 2, Ⅲ and 5[J]. Analyst, 2013, 138(21): 6582-6588. |
| 14 | CHOI E, KANG Y G, HWANG S H, et al. In vitro effects of dehydrotrametenolic acid on skin barrier function[J]. Molecules, 2019, 24(24): 4583. |
| 15 | JIA T H, QIAO W, YAO Q F, et al. Treatment with docosahexaenoic acid improves epidermal keratinocyte differentiation and ameliorates inflammation in human keratinocytes and reconstructed human epidermis models[J]. Molecules, 2019, 24(17): 3156. |
| 16 | MARINI A, REINELT K, KRUTMANN J, et al. Ectoine-containing cream in the treatment of mild to moderate atopic dermatitis: a randomised, comparator-controlled, intra-individual double-blind, multi-center trial[J]. Skin Pharmacology and Physiology, 2014, 27(2): 57-65. |
| 17 | KUMTORNRUT C, MANABE S D, NAVAPONGSIRI M, et al. A cleanser formulated with Tris (hydroxymethyl) aminomethane and L-arginine significantly improves facial acne in male Thai subjects[J]. Journal of Cosmetic Dermatology, 2020, 19(4): 901-909. |
| 18 | OLKOWSKA E, POLKOWSKA Ż, NAMIEŚNIK J. Analytics of surfactants in the environment: problems and challenges[J]. Chemical Reviews, 2011, 111(9): 5667-5700. |
| 19 | 石莹莹. N-酰基氨基酸表面活性剂的性能及应用研究进展[J]. 河南化工, 2016, 33(2): 16-18, 28. |
| SHI Y Y. Research progress on properties and application of N-acyl amino acid surfactants[J]. Henan Chemical Industry, 2016, 33(2): 16-18, 28. | |
| 20 | 王普兵, 谭晓延, 王雪敏. 化妆品用氨基酸表面活性剂的分类及应用[J]. 广东化工, 2019, 46(6): 125, 124. |
| WANG P B, TAN X Y, WANG X M. The classification and application of amino acid surfactants for cosmetics[J]. Guangdong Chemical Industry, 2019, 46(6): 125, 124. | |
| 21 | 蔡昌武, 柏新喜, 曾银凤. 氨基酸表面活性剂的合成研究进展[J]. 产业创新研究, 2022(24): 81-83. |
| CAI C W, BAI X X, ZENG Y F. Research progress in synthesis of amino acid surfactants[J]. Industrial Innovation, 2022(24): 81-83. | |
| 147 | YUAN S F, YI X N, JOHNSTON T G, et al. De novo resveratrol production through modular engineering of an Escherichia coli-Saccharomyces cerevisiae co-culture[J]. Microbial Cell Factories, 2020, 19(1): 143. |
| 148 | HONG J, IM D K, OH M K. Investigating E. coli coculture for resveratrol production with 13C metabolic flux analysis[J]. Journal of Agricultural and Food Chemistry, 2020, 68(11): 3466-3473. |
| 149 | LA TORRE G L, LAGANÀ G, BELLOCCO E, et al. Improvement on enzymatic hydrolysis of resveratrol glucosides in wine[J]. Food Chemistry, 2004, 85(2): 259-266. |
| 150 | ZHANG C Z, LI D, YU H S, et al. Purification and characterization of piceid-β-D-glucosidase from Aspergillus oryzae [J]. Process Biochemistry, 2007, 42(1): 83-88. |
| 151 | LI F L, TANG H, XIAO F R, et al. Protective effect of salidroside from Rhodiolae radix on diabetes-induced oxidative stress in mice[J]. Molecules, 2011, 16(12): 9912-9924. |
| 152 | HAZELWOOD L A, DARAN J M, VAN MARIS A J A, et al. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism[J]. Applied and Environmental Microbiology, 2008, 74(8): 2259-2266. |
| 153 | CHUNG D, KIM S Y, AHN J H. Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli [J]. Scientific Reports, 2017, 7(1): 2578. |
| 154 | GUO W, HUANG Q L, FENG Y H, et al. Rewiring central carbon metabolism for tyrosol and salidroside production in Saccharomyces cerevisiae [J]. Biotechnology and Bioengineering, 2020, 117(8): 2410-2419. |
| 155 | RÖSLER J, KREKEL F, AMRHEIN N, et al. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity[J]. Plant Physiology, 1997, 113(1): 175-179. |
| 156 | FURUYA T, ARAI Y, KINO K. Biotechnological production of caffeic acid by bacterial cytochrome P450 CYP199A2[J]. Applied and Environmental Microbiology, 2012, 78(17): 6087-6094. |
| 157 | CHOI O S, WU C Z, KANG S Y, et al. Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(10): 1657-1665. |
| 158 | BERNER M, KRUG D, BIHLMAIER C, et al. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis [J]. Journal of Bacteriology, 2006, 188(7): 2666-2673. |
| 22 | WESCHAWALIT S, THONGTHIP S, PHUTRAKOOL P, et al. Glutathione and its antiaging and antimelanogenic effects[J]. Clinical, Cosmetic and Investigational Dermatology, 2017, 10: 147-153. |
| 23 | WIRAGUNA A A G P, HARI E D, PRAHARSINI I G A A. Correlation between glutathione plasma with degree severity of Melasma in Balinese women[J]. Clinical, Cosmetic and Investigational Dermatology, 2020, 13: 455-459. |
| 24 | ARJINPATHANA N, ASAWANONDA P. Glutathione as an oral whitening agent: a randomized, double-blind, placebo-controlled study[J]. Journal of Dermatological Treatment, 2012, 23(2): 97-102. |
| 25 | PARK H J, CHO J H, HONG S H, et al. Whitening and anti-wrinkle activities of ferulic acid isolated from Tetragonia tetragonioides in B16F10 melanoma and CCD-986sk fibroblast cells[J]. Journal of Natural Medicines, 2018, 72(1): 127-135. |
| 26 | KIM D S, PARK S H, KWON S B, et al. (-)-Epigallocatechin-3-gallate and hinokitiol reduce melanin synthesis via decreased MITF production[J]. Archives of Pharmacal Research, 2004, 27(3): 334-339. |
| 27 | FAN M H, ZHANG G W, HU X, et al. Quercetin as a tyrosinase inhibitor: inhibitory activity, conformational change and mechanism[J]. Food Research International, 2017, 100: 226-233. |
| 28 | SOLANO F, BRIGANTI S, PICARDO M, et al. Hypopigmenting agents: an updated review on biological, chemical and clinical aspects[J]. Pigment Cell Research, 2006, 19(6): 550-571. |
| 29 | MA Q M, DAVIDSON P M, ZHONG Q X. Properties and potential food applications of lauric arginate as a cationic antimicrobial[J]. International Journal of Food Microbiology, 2020, 315: 108417. |
| 30 | CHEN S J, HUANG S T, LI Y, et al. Recent advances in epsilon-poly-L-lysine and L-lysine-based dendrimer synthesis, modification, and biomedical applications[J]. Frontiers in Chemistry, 2021, 9: 659304. |
| 31 | WANG Y, WANG L M, HU Y F, et al. Design and optimization of ε-poly-L-lysine with specific functions for diverse applications[J]. International Journal of Biological Macromolecules, 2024, 262: 129513. |
| 32 | ATALAH J, CÁCERES-MORENO P, ESPINA G, et al. Thermophiles and the applications of their enzymes as new biocatalysts[J]. Bioresource Technology, 2019, 280: 478-488. |
| 33 | YIN D Y, PAN J, ZHU J, et al. A green-by-design bioprocess for L-carnosine production integrating enzymatic synthesis with membrane separation[J]. Catalysis Science & Technology, 2019, 9(21): 5971-5978. |
| 159 | LIN Y H, YAN Y J. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex[J]. Microbial Cell Factories, 2012, 11: 42. |
| 160 | ZHOU P P, YUE C L, SHEN B, et al. Metabolic engineering of Saccharomyces cerevisiae for enhanced production of caffeic acid[J]. Applied Microbiology and Biotechnology, 2021, 105(14-15): 5809-5819. |
| 161 | LIU L Q, LIU H, ZHANG W, et al. Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations[J]. Engineering, 2019, 5(2): 287-295. |
| 162 | HUANG Q, LIN Y H, YAN Y J. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain[J]. Biotechnology and Bioengineering, 2013, 110(12): 3188-3196. |
| 163 | RODRIGUES J L, ARAÚJO R G, PRATHER K L J, et al. Heterologous production of caffeic acid from tyrosine in Escherichia coli [J]. Enzyme and Microbial Technology, 2015, 71: 36-44. |
| 164 | BROOKS S M, MARSAN C, REED K B, et al. A tripartite microbial co-culture system for de novo biosynthesis of diverse plant phenylpropanoids[J]. Nature Communications, 2023, 14(1): 4448. |
| 165 | HUANG K Y, LI M, LIU Y J, et al. Functional analysis of 3-dehydroquinate dehydratase/shikimate dehydrogenases involved in shikimate pathway in Camellia sinensis [J]. Frontiers in Plant Science, 2019, 10: 1268. |
| 166 | BONTPART T, MARLIN T, VIALET S, et al. Two shikimate dehydrogenases, VvSDH3 and VvSDH4, are involved in gallic acid biosynthesis in grapevine[J]. Journal of Experimental Botany, 2016, 67(11): 3537-3550. |
| 167 | KAMBOURAKIS S, DRATHS K M, FROST J W. Synthesis of gallic acid and pyrogallol from glucose: replacing natural product isolation with microbial catalysis[J]. Journal of the American Chemical Society, 2000, 122(37): 9042-9043. |
| 168 | PAN J, WANG N N, YIN X J, et al. Characterization of a robust and pH-stable tannase from mangrove-derived yeast Rhodosporidium diobovatum Q95[J]. Marine Drugs, 2020, 18(11): 546. |
| 169 | ZHANG K Q, LIN L L, XU H J. Research on antioxidant performance of diglucosyl gallic acid and its application in emulsion cosmetics[J]. International Journal of Cosmetic Science, 2022, 44(2): 177-188. |
| 170 | KHMALADZE I, OSTERLUND C, SMILJANIC S, et al. A novel multifunctional skin care formulation with a unique blend of antipollution, brightening and antiaging active complexes[J]. Journal of Cosmetic Dermatology, 2020, 19(6): 1415-1425. |
| 34 | SHE J J, FU L H, ZHENG X W, et al. Characterization of a new L-carnosine synthase mined from deep-sea sediment metagenome[J]. Microbial Cell Factories, 2022, 21(1): 129. |
| 35 | GUAN B H, YIN W T, CAO B H, et al. Characterization and mutagenesis of a high-activity and highly substrate-tolerant dipeptidase for L-carnosine biosynthesis via reversed hydrolysis[J]. Molecular Catalysis, 2023, 549: 113500. |
| 36 | KAPOOR S, RAFIQ A, SHARMA S. Protein engineering and its applications in food industry[J]. Critical Reviews in Food Science and Nutrition, 2017, 57(11): 2321-2329. |
| 37 | WHITEHURST R J, VAN OORT M. Enzymes in food technology[M/OL]. New York: Wiley Online Library, 2010. [2024-07-01]. . |
| 38 | CHEN K Q, ARNOLD F H. Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide[J]. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90(12): 5618-5622. |
| 39 | SONG W, SUN X, CHEN X L, et al. Enzymatic production of L-citrulline by hydrolysis of the guanidinium group of L-arginine with recombinant arginine deiminase[J]. Journal of Biotechnology, 2015, 208: 37-43. |
| 40 | XIONG D D, LU S K, WU J Y, et al. Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor[J]. Metabolic Engineering, 2017, 40: 115-123. |
| 41 | PENNACCHIETTI E, LAMMENS T M, CAPITANI G, et al. Mutation of His465 alters the pH-dependent spectroscopic properties of Escherichia coli glutamate decarboxylase and broadens the range of its activity toward more alkaline pH[J]. Journal of Biological Chemistry, 2009, 284(46): 31587-31596. |
| 42 | THU HO N A, HOU C Y, KIM W H, et al. Expanding the active pH range of Escherichia coli glutamate decarboxylase by breaking the cooperativeness[J]. Journal of Bioscience and Bioengineering, 2013, 115(2): 154-158. |
| 43 | KANG T J, HO N A T, PACK S P. Buffer-free production of gamma-aminobutyric acid using an engineered glutamate decarboxylase from Escherichia coli [J]. Enzyme and Microbial Technology, 2013, 53(3): 200-205. |
| 44 | YU K, LIN L, HU S, et al. C-terminal truncation of glutamate decarboxylase from Lactobacillus brevis CGMCC 1306 extends its activity toward near-neutral pH[J]. Enzyme and Microbial Technology, 2012, 50(4-5): 263-269. |
| 45 | JUN C H, JOO J C, LEE J H, et al. Thermostabilization of glutamate decarboxylase B from Escherichia coli by structure-guided design of its pH-responsive N-terminal interdomain[J]. Journal of Biotechnology, 2014, 174: 22-28. |
| 46 | 方卉, 吕常江, 花雨娇, 等. 利用脯氨酸效应提高短乳杆菌谷氨酸脱羧酶的热稳定性[J]. 生物工程学报, 2019, 35(4): 636-646. |
| 171 | 崔树梅, 曹孟岑, 杨雪晨, 等. 根皮素在化妆品中的应用[J]. 日用化学工业, 2018, 48(2): 113-118. |
| CUI S M, CAO M C, YANG X C, et al. Applications of phloretin in cosmetics[J]. China Surfactant Detergent & Cosmetics, 2018, 48(2): 113-118. | |
| 172 | LIN Y P, HSU F L, CHEN C S, et al. Constituents from the Formosan apple reduce tyrosinase activity in human epidermal melanocytes[J]. Phytochemistry, 2007, 68(8): 1189-1199. |
| 173 | 夏琛, 刘建华, 崔心禹, 等. 根皮素的生理功能及增溶方法研究进展[J]. 食品科学, 2022, 43(9): 383-390. |
| XIA C, LIU J H, CUI X Y, et al. Recent progress in physiological functions and solubilization methods of phloretin[J]. Food Science, 2022, 43(9): 383-390. | |
| 174 | BOTTA C, DI GIORGIO C, SABATIER A S, et al. Genotoxicity of visible light (400-800 nm) and photoprotection assessment of ectoin, L-ergothioneine and mannitol and four sunscreens[J]. Journal of Photochemistry and Photobiology B: Biology, 2008, 91(1): 24-34. |
| 175 | 董怡麟, 张浩, 陈金龙. 化妆品级聚天冬氨酸钠的制备[J]. 应用化学, 2020, 37(8): 883-888. |
| DONG Y L, ZHANG H, CHEN J L. Preparation of cosmetic-grade sodium polyaspartate[J]. Chinese Journal of Applied Chemistry, 2020, 37(8): 883-888. | |
| 176 | 沈翠云, 梁超群, 喻丹丹, 等. 聚天冬氨酸钠在护肤品中的应用[J]. 日用化学工业, 2017, 47(2): 82-86. |
| SHEN C Y, LIANG C Q, YU D D, et al. Application of sodium polyaspartate in skin care products[J]. China Surfactant Detergent & Cosmetics, 2017, 47(2): 82-86. | |
| 177 | LI Y, ZHANG S Y, LI H D, et al. Metabolic engineering for improving ectoine production in Escherichia coli [J]. Systems Microbiology and Biomanufacturing, 2024, 4(1): 337-347. |
| 178 | MA Z, CHANG R J, ZHU L J, et al. Metabolic engineering of Corynebacterium glutamicum for highly efficient production of ectoine[J]. ACS Synthetic Biology, 2024, 13(7): 2081-2090. |
| 46 | FANG H, LÜ C J, HUA Y J, et al. Increasing the thermostability of glutamate decarboxylase from Lactobacillus brevis by introducing proline[J]. Chinese Journal of Biotechnology, 2019, 35(4): 636-646. |
| 47 | LIU W J, HU X X, YAN Y, et al. Rational engineering of homospermidine synthase for enhanced catalytic efficiency toward spermidine synthesis[J]. Synthetic and Systems Biotechnology, 2024, 9(3): 549-557. |
| 48 | LIU Y R, PAN X W, ZHANG H W, et al. Combinatorial protein engineering and transporter engineering for efficient synthesis of L-carnosine in Escherichia coli [J]. Bioresource Technology, 2023, 387: 129628. |
| 49 | LU J W, NIE M F, LI Y R, et al. Design of composite nanosupports and applications thereof in enzyme immobilization: a review[J]. Colloids and Surfaces B: Biointerfaces, 2022, 217: 112602. |
| 50 | ZHANG W, ZHANG Z, JI L R, et al. Laccase immobilized on nanocomposites for wastewater pollutants degradation: current status and future prospects[J]. Bioprocess and Biosystems Engineering, 2023, 46(11): 1513-1531. |
| 51 | MAGHRABY Y R, EL-SHABASY R M, IBRAHIM A H, et al. Enzyme immobilization technologies and industrial applications[J]. ACS Omega, 2023, 8(6): 5184-5196. |
| 52 | LI L, LI Z M, WANG C H, et al. The electrostatic driving force for nucleophilic catalysis in L-arginine deiminase: a combined experimental and theoretical study[J]. Biochemistry, 2008, 47(16): 4721-4732. |
| 53 | KIM J E, JEONG D W, LEE H J. Expression, purification, and characterization of arginine deiminase from Lactococcus lactis ssp. lactis ATCC 7962 in Escherichia coli BL21[J]. Protein Expression and Purification, 2007, 53(1): 9-15. |
| 54 | 赵艳杰, 曾倡, 张淑荣, 等. 固定化粪链球菌酶法连续生产L-瓜氨酸[J]. 北京化工大学学报(自然科学版), 2010, 37(4): 98-102. |
| ZHAO Y J, ZENG C, ZHANG S R, et al. Enzymatic and continuous production of L-citrulline by immobilized Streptococcus faecalis cells[J]. Journal of Beijing University of Chemical Technology (Natural Science Edition), 2010, 37(4): 98-102. | |
| 55 | MOHAMMADI N S, KHIABANI M S, GHANBARZADEH B, et al. Improvement of lipase biochemical properties via a two-step immobilization method: adsorption onto silicon dioxide nanoparticles and entrapment in a polyvinyl alcohol/alginate hydrogel[J]. Journal of Biotechnology, 2020, 323: 189-202. |
| 56 | SHEN F, ARSHI S, MAGNER E, et al. One-step electrochemical approach of enzyme immobilization for bioelectrochemical applications[J]. Synthetic Metals, 2022, 291: 117205. |
| 179 | MA Q, XIA L, WU H Y, et al. Metabolic engineering of Escherichia coli for efficient osmotic stress-free production of compatible solute hydroxyectoine[J]. Biotechnology and Bioengineering, 2022, 119(1): 89-101. |
| 180 | JUNGMANN L, HOFFMANN S L, LANG C, et al. High-efficiency production of 5-hydroxyectoine using metabolically engineered Corynebacterium glutamicum [J]. Microbial Cell Factories, 2022, 21(1): 274. |
| 181 | WANG L, LI S, ZHAO J J, et al. Efficiently activated ε-poly-L-lysine production by multiple antibiotic-resistance mutations and acidic pH shock optimization in Streptomyces albulus [J]. MicrobiologyOpen, 2019, 8(5): e00728. |
| 182 | 朱道君, 刁文娇, 张佳微, 等. 小白链霉菌全细胞转化L-赖氨酸合成ε-聚赖氨酸的体系构建与优化[J]. 食品与发酵工业, 2024, 50(1): 29-36. |
| ZHU D J, DIAO W J, ZHANG J W, et al. Construction and optimization of whole-cell transformation method for ε-poly-L-lysine production from L-lysine by Streptomyces albulus [J]. Food and Fermentation Industries, 2024, 50(1): 29-36. | |
| 183 | ZHANG L W, TANG J W, FENG M Q, et al. Engineering methyltransferase and sulfoxide synthase for high-yield production of ergothioneine[J]. Journal of Agricultural and Food Chemistry, 2023, 71(1): 671-679. |
| 184 | 张山, 丁利平, 焦银山. 一种麦角硫因生产工艺及其应用: CN114854659B [P]. 2024-03-26. |
| ZHANG S, DING L P, JIAO Y S. A ergothionein production process and its application: CN114854659B [P]. 2024-03-26. | |
| 185 | 吴鹤云, 刘万才, 谢希贤, 等. 一种生产麦角硫因的基因工程菌及其构建方法与应用: CN116121161A[P]. 2023-05-16. |
| WU H Y, LIU W C, XIE X X, et al. A genetic engineering bacteria producing ergothioneine and its construction method and application: CN116121161A[P]. 2023-05-16. | |
| 186 | ZHOU L Q, XIANG T, YANG M X, et al. Yeast strain and use thereof and preparation method of ergothioneine: US20230220428[P]. 2023-07-13. |
| 187 | XING B, LI Z W, CHANG J Z, et al. Molecular modification based on site-directed mutagenesis improves the substrate specificity of β-ester acyltransferase for L-carnosine synthesis[J]. Process Biochemistry, 2024, 137: 1-9. |
| 57 | LIAN J Z, MISHRA S, ZHAO H M. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications[J]. Metabolic Engineering, 2018, 50: 85-108. |
| 58 | GIEßELMANN G, DIETRICH D, JUNGMANN L, et al. Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway[J]. Biotechnology Journal, 2019, 14(9): 1800417. |
| 59 | XU S Q, ZHANG B, CHEN W H, et al. Highly efficient production of ectoine via an optimized combination of precursor metabolic modules in Escherichia coli BL21[J]. Bioresource Technology, 2023, 390: 129803. |
| 60 | ZHOU D D, LUO M, HUANG S Y, et al. Effects and mechanisms of resveratrol on aging and age-related diseases[J]. Oxidative Medicine and Cellular Longevity, 2021, 2021(1): 9932218. |
| 61 | LIU M S, WANG C, REN X F, et al. Remodelling metabolism for high-level resveratrol production in Yarrowia lipolytica [J]. Bioresource Technology, 2022, 365: 128178. |
| 62 | 马倩, 夏利, 谭淼, 等. 氨基酸生产的代谢工程研究进展与发展趋势[J]. 生物工程学报, 2021, 37(5): 1677-1696. |
| MA Q, XIA L, TAN M, et al. Advances and prospects in metabolic engineering for the production of amino acids[J]. Chinese Journal of Biotechnology, 2021, 37(5): 1677-1696. | |
| 63 | 叶健文, 陈江楠, 张旭, 等. 动态调控: 一种高效的细胞工厂工程化代谢改造策略[J]. 生物技术通报, 2020, 36(6): 1-12. |
| YE J W, CHEN J N, ZHANG X, et al. Dynamic control: an efficient strategy for metabolically engineering microbial cell factories[J]. Biotechnology Bulletin, 2020, 36(6): 1-12. | |
| 64 | QIAN S, CIRINO P C. Using metabolite-responsive gene regulators to improve microbial biosynthesis[J]. Current Opinion in Chemical Engineering, 2016, 14: 93-102. |
| 65 | JIANG S, WANG D H, WANG R R, et al. Reconstructing a recycling and nonauxotroph biosynthetic pathway in Escherichia coli toward highly efficient production of L-citrulline[J]. Metabolic Engineering, 2021, 68: 220-231. |
| 66 | HU L X, ZHAO M, HU W S, et al. Poly-γ-glutamic acid production by engineering a DegU quorum-sensing circuit in Bacillus subtilis [J]. ACS Synthetic Biology, 2022, 11(12): 4156-4170. |
| 188 | KIM M H, KO Y J, JEONG D W, et al. Ecofriendly synthesis of L-carnosine in metabolically engineered Corynebacterium glutamicum by reinforcing precursor accumulation[J]. ACS Synthetic Biology, 2021, 10(6): 1553-1562. |
| 189 | PATZSCHKE A, STEIGER M G, HOLZ C, et al. Enhanced glutathione production by evolutionary engineering of Saccharomyces cerevisiae strains[J]. Biotechnology Journal, 2015, 10(11): 1719-1726. |
| 190 | CHEN H L, CAO X T, ZHU N Q, et al. A stepwise control strategy for glutathione synthesis in Saccharomyces cerevisiae based on oxidative stress and energy metabolism[J]. World Journal of Microbiology & Biotechnology, 2020, 36(8): 117. |
| 191 | WANG C, ZHANG J, WU H, et al. Heterologous gshF gene expression in various vector systems in Escherichia coli for enhanced glutathione production[J]. Journal of Biotechnology, 2015, 214: 63-68. |
| 192 | KUHLMANN A U, BREMER E. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp[J]. Applied and Environmental Microbiology, 2002, 68(2): 772-783. |
| 193 | GALINSKI E A, PFEIFFER H P, TRÜPER H G. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira [J]. European Journal of Biochemistry, 1985, 149(1): 135-139. |
| 194 | INBAR L, LAPIDOT A. The structure and biosynthesis of new tetrahydropyrimidine derivatives in actinomycin D producer Streptomyces parvulus. Use of 13C- and 15N-labeled L-glutamate and 13C and 15N NMR spectroscopy[J]. Journal of Biological Chemistry, 1988, 263(31): 16014-16022. |
| 195 | KUNTE H J, LENTZEN G, GALINSKI E. Industrial production of the cell protectant ectoine: protection mechanisms, processes, and products[J]. Current Biotechnology, 2014, 3(1): 10-25. |
| 196 | RESHETNIKOV A S, ROZOVA O N, TROTSENKO Y A, et al. Ectoine degradation pathway in halotolerant methylotrophs[J]. PLoS One, 2020, 15(4): e0232244. |
| 197 | PASTOR J M, SALVADOR M, ARGANDOÑA M, et al. Ectoines in cell stress protection: uses and biotechnological production[J]. Biotechnology Advances, 2010, 28(6): 782-801. |
| 198 | STÖVEKEN N, PITTELKOW M, SINNER T, et al. A specialized aspartokinase enhances the biosynthesis of the osmoprotectants ectoine and hydroxyectoine in Pseudomonas stutzeri A1501[J]. Journal of Bacteriology, 2011, 193(17): 4456-4468. |
| 199 | HILLIER H T, ALTERMARK B, LEIROS I. The crystal structure of the tetrameric DABA-aminotransferase EctB, a rate-limiting enzyme in the ectoine biosynthesis pathway[J]. The FEBS Journal, 2020, 287(21): 4641-4658. |
| 67 | 李强, 韩亚昆, 蒋帅, 等. 代谢工程改造大肠杆菌合成反式-4-羟基-L-脯氨酸[J]. 食品科学, 2020, 41(2): 202-207. |
| LI Q, HAN Y K, JIANG S, et al. Metabolic engineering of Escherichia coli for production of trans-4-hydroxy-L-proline[J]. Food Science, 2020, 41(2): 202-207. | |
| 68 | XU M J, RAO Z M, DOU W F, et al. Site-directed mutagenesis and feedback-resistant N-acetyl-L-glutamate kinase (NAGK) increase Corynebacterium crenatum L-arginine production[J]. Amino Acids, 2012, 43(1): 255-266. |
| 69 | IKEDA M, MITSUHASHI S, TANAKA K, et al. Reengineering of a Corynebacterium glutamicum L-arginine and L-citrulline producer[J]. Applied and Environmental Microbiology, 2009, 75(6): 1635-1641. |
| 70 | PARK S H, KIM H U, KIM T Y, et al. Metabolic engineering of Corynebacterium glutamicum for L-arginine production[J]. Nature Communications, 2014, 5: 4618. |
| 71 | LIU H Y, TIAN Y J, ZHOU Y, et al. Multi-modular engineering of Saccharomyces cerevisiae for high-titre production of tyrosol and salidroside[J]. Microbial Biotechnology, 2021, 14(6): 2605-2616. |
| 72 | WANG L, LI N, YU S Q, et al. Enhancing caffeic acid production in Escherichia coli by engineering the biosynthesis pathway and transporter[J]. Bioresource Technology, 2023, 368: 128320. |
| 73 | GONG Y, WANG R Q, MA L, et al. Optimization of trans-4-hydroxyproline synthesis pathway by rearrangement center carbon metabolism in Escherichia coli [J]. Microbial Cell Factories, 2023, 22(1): 240. |
| 74 | QIU C, WANG X G, ZUO J J, et al. Systems engineering Escherichia coli for efficient production p-coumaric acid from glucose[J]. Biotechnology and Bioengineering, 2024, 121(7): 2147-2162. |
| 75 | CHEN R B, GAO J Q, YU W, et al. Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast[J]. Nature Chemical Biology, 2022, 18(5): 520-529. |
| 76 | ZHANG X, ZHANG X F, LI H P, et al. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool[J]. Applied Microbiology and Biotechnology, 2014, 98(12): 5387-5396. |
| 77 | OTTENHEIM C, NAWRATH M, WU J C. Microbial mutagenesis by atmospheric and room-temperature plasma (ARTP): the latest development[J]. Bioresources and Bioprocessing, 2018, 5(1): 12. |
| 200 | RICHTER A A, KOBUS S, CZECH L, et al. The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine[J]. Journal of Biological Chemistry, 2020, 295(9): 2822-2838. |
| 201 | 李莉, 左甜甜, 董亚蕾, 等. 化妆品中替代性防腐成分使用情况分析[J]. 香料香精化妆品, 2023(1): 92-98. |
| LI L, ZUO T T, DONG Y L, et al. Use status of alternative preservatives in cosmetics[J]. Flavour Fragrance Cosmetics, 2023(1): 92-98. | |
| 202 | 郑中博, 丛远华, 冯春波. 聚赖氨酸在化妆品中的防腐效能研究[J]. 广东化工, 2021, 48(1): 25-26, 13. |
| ZHENG Z B, CONG Y H, FENG C B. Study on a naturally derived preservative: ε-polylysine[J]. Guangdong Chemical Industry, 2021, 48(1): 25-26, 13. | |
| 203 | CHEAH I K, HALLIWELL B. Ergothioneine; antioxidant potential, physiological function and role in disease[J]. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 2012, 1822(5): 784-793. |
| 204 | KITSANAYANYONG L, OHSHIMA T. Ergothioneine: a potential antioxidative and antimelanosis agent for food quality preservation[J]. FEBS Letters, 2022, 596(10): 1330-1347. |
| 205 | LIU H M, TANG W, WANG X Y, et al. Safe and effective antioxidant: the biological mechanism and potential pathways of ergothioneine in the skin[J]. Molecules, 2023, 28(4): 1648. |
| 206 | GRÜNDEMANN D, HARLFINGER S, GOLZ S, et al. Discovery of the ergothioneine transporter[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(14): 5256-5261. |
| 207 | LIAO W C, WU W H, TSAI P C, et al. Kinetics of ergothioneine inhibition of mushroom tyrosinase[J]. Applied Biochemistry and Biotechnology, 2012, 166(2): 259-267. |
| 208 | OBAYASHI K, KURIHARA K, OKANO Y, et al. L-ergothioneine scavenges superoxide and singlet oxygen and suppresses TNF-α and MMP-1 expression in UV-irradiated human dermal fibroblasts[J]. International Journal of Cosmetic Science, 2005, 27(3): 191. |
| 209 | STAMPFLI A R, SEEBECK F P. The catalytic mechanism of sulfoxide synthases[J]. Current Opinion in Chemical Biology, 2020, 59: 111-118. |
| 210 | JONES G W, DOYLE S, FITZPATRICK D A. The evolutionary history of the genes involved in the biosynthesis of the antioxidant ergothioneine[J]. Gene, 2014, 549(1): 161-170. |
| 78 | LV Q L, HU M K, TIAN L Z, et al. Enhancing L-glutamine production in Corynebacterium glutamicum by rational metabolic engineering combined with a two-stage pH control strategy[J]. Bioresource Technology, 2021, 341: 125799. |
| 79 | ZHAO Z Q, CAI M M, LIU Y R, et al. Genomics and transcriptomics-guided metabolic engineering Corynebacterium glutamicum for L-arginine production[J]. Bioresource Technology, 2022, 364: 128054. |
| 80 | JIANG S, WANG R R, WANG D H, et al. Metabolic reprogramming and biosensor-assisted mutagenesis screening for high-level production of L-arginine in Escherichia coli [J]. Metabolic Engineering, 2023, 76: 146-157. |
| 81 | 罗丽娟, 王刚, 万玉军, 等. γ-聚谷氨酸高产菌株选育及发酵条件优化[J]. 食品与发酵科技, 2021, 57(6): 35-42. |
| LUO L J, WANG G, WAN Y J, et al. Breeding of high-yield γ-polyglutamic acid strains and optimization of fermentation conditions[J]. Food and Fermentation Sciences & Technology, 2021, 57(6): 35-42. | |
| 82 | 柳天一, 张越, 王靓, 等. 基于低pH适应性进化策略提高小白链霉菌ε-聚赖氨酸合成能力[J]. 食品与发酵工业, 2024, 50(1): 14-21. |
| LIU T Y, ZHANG Y, WANG L, et al. Improvement of ε-poly-L-lysine production by Streptomyces albulus based on low-pH adaptive evolution strategy[J]. Food and Fermentation Industries, 2024, 50(1): 14-21. | |
| 83 | MAVROMMATI M, DASKALAKI A, PAPANIKOLAOU S, et al. Adaptive laboratory evolution principles and applications in industrial biotechnology[J]. Biotechnology Advances, 2022, 54: 107795. |
| 84 | MUNDHADA H, SEOANE J M, SCHNEIDER K, et al. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution[J]. Metabolic Engineering, 2017, 39: 141-150. |
| 85 | CIOBANU C P, BLAGA A C, FROIDEVAUX R, et al. Enhanced growth and β-galactosidase production on Escherichia coli using oxygen vectors[J]. 3 Biotech, 2020, 10(7): 298. |
| 86 | ZHANG R Z, YANG T W, RAO Z M, et al. Efficient one-step preparation of γ-aminobutyric acid from glucose without an exogenous cofactor by the designed Corynebacterium glutamicum [J]. Green Chemistry, 2014, 16(9): 4190-4197. |
| 87 | WEN J B, SUN W L, LENG G H, et al. Enhanced fermentative γ-aminobutyric acid production by a metabolic engineered Corynebacterium glutamicum [J]. Biotechnology and Bioprocess Engineering, 2024, 29(1): 129-140. |
| 211 | CHEN Z H, HE Y Z, WU X Y, et al. Toward more efficient ergothioneine production using the fungal ergothioneine biosynthetic pathway[J]. Microbial Cell Factories, 2022, 21(1): 76. |
| 212 | 王丽, 王阳, 李江华, 等. 产麦角硫因大肠杆菌工程菌株的构建与优化[J]. 生物工程学报, 2022, 38(2): 796-806. |
| WANG L, WANG Y, LI J H, et al. Construction and optimization of ergothioneine-producing Escherichia coli [J]. Chinese Journal of Biotechnology, 2022, 38(2): 796-806. | |
| 213 | 陈佳敏, 王阳, 堵国成, 等. 优化前体供给与细胞膜通透性强化大肠杆菌合成麦角硫因[J]. 食品与生物技术学报, 2022, 41(8): 43-52. |
| CHEN J M, WANG Y, DU G C, et al. Enhancement of ergothioneine synthesis in Escherichia coli via optimization of precursor supply and cell membrane permeability[J]. Journal of Food Science and Biotechnology, 2022, 41(8): 43-52. | |
| 214 | 马倩, 田道光, 谢希贤, 等. 一种生产麦角硫因的基因工程菌株及其应用: CN112251392B[P]. 2022-09-09. |
| MA Q, TIAN D G, XIE X X, et al. Genetically engineered strain for producing ergothioneine and application: CN112251392B [P]. 2022-09-09. | |
| 215 | PLUSKAL T, UENO M, YANAGIDA M. Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system[J]. PLoS One, 2014, 9(5): e97774. |
| 216 | QIU Y B, CHEN Z L, SU E Z, et al. Recent strategies for the biosynthesis of ergothioneine[J]. Journal of Agricultural and Food Chemistry, 2021, 69(46): 13682-13690. |
| 217 | HADMED H H EL, CASTILLO R F. Cosmeceuticals: peptides, proteins, and growth factors[J]. Journal of Cosmetic Dermatology, 2016, 15(4): 514-519. |
| 218 | HECK T, KOHLER H P E, LIMBACH M, et al. Enzyme-catalyzed formation of β-peptides: β-peptidyl aminopeptidases BapA and DmpA acting as β-peptide-synthesizing enzymes[J]. Chemistry & Biodiversity, 2007, 4(9): 2016-2030. |
| 219 | HEYLAND J, ANTWEILER N, LUTZ J, et al. Simple enzymatic procedure for L-carnosine synthesis: whole-cell biocatalysis and efficient biocatalyst recycling[J]. Microbial Biotechnology, 2010, 3(1): 74-83. |
| 88 | MORI H, MATSUI M, BAMBA T, et al. Engineering Escherichia coli for efficient glutathione production[J]. Metabolic Engineering, 2024, 84: 180-190. |
| 89 | HAO N, MU J R, HU N, et al. Improvement of L-citrulline production in Corynebacterium glutamicum by ornithine acetyltransferase[J]. Journal of Industrial Microbiology & Biotechnology, 2015, 42(2): 307-313. |
| 90 | CAI D B, CHEN Y Z, HE P H, et al. Enhanced production of poly-γ-glutamic acid by improving ATP supply in metabolically engineered Bacillus licheniformis [J]. Biotechnology and Bioengineering, 2018, 115(10): 2541-2553. |
| 91 | WANG D X, FU X P, GAO J Q, et al. Enhancing poly-γ- glutamic acid production in Bacillus tequilensis BL01 through a multienzyme assembly strategy and expression features of glutamate synthesis from Corynebacterium glutamicum [J]. Journal of Agricultural and Food Chemistry, 2024, 72(15): 8674-8683. |
| 92 | XU G Q, WANG J Y, SHEN J C, et al. Enhanced poly-γ- glutamic acid synthesis in Corynebacterium glutamicum by reconstituting PgsBCA complex and fermentation optimization[J]. Metabolic Engineering, 2024, 81: 238-248. |
| 93 | KE C R, YANG X W, RAO H X, et al. Whole-cell conversion of L-glutamic acid into gamma-aminobutyric acid by metabolically engineered Escherichia coli [J]. SpringerPlus, 2016, 5: 591. |
| 94 | YANG X W, HUO X J, TANG Y Q, et al. Integrating enzyme evolution and metabolic engineering to improve the productivity of γ-aminobutyric acid by whole-cell biosynthesis in Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2023, 71(11): 4656-4664. |
| 95 | WEN J B, BAO J. Improved fermentative γ-aminobutyric acid production by secretory expression of glutamate decarboxylase by Corynebacterium glutamicum [J]. Journal of Biotechnology, 2021, 331: 19-25. |
| 96 | WEI L, ZHAO J H, WANG Y R, et al. Engineering of Corynebacterium glutamicum for high-level γ-aminobutyric acid production from glycerol by dynamic metabolic control[J]. Metabolic Engineering, 2022, 69: 134-146. |
| 97 | JIA M Y, ZHU Y S, WANG L Q, et al. pH auto-sustain-based fermentation supports efficient gamma-aminobutyric acid production by Lactobacillus brevis CD0817[J]. Fermentation, 2022, 8(5): 208. |
| 98 | WANG X C, LIU J, ZHAO J, et al. Efficient production of trans-4-hydroxy-L-proline from glucose using a new trans-proline 4-hydroxylase in Escherichia coli [J]. Journal of Bioscience and Bioengineering, 2018, 126(4): 470-477. |
| 99 | ZOU D, LI L, MIN Y, et al. Biosynthesis of a novel bioactive metabolite of spermidine from Bacillus amyloliquefaciens: gene mining, sequence analysis, and combined expression[J]. Journal of Agricultural and Food Chemistry, 2021, 69(1): 267-274. |
| 220 | HECK T, MAKAM V S, LUTZ J, et al. Kinetic analysis of L-carnosine formation by β-aminopeptidases[J]. Advanced Synthesis & Catalysis, 2010, 352(2-3): 407-415. |
| 221 | TEUFEL M, SAUDEK V, LEDIG J P, et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase[J]. Journal of Biological Chemistry, 2003, 278(8): 6521-6531. |
| 222 | INABA C, HIGUCHI S, MORISAKA H, et al. Synthesis of functional dipeptide carnosine from nonprotected amino acids using carnosinase-displaying yeast cells[J]. Applied Microbiology and Biotechnology, 2010, 86(6): 1895-1902. |
| 223 | BAHUT F, ROMANET R, SIECZKOWSKI N, et al. Antioxidant activity from inactivated yeast: expanding knowledge beyond the glutathione-related oxidative stability of wine[J]. Food Chemistry, 2020, 325: 126941. |
| 224 | MALAIRUANG K, KRAJANG M, SUKNA J, et al. High cell density cultivation of Saccharomyces cerevisiae with intensive multiple sequential batches together with a novel technique of fed-batch at cell level (FBC)[J]. Processes, 2020, 8(10): 1321. |
| 225 | ZHU Y B, SUN J, ZHU Y Y, et al. Endogenic oxidative stress response contributes to glutathione over-accumulation in mutant Saccharomyces cerevisiae Y518[J]. Applied Microbiology and Biotechnology, 2015, 99(17): 7069-7078. |
| 226 | WEN S H, ZHANG T, TAN T W. Utilization of amino acids to enhance glutathione production in Saccharomyces cerevisiae [J]. Enzyme and Microbial Technology, 2004, 35(6-7): 501-507. |
| 227 | KOBAYASHI J, SASAKI D, HARA K Y, et al. Metabolic engineering of the L-serine biosynthetic pathway improves glutathione production in Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2022, 21(1): 153. |
| 228 | LORENZ E, SCHMACHT M, STAHL U, et al. Enhanced incorporation yield of cysteine for glutathione overproduction by fed-batch fermentation of Saccharomyces cerevisiae [J]. Journal of Biotechnology, 2015, 216: 131-139. |
| 229 | HU X Y, SHEN X L, ZHU S, et al. Optimization of glutathione production in Saccharomyces cerevisiae HBSD-W08 using Plackett-Burman and central composite rotatable designs[J]. BMC Microbiology, 2023, 23(1): 11. |
| 230 | ZHANG J, QUAN C, WANG C, et al. Systematic manipulation of glutathione metabolism in Escherichia coli for improved glutathione production[J]. Microbial Cell Factories, 2016, 15: 38. |
| 231 | CUI X W, WAN J X, ZHANG X, et al. Efficient glutathione production in metabolically engineered Escherichia coli strains using constitutive promoters[J]. Journal of Biotechnology, 2019, 289: 39-45. |
| 100 | QIN J F, KRIVORUCHKO A, JI B Y, et al. Engineering yeast metabolism for the discovery and production of polyamines and polyamine analogues[J]. Nature Catalysis, 2021, 4: 498-509. |
| 101 | LIU Y, GUO X, WANG X, et al. A two-enzyme cascade system for the bio-production of spermidine from putrescine[J]. Molecular Catalysis, 2021, 504: 111439. |
| 102 | GINESY M, BELOTSERKOVSKY J, ENMAN J, et al. Metabolic engineering of Escherichia coli for enhanced arginine biosynthesis[J]. Microbial Cell Factories, 2015, 14: 29. |
| 103 | JENSEN J V K, EBERHARDT D, WENDISCH V F. Modular pathway engineering of Corynebacterium glutamicum for production of the glutamate-derived compounds ornithine, proline, putrescine, citrulline, and arginine[J]. Journal of Biotechnology, 2015, 214: 85-94. |
| 104 | WANG H D, XU J Z, ZHANG W G. Metabolic engineering of Escherichia coli for efficient production of L-arginine[J]. Applied Microbiology and Biotechnology, 2022, 106(17): 5603-5613. |
| 105 | NGUYEN A Q D, SCHNEIDER J, REDDY G K, et al. Fermentative production of the diamine putrescine: system metabolic engineering of Corynebacterium glutamicum [J]. Metabolites, 2015, 5(2): 211-231. |
| 106 | SCHRAMM T, LEMPP M, BEUTER D, et al. High-throughput enrichment of temperature-sensitive argininosuccinate synthetase for two-stage citrulline production in E. coli [J]. Metabolic Engineering, 2020, 60: 14-24. |
| 107 | 胥琳峰, 于文文, 朱学文, 等. 代谢工程改造大肠杆菌高效合成L-瓜氨酸[J]. 生物工程学报, 2025, 41(1): 242-255. |
| XU L F, YU W W, ZHU X W, et al. Metabolic engineering of Escherichia coli for efficient biosynthesis of L-citrulline[J]. Chinese Journal of Biotechnology, 2025, 41(1): 242-255. | |
| 108 | CANDELA T, FOUET A. Poly-gamma-glutamate in bacteria[J]. Molecular Microbiology, 2006, 60(5): 1091-1098. |
| 109 | DAHIYA D, CHETTRI R, NIGAM P S. Chapter 25-Biosynthesis of polyglutamic acid (γ-PGA), a biodegradable and economical polyamide biopolymer for industrial applications[M/OL]//Microbial and natural macromolecules, 2021: 681-688. (2020-09-18)[2024-06-01]. . |
| 110 | LUO Z T, GUO Y, LIU J D, et al. Microbial synthesis of poly-γ-glutamic acid: current progress, challenges, and future perspectives[J]. Biotechnology for Biofuels, 2016, 9: 134. |
| [1] | WU Ke, LUO Jiahao, LI Feiran. Applications of machine learning in the reconstruction and curation of genome-scale metabolic models [J]. Synthetic Biology Journal, 2025, 6(3): 566-584. |
| [2] | TIAN Xiao-jun, ZHANG Rixin. “Economics Paradox” with cells in synthetic gene circuits [J]. Synthetic Biology Journal, 2025, 6(3): 532-546. |
| [3] | ZHANG Yiqing, LIU Gaowen. Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(3): 685-700. |
| [4] | YANG Ying, LI Xia, LIU Lizhong. Applications of synthetic biology to stem-cell-derived modeling of early embryonic development [J]. Synthetic Biology Journal, 2025, 6(3): 669-684. |
| [5] | HUANG Yi, SI Tong, LU Anjing. Standardization for biomanufacturing: global landscape, critical challenges, and pathways forward [J]. Synthetic Biology Journal, 2025, 6(3): 701-714. |
| [6] | SONG Chengzhi, LIN Yihan. AI-enabled directed evolution for protein engineering and optimization [J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [7] | ZHANG Mengyao, CAI Peng, ZHOU Yongjin. Synthetic biology drives the sustainable production of terpenoid fragrances and flavors [J]. Synthetic Biology Journal, 2025, 6(2): 334-356. |
| [8] | ZHANG Lu’ou, XU Li, HU Xiaoxu, YANG Ying. Synthetic biology ushers cosmetic industry into the “bio-cosmetics” era [J]. Synthetic Biology Journal, 2025, 6(2): 479-491. |
| [9] | WEI Lingzhen, WANG Jia, SUN Xinxiao, YUAN Qipeng, SHEN Xiaolin. Biosynthesis of flavonoids and their applications in cosmetics [J]. Synthetic Biology Journal, 2025, 6(2): 373-390. |
| [10] | XIAO Sen, HU Litao, SHI Zhicheng, WANG Fayin, YU Siting, DU Guocheng, CHEN Jian, KANG Zhen. Research advances in biosynthesis of hyaluronic acid with controlled molecular weights [J]. Synthetic Biology Journal, 2025, 6(2): 445-460. |
| [11] | WANG Qian, GUO Shiting, XIN Bo, ZHONG Cheng, WANG Yu. Advances in biosynthesis of L-arginine using engineered microorganisms [J]. Synthetic Biology Journal, 2025, 6(2): 290-305. |
| [12] | ZUO Yimeng, ZHANG Jiaojiao, LIAN Jiazhang. Enabling technology for the biosynthesis of cosmetic raw materials with Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2025, 6(2): 233-253. |
| [13] | TANG Chuan′gen, WANG Jing, ZHANG Shuo, ZHANG Haoning, KANG Zhen. Advances in synthesis and mining strategies for functional peptides [J]. Synthetic Biology Journal, 2025, 6(2): 461-478. |
| [14] | GUO Tingting, HAN Xiangning, HUANG Xiting, ZHANG Tingting, KONG Jian. Advances in synthetic biology tools for lactic acid bacteria and their application in the development of skin beneficial products [J]. Synthetic Biology Journal, 2025, 6(2): 320-333. |
| [15] | ZHANG Ping, ZHANG Weijiao, XU Ruirui, LI Jianghua, CHEN Jian, KANG Zhen. Research advances on the biosynthesis of mycosporine-like amino acids [J]. Synthetic Biology Journal, 2025, 6(2): 306-319. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||