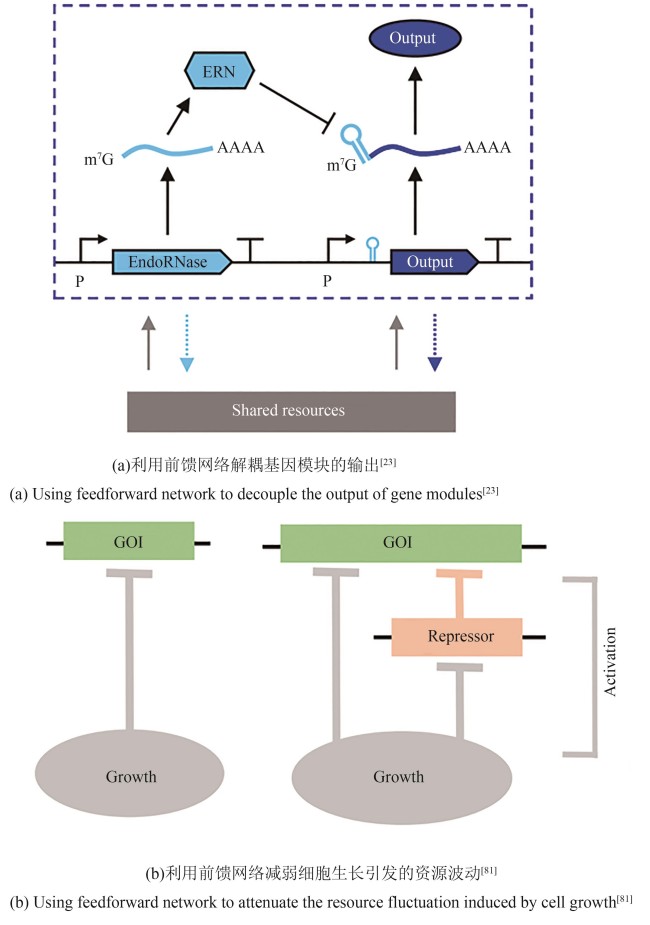
Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (3): 532-546.DOI: 10.12211/2096-8280.2024-083
• Invited Review • Previous Articles Next Articles
“Economics Paradox” with cells in synthetic gene circuits
TIAN Xiao-jun, ZHANG Rixin
- School of Biological and Health Systems Engineering,Arizona State University,Tempe 85281,Arizona,USA
-
Received:2024-11-27Revised:2025-02-19Online:2025-06-27Published:2025-06-30 -
Contact:TIAN Xiao-jun
合成基因回路面临的细胞“经济学窘境”
田晓军, 张日新
- 亚利桑那州立大学生物与健康系统工程学院,美国 亚利桑那州 坦佩 85281
-
通讯作者:田晓军 -
作者简介:田晓军 (1984—),男,博士生导师。研究方向为定量生物学、系统生物学、合成生物学,目前研究是优化人工合成基因回路设计及其应用。E-mail:Xiaojun.Tian@asu.edu
CLC Number:
Cite this article
TIAN Xiao-jun, ZHANG Rixin. “Economics Paradox” with cells in synthetic gene circuits[J]. Synthetic Biology Journal, 2025, 6(3): 532-546.
田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-083
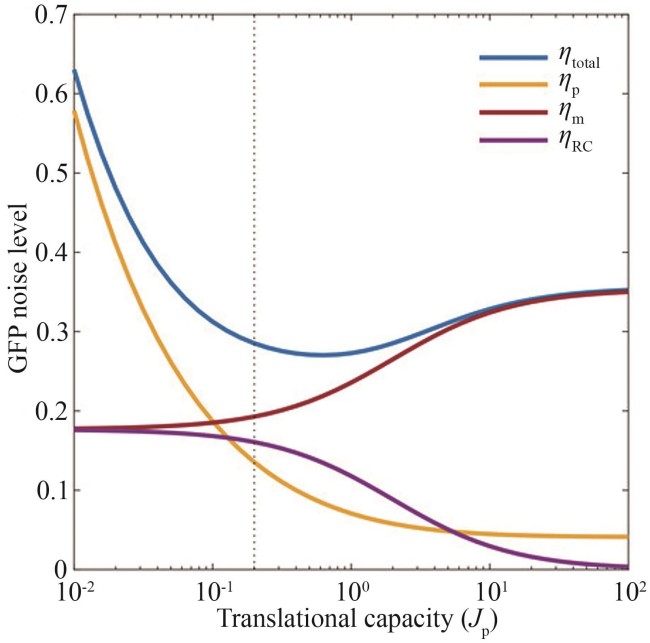
Fig. 2 Analytical solutions of the total protein noise[34](ηtotal—total protein noise; ηp—birth/death of protein noise; ηm—mRNA fluctuating noise; ηRC—resource competition noise)
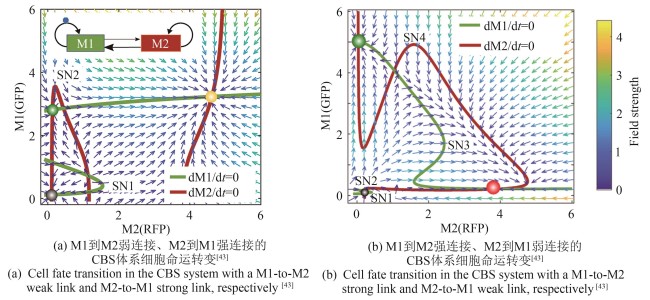
Fig. 4 Cascaded bistable switch circuit demonstrates two different paths for cell fate transition associated with the strength of links between the two modules[43]
| 1 | MOE-BEHRENS G H G, DAVIS R, HAYNES K A. Preparing synthetic biology for the world[J]. Frontiers in Microbiology, 2013, 4: 5. |
| 2 | WURTZEL E T, VICKERS C E, HANSON A D, et al. Revolutionizing agriculture with synthetic biology[J]. Nature Plants, 2019, 5(12): 1207-1210. |
| 3 | MENG F K, ELLIS T. The second decade of synthetic biology: 2010—2020[J]. Nature Communications, 2020, 11(1): 5174. |
| 4 | NGUYEN P Q, HUANG X N, COLLINS D S, et al. Harnessing synthetic biology to enhance ocean health[J]. Trends in Biotechnology, 2023, 41(7): 860-874. |
| 5 | TANG T C, AN B L, HUANG Y Y, et al. Materials design by synthetic biology[J]. Nature Reviews Materials, 2021, 6(4): 332-350. |
| 6 | BORKOWSKI O, CERONI F, STAN G B, et al. Overloaded and stressed: whole-cell considerations for bacterial synthetic biology[J]. Current Opinion in Microbiology, 2016, 33: 123-130. |
| 7 | SHAKIBA N, JONES R D, WEISS R, et al. Context-aware synthetic biology by controller design: engineering the mammalian cell[J]. Cell Systems, 2021, 12(6): 561-592. |
| 8 | LIAO C, BLANCHARD A E, LU T. An integrative circuit-host modelling framework for predicting synthetic gene network behaviours[J]. Nature Microbiology, 2017, 2(12): 1658-1666. |
| 9 | DEL VECCHIO D. Modularity, context-dependence, and insulation in engineered biological circuits[J]. Trends in Biotechnology, 2015, 33(2): 111-119. |
| 10 | BOO A, ELLIS T, STAN G B. Host-aware synthetic biology[J]. Current Opinion in Systems Biology, 2019, 14: 66-72. |
| 11 | ILIA K, DEL VECCHIO D. Squaring a circle: to what extent are traditional circuit analogies impeding synthetic biology?[J]. GEN Biotechnology, 2022, 1(2): 150-155. |
| 12 | ŞIMŞEK E, YAO Y, LEE D, et al. Toward predictive engineering of gene circuits[J]. Trends in Biotechnology, 2023, 41(6): 760-768. |
| 13 | BREMER H, DENNIS P P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates[J]. EcoSal Plus, 2008, 3(1): 10.1128/ecosal.5.2.3. |
| 14 | VIND J, SØRENSEN M A, RASMUSSEN M D, et al. Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. Expression from reporter genes does not always reflect functional mRNA levels[J]. Journal of Molecular Biology, 1993, 231(3): 678-688. |
| 15 | CHURCHWARD G, BREMER H, YOUNG R. Transcription in bacteria at different DNA concentrations[J]. Journal of Bacteriology, 1982, 150(2): 572-581. |
| 16 | STOEBEL D M, DEAN A M, DYKHUIZEN D E. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products[J]. Genetics, 2008, 178(3): 1653-1660. |
| 17 | CARBONELL-BALLESTERO M, GARCIA-RAMALLO E, MONTAÑEZ R, et al. Dealing with the genetic load in bacterial synthetic biology circuits: convergences with the Ohm's law[J]. Nucleic Acids Research, 2016, 44(1): 496-507. |
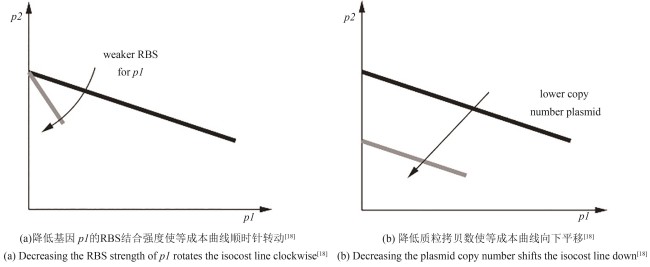
| 18 | GYORGY A, JIMÉNEZ J I, YAZBEK J, et al. Isocost lines describe the cellular economy of genetic circuits[J]. Biophysical Journal, 2015, 109(3): 639-646. |
| 19 | DEL VECCHIO D, QIAN Y L, MURRAY R M, et al. Future systems and control research in synthetic biology[J]. Annual Reviews in Control, 2018, 45: 5-17. |
| 20 | BASHOR C J, COLLINS J J. Insulating gene circuits from context by RNA processing[J]. Nature Biotechnology, 2012, 30(11): 1061-1062. |
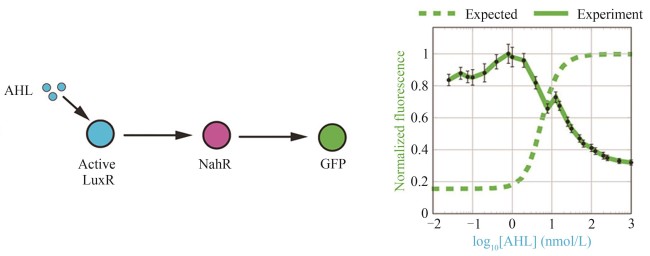
| 21 | QIAN Y L, HUANG H H, JIMÉNEZ J I, et al. Resource competition shapes the response of genetic circuits[J]. ACS Synthetic Biology, 2017, 6(7): 1263-1272. |
| 22 | CERONI F, ALGAR R, STAN G B, et al. Quantifying cellular capacity identifies gene expression designs with reduced burden[J]. Nature Methods, 2015, 12(5): 415-418. |
| 23 | JONES R D, QIAN Y L, SICILIANO V, et al. An endoribonuclease-based feedforward controller for decoupling resource-limited genetic modules in mammalian cells[J]. Nature Communications, 2020, 11(1): 5690. |
| 24 | DI BLASI R, PISANI M, TEDESCHI F, et al. Resource-aware construct design in mammalian cells[J]. Nature Communications, 2023, 14(1): 3576. |
| 25 | MORIYA T, YAMAOKA T, WAKAYAMA Y, et al. Comparison between effects of retroactivity and resource competition upon change in downstream reporter genes of synthetic genetic circuits[J]. Life, 2019, 9(1): 30. |
| 26 | CAMERON D E, COLLINS J J. Tunable protein degradation in bacteria[J]. Nature Biotechnology, 2014, 32(12): 1276-1281. |
| 27 | HERMSEN R, TANS S, WOLDE P R TEN. Transcriptional regulation by competing transcription factor modules[J]. PLoS Computational Biology, 2006, 2(12): e164. |
| 28 | DONG H J, NILSSON L, KURLAND C G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates[J]. Journal of Molecular Biology, 1996, 260(5): 649-663. |
| 29 | LOVE A M, NAIR N U. Specific codons control cellular resources and fitness[J]. Science Advances, 2024, 10(8): eadk3485. |
| 30 | COOKSON N A, MATHER W H, DANINO T, et al. Queueing up for enzymatic processing: correlated signaling through coupled degradation[J]. Molecular Systems Biology, 2011, 7: 561. |
| 31 | BUTZIN N C, HOCHENDONER P, OGLE C T, et al. Entrainment of a bacterial synthetic gene oscillator through proteolytic queueing[J]. ACS Synthetic Biology, 2017, 6(3): 455-462. |
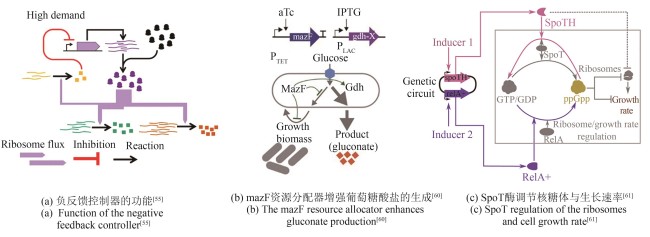
| 32 | PAULSSON J. Models of stochastic gene expression[J]. Physics of Life Reviews, 2005, 2(2): 157-175. |
| 33 | PAULSSON J. Summing up the noise in gene networks[J]. Nature, 2004, 427(6973): 415-418. |
| 34 | GOETZ H, STONE A, ZHANG R, et al. Double-edged role of resource competition in gene expression noise and control[J]. Advanced Genetics, 2022, 3(1): 2100050. |
| 35 | ALON U. Network motifs: theory and experimental approaches[J]. Nature Reviews Genetics, 2007, 8(6): 450-461. |
| 36 | SHEN-ORR S S, MILO R, MANGAN S, et al. Network motifs in the transcriptional regulation network of Escherichia coli [J]. Nature Genetics, 2002, 31(1): 64-68. |
| 37 | MILO R, SHEN-ORR S, ITZKOVITZ S, et al. Network motifs: simple building blocks of complex networks[J]. Science, 2002, 298(5594): 824-827. |
| 38 | WANG L, WALKER B L, IANNACCONE S, et al. Bistable switches control memory and plasticity in cellular differentiation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(16): 6638-6643. |
| 39 | VEENING J W, SMITS W K, KUIPERS O P. Bistability, epigenetics, and bet-hedging in bacteria[J]. Annual Review of Microbiology, 2008, 62: 193-210. |
| 40 | CHAKRABORTY P, GHOSH S. Emergent correlations in gene expression dynamics as footprints of resource competition[J]. The European Physical Journal E, 2021, 44(10): 131. |
| 41 | PARTCH C L, GREEN C B, TAKAHASHI J S. Molecular architecture of the mammalian circadian clock[J]. Trends in Cell Biology, 2014, 24(2): 90-99. |
| 42 | ITO H, MUTSUDA M, MURAYAMA Y, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus [J]. Proceedingsof the National Academy of Sciences of the United States of America, 2009, 106(33): 14168-14173. |
| 43 | ZHANG R, GOETZ H, MELENDEZ-ALVAREZ J, et al. Winner-takes-all resource competition redirects cascading cell fate transitions[J]. Nature Communications, 2021, 12(1): 853. |
| 44 | LIANG S T, XU Y C, DENNIS P, et al. mRNA composition and control of bacterial gene expression[J]. Journal of Bacteriology, 2000, 182(11): 3037-3044. |
| 45 | LIANG S T, BIPATNATH M, XU Y C, et al. Activities of constitutive promoters in Escherichia coli [J]. Journal of Molecular Biology, 1999, 292(1): 19-37. |
| 46 | KLUMPP S, HWA T. Growth-rate-dependent partitioning of RNA polymerases in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(51): 20245-20250. |
| 47 | KLUMPP S, ZHANG Z G, HWA T. Growth rate-dependent global effects on gene expression in bacteria[J]. Cell, 2009, 139(7): 1366-1375. |
| 48 | ALEXANDER W A, MOSS B, FUERST T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor[J]. Journal of Virology, 1992, 66(5): 2934-2942. |
| 49 | CHAMBERLIN M, MCGRATH J, WASKELL L. New RNA polymerase from Escherichia coli infected with bacteriophage T7[J]. Nature, 1970, 228(5268): 227-231. |
| 50 | STUDIER F W, MOFFATT B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes[J]. Journal of Molecular Biology, 1986, 189(1): 113-130. |
| 51 | KUSHWAHA M, SALIS H M. A portable expression resource for engineering cross-species genetic circuits and pathways[J]. Nature Communications, 2015, 6: 7832. |
| 52 | EL-SAMAD H, KURATA H, DOYLE J C, et al. Surviving heat shock: control strategies for robustness and performance[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(8): 2736-2741. |
| 53 | SEGALL-SHAPIRO T H, MEYER A J, ELLINGTON A D, et al. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase[J]. Molecular Systems Biology, 2014, 10(7): 742. |
| 54 | DARLINGTON A P S, BATES D G. Architectures for combined transcriptional and translational resource allocation controllers[J]. Cell Systems, 2020, 11(4): 382-392. e9. |
| 55 | DARLINGTON A P S, KIM J, JIMÉNEZ J I, et al. Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes[J]. Nature Communications, 2018, 9(1): 695. |
| 56 | ORELLE C, CARLSON E D, SZAL T, et al. Protein synthesis by ribosomes with tethered subunits[J]. Nature, 2015, 524(7563): 119-124. |
| 57 | ALEKSASHIN N A, SZAL T, D’AQUINO A E, et al. A fully orthogonal system for protein synthesis in bacterial cells[J]. Nature Communications, 2020, 11(1): 1858. |
| 58 | DE JONG H, GEISELMANN J, ROPERS D. Resource reallocation in bacteria by reengineering the gene expression machinery[J]. Trends in Microbiology, 2017, 25(6): 480-493. |
| 59 | DARLINGTON A P S, KIM J, JIMÉNEZ J I, et al. Engineering translational resource allocation controllers: mechanistic models, design guidelines, and potential biological implementations[J]. ACS Synthetic Biology, 2018, 7(11): 2485-2496. |
| 60 | VENTURELLI O S, TEI M, BAUER S, et al. Programming mRNA decay to modulate synthetic circuit resource allocation[J]. Nature Communications, 2017, 8: 15128. |
| 61 | BARAJAS C, HUANG H H, GIBSON J, et al. Feedforward growth rate control mitigates gene activation burden[J]. Nature Communications, 2022, 13(1): 7054. |
| 62 | ZHU M L, DAI X F. Growth suppression by altered (p)ppGpp levels results from non-optimal resource allocation in Escherichia coli [J]. Nucleic Acids Research, 2019, 47(9): 4684-4693. |
| 63 | BÜKE F, GRILLI J, COSENTINO LAGOMARSINO M, et al. ppGpp is a bacterial cell size regulator[J]. Current Biology, 2022, 32(4): 870-877. e5. |
| 64 | MU H Y, HAN F, WANG Q, et al. Recent functional insights into the magic role of (p)ppGpp in growth control[J]. Computational and Structural Biotechnology Journal, 2023, 21: 168-175. |
| 65 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor[J]. Microbial Cell Factories, 2019, 18(1): 35. |
| 66 | TSOI R, WU F L, ZHANG C, et al. Metabolic division of labor in microbial systems[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2526-2531. |
| 67 | OVÁDI J, SAKS V. On the origin of intracellular compartmentation and organized metabolic systems[J]. Molecular and Cellular Biochemistry, 2004, 256-257(1-2): 5-12. |
| 68 | MAMPEL J, BUESCHER J M, MEURER G, et al. Coping with complexity in metabolic engineering[J]. Trends in Biotechnology, 2013, 31(1): 52-60. |
| 69 | CHOWDHURY C, SINHA S, CHUN S, et al. Diverse bacterial microcompartment organelles[J]. Microbiology and Molecular Biology Reviews, 2014, 78(3): 438-468. |
| 70 | YEATES T O, JORDA J, BOBIK T A. The shells of BMC-type microcompartment organelles in bacteria[J]. Journal of Molecular Microbiology and Biotechnology, 2013, 23(4-5): 290-299. |
| 71 | KERFELD C A, HEINHORST S, CANNON G C. Bacterial microcompartments[J]. Annual Review of Microbiology, 2010, 64: 391-408. |
| 72 | THOMMES M, WANG T Y, ZHAO Q, et al. Designing metabolic division of labor in microbial communities[J]. mSystems, 2019, 4(2): e00263-18. |
| 73 | LINDEMANN S R. A piece of the pie: engineering microbiomes by exploiting division of labor in complex polysaccharide consumption[J]. Current Opinion in Chemical Engineering, 2020, 30: 96-102. |
| 74 | ALNAHHAS R N, WINKLE J J, HIRNING A J, et al. Spatiotemporal dynamics of synthetic microbial consortia in microfluidic devices[J]. ACS Synthetic Biology, 2019, 8(9): 2051-2058. |
| 75 | KIM H J, BOEDICKER J Q, CHOI J W, et al. Defined spatial structure stabilizes a synthetic multispecies bacterial community[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(47): 18188-18193. |
| 76 | XU P. Dynamics of microbial competition, commensalism, and cooperation and its implications for coculture and microbiome engineering[J]. Biotechnology and Bioengineering, 2021, 118(1): 199-209. |
| 77 | SHOPERA T, HE L, OYETUNDE T, et al. Decoupling resource-coupled gene expression in living cells[J]. ACS Synthetic Biology, 2017, 6(8): 1596-1604. |
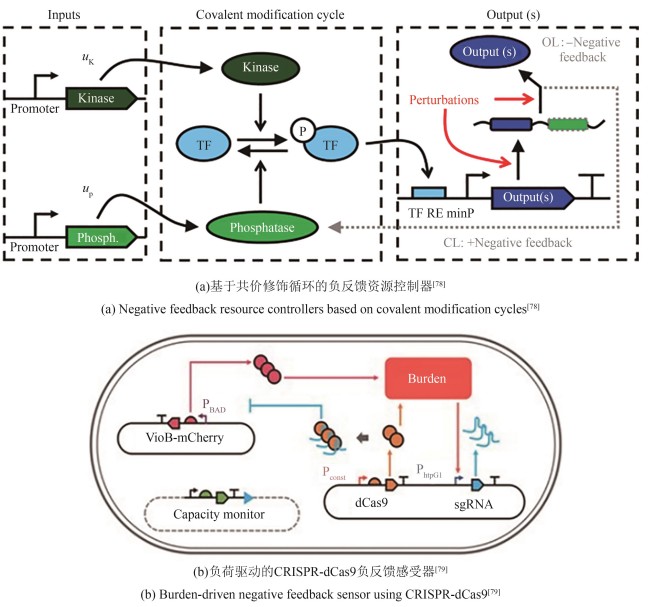
| 78 | JONES R D, QIAN Y L, ILIA K, et al. Robust and tunable signal processing in mammalian cells via engineered covalent modification cycles[J]. Nature Communications, 2022, 13(1): 1720. |
| 79 | CERONI F, BOO A, FURINI S, et al. Burden-driven feedback control of gene expression[J]. Nature Methods, 2018, 15(5): 387-393. |
| 80 | FREI T, CELLA F, TEDESCHI F, et al. Characterization and mitigation of gene expression burden in mammalian cells[J]. Nature Communications, 2020, 11(1): 4641. |
| 81 | STONE A, RIJAL S, ZHANG R, et al. Enhancing circuit stability under growth feedback with supplementary repressive regulation[J]. Nucleic Acids Research, 2024, 52(3): 1512-1521. |
| 82 | HUANG H H, QIAN Y L, DEL VECCHIO D. A quasi-integral controller for adaptation of genetic modules to variable ribosome demand[J]. Nature Communications, 2018, 9(1): 5415. |
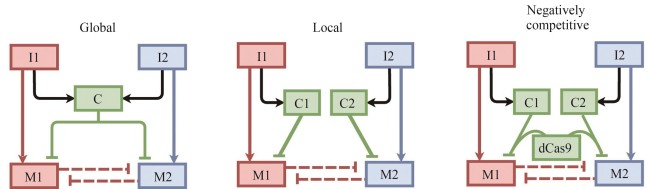
| 83 | STONE A, RYAN J, TANG X, et al. Negatively competitive incoherent feedforward loops mitigate winner-take-all resource competition[J]. ACS Synthetic Biology, 2022, 11(12): 3986-3995. |
| 84 | STONE A, ZHANG R, TIAN X J. Coupling shared and tunable negative competition against winner-take-all resource competition via CRISPRi moieties[C/OL]//2021 American Control Conference (ACC). IEEE, 2021: 1-6. (2021-07-28)[2024-11-01]. . |
| 85 | CHAKRAVARTY S, ZHANG R, TIAN X J. Noise reduction in resource-coupled multi-module gene circuits through antithetic feedback control[J]. bioRxiv, 2024: 2024. 05. 24. 595570. |
| 86 | CHAKRAVARTY S, GUTTAL R, ZHANG R, et al. Mitigating winner-take-all resource competition through antithetic control mechanism[J]. ACS Synthetic Biology, 2024, 13(12): 4050-4060. |
| 87 | RAI K, WANG Y D, O’CONNELL R W, et al. Using machine learning to enhance and accelerate synthetic biology[J]. Current Opinion in Biomedical Engineering, 2024, 31: 100553. |
| [1] | WU Ke, LUO Jiahao, LI Feiran. Applications of machine learning in the reconstruction and curation of genome-scale metabolic models [J]. Synthetic Biology Journal, 2025, 6(3): 566-584. |
| [2] | ZHANG Yiqing, LIU Gaowen. Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(3): 685-700. |
| [3] | YANG Ying, LI Xia, LIU Lizhong. Applications of synthetic biology to stem-cell-derived modeling of early embryonic development [J]. Synthetic Biology Journal, 2025, 6(3): 669-684. |
| [4] | HUANG Yi, SI Tong, LU Anjing. Standardization for biomanufacturing: global landscape, critical challenges, and pathways forward [J]. Synthetic Biology Journal, 2025, 6(3): 701-714. |
| [5] | SONG Chengzhi, LIN Yihan. AI-enabled directed evolution for protein engineering and optimization [J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [6] | ZHANG Mengyao, CAI Peng, ZHOU Yongjin. Synthetic biology drives the sustainable production of terpenoid fragrances and flavors [J]. Synthetic Biology Journal, 2025, 6(2): 334-356. |
| [7] | ZHANG Lu’ou, XU Li, HU Xiaoxu, YANG Ying. Synthetic biology ushers cosmetic industry into the “bio-cosmetics” era [J]. Synthetic Biology Journal, 2025, 6(2): 479-491. |
| [8] | YI Jinhang, TANG Yulin, LI Chunyu, WU Heyun, MA Qian, XIE Xixian. Applications and advances in the research of biosynthesis of amino acid derivatives as key ingredients in cosmetics [J]. Synthetic Biology Journal, 2025, 6(2): 254-289. |
| [9] | WEI Lingzhen, WANG Jia, SUN Xinxiao, YUAN Qipeng, SHEN Xiaolin. Biosynthesis of flavonoids and their applications in cosmetics [J]. Synthetic Biology Journal, 2025, 6(2): 373-390. |
| [10] | XIAO Sen, HU Litao, SHI Zhicheng, WANG Fayin, YU Siting, DU Guocheng, CHEN Jian, KANG Zhen. Research advances in biosynthesis of hyaluronic acid with controlled molecular weights [J]. Synthetic Biology Journal, 2025, 6(2): 445-460. |
| [11] | WANG Qian, GUO Shiting, XIN Bo, ZHONG Cheng, WANG Yu. Advances in biosynthesis of L-arginine using engineered microorganisms [J]. Synthetic Biology Journal, 2025, 6(2): 290-305. |
| [12] | ZUO Yimeng, ZHANG Jiaojiao, LIAN Jiazhang. Enabling technology for the biosynthesis of cosmetic raw materials with Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2025, 6(2): 233-253. |
| [13] | TANG Chuan′gen, WANG Jing, ZHANG Shuo, ZHANG Haoning, KANG Zhen. Advances in synthesis and mining strategies for functional peptides [J]. Synthetic Biology Journal, 2025, 6(2): 461-478. |
| [14] | GUO Tingting, HAN Xiangning, HUANG Xiting, ZHANG Tingting, KONG Jian. Advances in synthetic biology tools for lactic acid bacteria and their application in the development of skin beneficial products [J]. Synthetic Biology Journal, 2025, 6(2): 320-333. |
| [15] | ZHANG Ping, ZHANG Weijiao, XU Ruirui, LI Jianghua, CHEN Jian, KANG Zhen. Research advances on the biosynthesis of mycosporine-like amino acids [J]. Synthetic Biology Journal, 2025, 6(2): 306-319. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||