Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (3): 669-684.DOI: 10.12211/2096-8280.2025-013
• Invited Review • Previous Articles Next Articles
Applications of synthetic biology to stem-cell-derived modeling of early embryonic development
YANG Ying1, LI Xia1, LIU Lizhong1,2,3
- 1.Westlake Laboratory of Life Sciences and Biomedicine,Hangzhou 310024,Zhejiang,China
2.School of Life Sciences,Westlake University,Hangzhou 310030,Zhejiang,China
3.Institute of Biology,Westlake Institute for Advanced Study,Hangzhou 310024,Zhejiang,China
-
Received:2025-03-03Revised:2025-05-12Online:2025-06-27Published:2025-06-30 -
Contact:LIU Lizhong
合成生物学在干细胞早期胚胎发育模型中的应用
杨莹1, 李霞1, 刘立中1,2,3
- 1.西湖实验室(生命科学和生物医学浙江省实验室),浙江 杭州 310024
2.西湖大学生命科学学院,浙江 杭州 310030
3.浙江西湖高等研究院生物学研究所,浙江 杭州 310024
-
通讯作者:刘立中 -
作者简介:杨莹 (1996—),女,博士后。研究方向为神经系统发育及干细胞命运调控。 E-mail:yangying@westlake.edu.cn刘立中 (1985—),男,研究员,博士生导师。研究方向为早期胚胎发育及干细胞生物学,类胚胎模型的构建与应用,合成生物学等。 E-mail:liulizhong@westlake.edu.cn -
基金资助:浙江省“尖兵”“领雁”研发攻关计划(2024SSYS0036)
CLC Number:
Cite this article
YANG Ying, LI Xia, LIU Lizhong. Applications of synthetic biology to stem-cell-derived modeling of early embryonic development[J]. Synthetic Biology Journal, 2025, 6(3): 669-684.
杨莹, 李霞, 刘立中. 合成生物学在干细胞早期胚胎发育模型中的应用[J]. 合成生物学, 2025, 6(3): 669-684.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-013
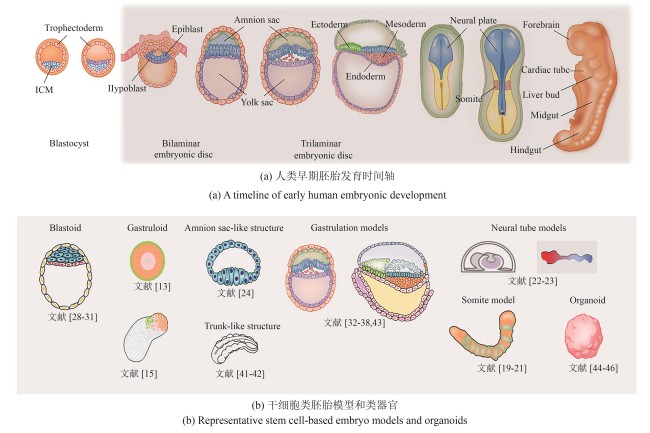
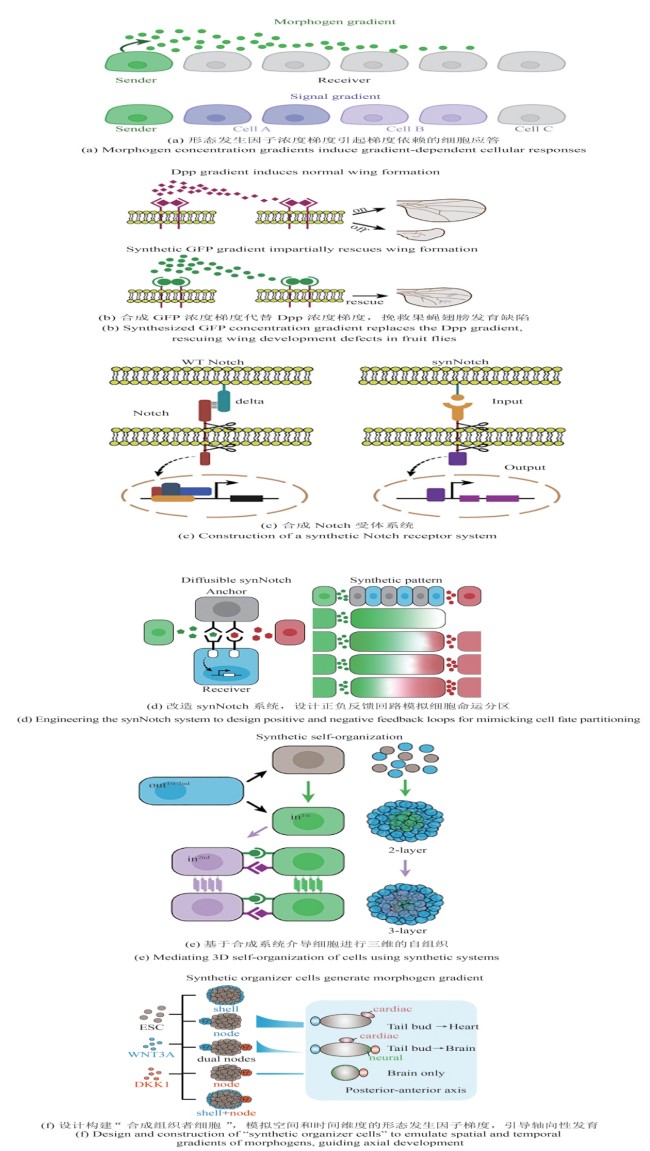
Fig.1 Early embryonic development models and organoids(Human embryonic development begins with the blastocyst implanting into the uterine wall, followed by the formation of the bilaminar disc composed of the epiblast and hypoblast. Gastrulation then transforms the disc into a trilaminar structure: ectoderm, mesoderm, and endoderm, laying the foundation for early organogenesis. Here are representative stem cell-based embryo models selected by the authors, which recapitulate key developmental stages ranging from pre-implantation and peri-implantation to gastrulation and early organogenesis. Due to space constraints, we regret that not all relevant studies could be included.)
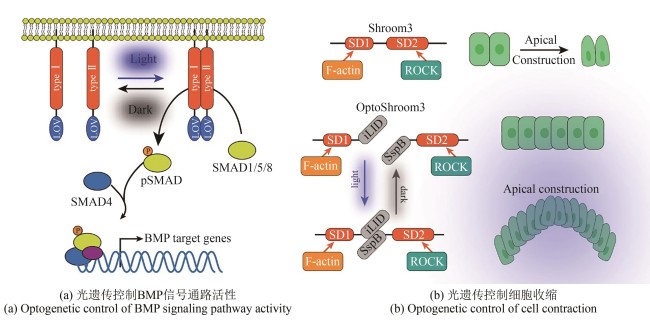
Fig. 3 Applications of optogenetic tools in developmental biology(Dimerization mediated by the light-oxygen-voltage (LOV) sensing domain regulates intracellular tyrosine kinase activity, leading to SMAD1/5 phosphorylation and their translocation into the nucleus to initiate downstream BMP target gene expression. Optogenetically activated Shroom3 drives apical constriction through ROCK recruitment at apical junctions.)
| 1 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403(6767): 335-338. |
| 2 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 3 | MARTÍNEZ-ARA G, STAPORNWONGKUL K S, EBISUYA M. Scaling up complexity in synthetic developmental biology[J]. Science, 2022, 378(6622): 864-868. |
| 4 | TRENTESAUX C, YAMADA T, KLEIN O D, et al. Harnessing synthetic biology to engineer organoids and tissues[J]. Cell Stem Cell, 2023, 30(1): 10-19. |
| 5 | SRIVATSAN S R, REGIER M C, BARKAN E, et al. Embryo-scale, single-cell spatial transcriptomics[J]. Science, 2021, 373(6550): 111-117. |
| 6 | PENG G D, CUI G Z, KE J C, et al. Using single-cell and spatial transcriptomes to understand stem cell lineage specification during early embryo development[J]. Annual Review of Genomics and Human Genetics, 2020, 21: 163-181. |
| 7 | GULATI G S, D’SILVA J P, LIU Y H, et al. Profiling cell identity and tissue architecture with single-cell and spatial transcriptomics[J]. Nature Reviews Molecular Cell Biology, 2025, 26(1): 11-31. |
| 8 | DU P, WU J. Hallmarks of totipotent and pluripotent stem cell states[J]. Cell Stem Cell, 2024, 31(3): 312-333. |
| 9 | SMITH A. Propagating pluripotency-the conundrum of self-renewal[J]. BioEssays, 2024, 46(12): 2400108. |
| 10 | ROSSANT J, TAM P P L. Opportunities and challenges with stem cell-based embryo models[J]. Stem Cell Reports, 2021, 16(5): 1031-1038. |
| 11 | MARTIN G R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells[J]. Proceedings of the National Academy of Sciences of the United States of America, 1981, 78(12): 7634-7638. |
| 12 | MARTIN G R, EVANS M J. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro [J]. Proceedings of the National Academy of Sciences of the United States of America, 1975, 72(4): 1441-1445. |
| 13 | WARMFLASH A, SORRE B, ETOC F, et al. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells[J]. Nature Methods, 2014, 11(8): 847-854. |
| 14 | VAN DEN BRINK S C, ALEMANY A, VAN BATENBURG V, et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids[J]. Nature, 2020, 582(7812): 405-409. |
| 15 | MORIS N, ANLAS K, VAN DEN BRINK S C, et al. An in vitro model of early anteroposterior organization during human development[J]. Nature, 2020, 582(7812): 410-415. |
| 16 | BECCARI L, MORIS N, GIRGIN M, et al. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids[J]. Nature, 2018, 562(7726): 272-276. |
| 17 | TURNER D A, GIRGIN M, ALONSO-CRISOSTOMO L, et al. Anteroposterior polarity and elongation in the absence of extra-embryonic tissues and of spatially localised signalling in gastruloids: mammalian embryonic organoids[J]. Development, 2017, 144(21): 3894-3906. |
| 18 | VAN DEN BRINK S C, BAILLIE-JOHNSON P, BALAYO T, et al. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells[J]. Development, 2014, 141(22): 4231-4242. |
| 19 | YAMANAKA Y, HAMIDI S, YOSHIOKA-KOBAYASHI K, et al. Reconstituting human somitogenesis in vitro [J]. Nature, 2023, 614(7948): 509-520. |
| 20 | SANAKI-MATSUMIYA M, MATSUDA M, GRITTI N, et al. Periodic formation of epithelial somites from human pluripotent stem cells[J]. Nature Communications, 2022, 13: 2325. |
| 21 | MIAO Y C, DJEFFAL Y, DE SIMONE A, et al. Reconstruction and deconstruction of human somitogenesis in vitro [J]. Nature, 2023, 614(7948): 500-508. |
| 22 | XUE X F, KIM Y S, PONCE-ARIAS A I, et al. A patterned human neural tube model using microfluidic gradients[J]. Nature, 2024, 628(8007): 391-399. |
| 23 | KARZBRUN E, KHANKHEL A H, MEGALE H C, et al. Human neural tube morphogenesis in vitro by geometric constraints[J]. Nature, 2021, 599(7884): 268-272. |
| 24 | ZHENG Y, XUE X F, SHAO Y, et al. Controlled modelling of human epiblast and amnion development using stem cells[J]. Nature, 2019, 573(7774): 421-425. |
| 25 | SHAO Y, TANIGUCHI K, TOWNSHEND R F, et al. A pluripotent stem cell-based model for post-implantation human amniotic sac development[J]. Nature Communications, 2017, 8: 208. |
| 26 | SHAO Y, TANIGUCHI K, GURDZIEL K, et al. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche[J]. Nature Materials, 2017, 16(4): 419-425. |
| 27 | RIVRON N C, FRIAS-ALDEGUER J, VRIJ E J, et al. Blastocyst-like structures generated solely from stem cells[J]. Nature, 2018, 557(7703): 106-111. |
| 28 | YU L Q, WEI Y L, DUAN J L, et al. Blastocyst-like structures generated from human pluripotent stem cells[J]. Nature, 2021, 591(7851): 620-626. |
| 29 | LIU X D, TAN J P, SCHRÖDER J, et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids[J]. Nature, 2021, 591(7851): 627-632. |
| 30 | YANAGIDA A, SPINDLOW D, NICHOLS J, et al. Naive stem cell blastocyst model captures human embryo lineage segregation[J]. Cell Stem Cell, 2021, 28(6): 1016-1022.e4. |
| 31 | KAGAWA H, JAVALI A, KHOEI H H, et al. Human blastoids model blastocyst development and implantation[J]. Nature, 2022, 601(7894): 600-605. |
| 32 | OKUBO T, RIVRON N, KABATA M, et al. Hypoblast from human pluripotent stem cells regulates epiblast development[J]. Nature, 2024, 626(7998): 357-366. |
| 33 | PEDROZA M, GASSALOGLU S I, DIAS N, et al. Self-patterning of human stem cells into post-implantation lineages[J]. Nature, 2023, 622(7983): 574-583. |
| 34 | LIU L Z, OURA S, MARKHAM Z, et al. Modeling post-implantation stages of human development into early organogenesis with stem-cell-derived peri-gastruloids[J]. Cell, 2023, 186(18): 3776-3792.e16. |
| 35 | AI Z Y, NIU B H, YIN Y, et al. Dissecting peri-implantation development using cultured human embryos and embryo-like assembloids[J]. Cell Research, 2023, 33(9): 661-678. |
| 36 | HISLOP J, SONG Q, KESHAVARZ F K, et al. Modelling post-implantation human development to yolk sac blood emergence[J]. Nature, 2024, 626(7998): 367-376. |
| 37 | WEATHERBEE B A T, GANTNER C W, IWAMOTO-STOHL L K, et al. Pluripotent stem cell-derived model of the post-implantation human embryo[J]. Nature, 2023, 622(7983): 584-593. |
| 38 | OLDAK B, WILDSCHUTZ E, BONDARENKO V, et al. Complete human day 14 post-implantation embryo models from naive ES cells[J]. Nature, 2023, 622(7983): 562-573. |
| 39 | 胡博文, 陈家斌, 刘晓东. 人类早期胚胎发育体外模型研究进展[J]. 合成生物学, 2024, 5(4): 719-733. |
| HU B W, TAN J P, LIU X D. Advances in the development of human embryo models[J]. Synthetic Biology Journal, 2024, 5(4): 719-733. | |
| 40 | 韩宜钊, 郭佳, 邵玥. 干细胞模拟发育: 细胞元件、 胚胎模型与工程方法[J]. 合成生物学, 2024, 5(4): 734-753. |
| HAN Y Z, GUO J, SHAO Y. Stem cell-based synthetic development: cellular components, embryonic models, and engineering approaches[J]. Synthetic Biology Journal, 2024, 5(4): 734-753. | |
| 41 | GRIBAUDO S, ROBERT R, VAN SAMBEEK B, et al. Self-organizing models of human trunk organogenesis recapitulate spinal cord and spine co-morphogenesis[J]. Nature Biotechnology, 2024, 42(8): 1243-1253. |
| 42 | HAMAZAKI N, YANG W, KUBO C A, et al. Retinoic acid induces human gastruloids with posterior embryo-like structures[J]. Nature Cell Biology, 2024, 26(10): 1790-1803. |
| 43 | YUAN G G, WANG J C, LIU Z D. Establishment of a novel non-integrated human pluripotent stem cell-based gastruloid model[EB/OL]. bioRxiv, 2023: 2023.06.28.546720. (2023-06-28)[2025-02-01]. . |
| 44 | LANCASTER M A, RENNER M, MARTIN C A, et al. Cerebral organoids model human brain development and microcephaly[J]. Nature, 2013, 501(7467): 373-379. |
| 45 | TAKASATO M, ER P X, CHIU H S, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis[J]. Nature, 2015, 526(7574): 564-568. |
| 46 | DRAKHLIS L, BISWANATH S, FARR C M, et al. Human heart-forming organoids recapitulate early heart and foregut development[J]. Nature Biotechnology, 2021, 39(6): 737-746. |
| 47 | CLEVERS H. Modeling development and disease with organoids[J]. Cell, 2016, 165(7): 1586-1597. |
| 48 | GEURTS M H, CLEVERS H. CRISPR engineering in organoids for gene repair and disease modelling[J]. Nature Reviews Bioengineering, 2023, 1(1): 32-45. |
| 49 | HOFER M, LUTOLF M P. Engineering organoids[J]. Nature Reviews Materials, 2021, 6(5): 402-420. |
| 50 | SCHUTGENS F, CLEVERS H. Human organoids: tools for understanding biology and treating diseases[J]. Annual Review of Pathology, 2020, 15: 211-234. |
| 51 | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| CHEN Z L, XIANG Y F. Integrated development of organoid technology and synthetic biology[J]. Synthetic Biology Journal, 2024, 5(4): 795-812. | |
| 52 | 洪源, 刘妍. 脑类器官在再生医学中的研究进展[J]. 合成生物学, 2024, 5(4): 754-769. |
| HONG Y, LIU Y. Research progress of brain organoids in regenerative medicine[J]. Synthetic Biology Journal, 2024, 5(4): 754-769. | |
| 53 | 胡可儿, 王汉奇, 黄儒麒, 等. 整合设计策略下的工程化类器官与类器官芯片技术[J]. 合成生物学, 2024, 5(4): 883-897. |
| HU K E, WANG H Q, HUANG R Q, et al. Integrated design strategies for engineered organoids and organ-on-a-chip technologies[J]. Synthetic Biology Journal, 2024, 5(4): 883-897. | |
| 54 | LANDER A D. Morpheus unbound: reimagining the morphogen gradient[J]. Cell, 2007, 128(2): 245-256. |
| 55 | LANDER A D. Pattern, growth, and control[J]. Cell, 2011, 144(6): 955-969. |
| 56 | LANDER A D, NIE Q, WAN F Y M. Do morphogen gradients arise by diffusion?[J]. Developmental Cell, 2002, 2(6): 785-796. |
| 57 | MÜLLER P, ROGERS K W, YU S R, et al. Morphogen transport[J]. Development, 2013, 140(8): 1621-1638. |
| 58 | ROGERS K W, SCHIER A F. Morphogen gradients: from generation to interpretation[J]. Annual Review of Cell and Developmental Biology, 2011, 27: 377-407. |
| 59 | STAPORNWONGKUL K S, DE GENNES M, COCCONI L, et al. Patterning and growth control in vivo by an engineered GFP gradient[J]. Science, 2020, 370(6514): 321-327. |
| 60 | ZHOU B H, LIN W L, LONG Y L, et al. Notch signaling pathway: architecture, disease, and therapeutics[J]. Signal Transduction and Targeted Therapy, 2022, 7: 95. |
| 61 | MORSUT L, ROYBAL K T, XIONG X, et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors[J]. Cell, 2016, 164(4): 780-791. |
| 62 | TODA S, MCKEITHAN W L, HAKKINEN T J, et al. Engineering synthetic morphogen systems that can program multicellular patterning[J]. Science, 2020, 370(6514): 327-331. |
| 63 | STEINBERG M S. Differential adhesion in morphogenesis: a modern view[J]. Current Opinion in Genetics & Development, 2007, 17(4): 281-286. |
| 64 | TODA S, BLAUCH L R, TANG S K Y, et al. Programming self-organizing multicellular structures with synthetic cell-cell signaling[J]. Science, 2018, 361(6398): 156-162. |
| 65 | YAMADA T, TRENTESAUX C, BRUNGER J M, et al. Synthetic organizer cells guide development via spatial and biochemical instructions[J]. Cell, 2025, 188(3): 778-795.e18. |
| 66 | NIAKAN K K, EGGAN K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse[J]. Developmental Biology, 2013, 375(1): 54-64. |
| 67 | FOGARTY N M E, MCCARTHY A, SNIJDERS K E, et al. Genome editing reveals a role for OCT4 in human embryogenesis[J]. Nature, 2017, 550(7674): 67-73. |
| 68 | ADIKUSUMA F, PILTZ S, CORBETT M A, et al. Large deletions induced by Cas9 cleavage[J]. Nature, 2018, 560(7717): E8-E9. |
| 69 | KOSICKI M, TOMBERG K, BRADLEY A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements[J]. Nature Biotechnology, 2018, 36(8): 765-771. |
| 70 | KORABLEV A, LUKYANCHIKOVA V, SEROVA I, et al. On-target CRISPR/Cas9 activity can cause undesigned large deletion in mouse zygotes[J]. International Journal of Molecular Sciences, 2020, 21(10): 3604. |
| 71 | CHEN S M, YAO Y F, ZHANG Y C, et al. CRISPR system: discovery, development and off-target detection[J]. Cellular Signalling, 2020, 70: 109577. |
| 72 | JOUNG J, MA S, TAY T, et al. A transcription factor atlas of directed differentiation[J]. Cell, 2023, 186(1): 209-229.e26. |
| 73 | APPLETON E, TAO J H, FONSECA G, et al. Machine-guided cell-fate engineering[EB/OL]. bioRxiv, 2023: 2022.10.14.512279. (2023-01-12)[2025-02-01]. . |
| 74 | CHAVEZ A, SCHEIMAN J, VORA S, et al. Highly efficient Cas9-mediated transcriptional programming[J]. Nature Methods, 2015, 12(4): 326-328. |
| 75 | SOZEN B, AMADEI G, COX A, et al. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures[J]. Nature Cell Biology, 2018, 20(8): 979-989. |
| 76 | HARRISON S E, SOZEN B, CHRISTODOULOU N, et al. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro [J]. Science, 2017, 356(6334): eaal1810. |
| 77 | AMADEI G, LAU K Y C, DE JONGHE J, et al. Inducible stem-cell-derived embryos capture mouse morphogenetic events in vitro [J]. Developmental Cell, 2021, 56(3): 366-382.e9. |
| 78 | LAU K Y C, RUBINSTEIN H, GANTNER C W, et al. Mouse embryo model derived exclusively from embryonic stem cells undergoes neurulation and heart development[J]. Cell Stem Cell, 2022, 29(10): 1445-1458.e8. |
| 79 | MCNAMARA H M, SOLLEY S C, ADAMSON B, et al. Recording morphogen signals reveals mechanisms underlying gastruloid symmetry breaking[J]. Nature Cell Biology, 2024, 26(11): 1832-1844. |
| 80 | BOYDEN E S, ZHANG F, BAMBERG E, et al. Millisecond-timescale, genetically targeted optical control of neural activity[J]. Nature Neuroscience, 2005, 8(9): 1263-1268. |
| 81 | FENNO L, YIZHAR O, DEISSEROTH K. The development and application of optogenetics[J]. Annual Review of Neuroscience, 2011, 34: 389-412. |
| 82 | LUO K, LODISH H F. Signaling by chimeric erythropoietin-TGF-beta receptors: homodimerization of the cytoplasmic domain of the type Ⅰ TGF-beta receptor and heterodimerization with the type Ⅱ receptor are both required for intracellular signal transduction[J]. The EMBO Journal, 1996, 15(17): 4485-4496. |
| 83 | SAKO K, PRADHAN S J, BARONE V, et al. Optogenetic control of nodal signaling reveals a temporal pattern of nodal signaling regulating cell fate specification during gastrulation[J]. Cell Reports, 2016, 16(3): 866-877. |
| 84 | HUMPHREYS P A, WOODS S, SMITH C A, et al. Optogenetic control of the BMP signaling pathway[J]. ACS Synthetic Biology, 2020, 9(11): 3067-3078. |
| 85 | SAWYER J M, HARRELL J R, SHEMER G, et al. Apical constriction: a cell shape change that can drive morphogenesis[J]. Developmental Biology, 2010, 341(1): 5-19. |
| 86 | MARTIN A C, GOLDSTEIN B. Apical constriction: themes and variations on a cellular mechanism driving morphogenesis[J]. Development (Cambridge, England), 2014, 141(10): 1987-1998. |
| 87 | BAUMSCHLAGER A, KHAMMASH M. Synthetic biological approaches for optogenetics and tools for transcriptional light-control in bacteria[J]. Advanced Biology, 2021, 5(5): e2000256. |
| 88 | ZHANG Z J, DENANS N, LIU Y F, et al. Optogenetic manipulation of cellular communication using engineered myosin motors[J]. Nature Cell Biology, 2021, 23(2): 198-208. |
| 89 | HARTMANN J, KRUEGER D, DE RENZIS S. Using optogenetics to tackle systems-level questions of multicellular morphogenesis[J]. Current Opinion in Cell Biology, 2020, 66: 19-27. |
| 90 | MUMFORD T R, ROTH L, BUGAJ L J. Reverse and forward engineering multicellular structures with optogenetics[J]. Current Opinion in Biomedical Engineering, 2020, 16: 61-71. |
| 91 | MARTÍNEZ-ARA G, TABERNER N, TAKAYAMA M, et al. Optogenetic control of apical constriction induces synthetic morphogenesis in mammalian tissues[J]. Nature Communications, 2022, 13: 5400. |
| 92 | OATES A C, GORFINKIEL N, GONZÁLEZ-GAITÁN M, et al. Quantitative approaches in developmental biology[J]. Nature Reviews Genetics, 2009, 10(8): 517-530. |
| 93 | 崔金明, 王力为, 常志广, 等. 合成生物学的医学应用研究进展[J]. 中国科学院院刊, 2018, 33(11): 1218-1227. |
| CUI J M, WANG L W, CHANG Z G, et al. Progress of synthetic biology research in medical applications[J]. Bulletin of Chinese Academy of Sciences, 2018, 33(11): 1218-1227. | |
| 94 | ZHU J W, CHU P, FU X F. Unbalanced response to growth variations reshapes the cell fate decision landscape[J]. Nature Chemical Biology, 2023, 19(9): 1097-1104. |
| 95 | WOLPERT L. Positional information and the spatial pattern of cellular differentiation[J]. Journal of Theoretical Biology, 1969, 25(1): 1-47. |
| 96 | WOLPERT L. Positional information and pattern formation[J]. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 1981, 295(1078): 441-450. |
| 97 | TURING A M. The chemical basis of morphogenesis[J]. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 1952, 237(641): 37-72. |
| 98 | NÁTR L. Murray, J.D.: mathematical biology. Ⅱ: spatial models and biomedical applications. 3rd ed[J]. Photosynthetica, 2003, 41(1): 42. |
| 99 | BALL P. Forging patterns and making waves from biology to geology: a commentary on Turing (1952) ‘The chemical basis of morphogenesis’[J]. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 2015, 370(1666): 20140218. |
| 100 | ERBAN R, CHAPMAN S J. Stochastic modelling of reaction-diffusion processes[M/OL]//Cambridge texts in applied mathematics. Cambridge: Cambridge University Press, 2020[2025-02-01]. . |
| 101 | ERBAN R, WINKELMANN S. Multi-grid reaction-diffusion master equation: applications to morphogen gradient modelling[J]. Bulletin of Mathematical Biology, 2024, 87(1): 6. |
| 102 | GORDON P V, SAMPLE C, BEREZHKOVSKII A M, et al. Local kinetics of morphogen gradients[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(15): 6157-6162. |
| 103 | GLIMM T, ZHANG J Y, SHEN Y Q, et al. Reaction-diffusion systems and external morphogen gradients: the two-dimensional case, with an application to skeletal pattern formation[J]. Bulletin of Mathematical Biology, 2012, 74(3): 666-687. |
| 104 | YUSTE S B, ABAD E, LINDENBERG K. Reaction-subdiffusion model of morphogen gradient formation[J]. Physical Review E, Statistical, Nonlinear, and Soft Matter Physics, 2010, 82(6 Pt 1): 061123. |
| 105 | DEWAR M A, KADIRKAMANATHAN V, OPPER M, et al. Parameter estimation and inference for stochastic reaction-diffusion systems: application to morphogenesis in D. Melanogaster [J]. BMC Systems Biology, 2010, 4: 21. |
| 106 | CHEN C, LIAO Y X, ZHU M, et al. Dual-nuclease single-cell lineage tracing by Cas9 and Cas12a[J]. Cell Reports, 2025, 44(1): 115105. |
| 107 | LI K R, YU P L, ZHENG Q Q, et al. Spatiotemporal and genetic cell lineage tracing of endodermal organogenesis at single-cell resolution[J]. Cell, 2025, 188(3): 796-813.e24. |
| 108 | PIJUAN-SALA B, GRIFFITHS J A, GUIBENTIF C, et al. A single-cell molecular map of mouse gastrulation and early organogenesis[J]. Nature, 2019, 566(7745): 490-495. |
| 109 | XIAO Z Y, CUI L N, YUAN Y, et al. 3D reconstruction of a gastrulating human embryo[J]. Cell, 2024, 187(11): 2855-2874.e19. |
| 110 | CUI L N, LIN S R, YANG X L, et al. Spatial transcriptomic characterization of a Carnegie stage 7 human embryo[J]. Nature Cell Biology, 2025, 27(2): 360-369. |
| 111 | TYSER R C V, MAHAMMADOV E, NAKANOH S, et al. Single-cell transcriptomic characterization of a gastrulating human embryo[J]. Nature, 2021, 600(7888): 285-289. |
| 112 | BERGMANN S, PENFOLD C A, SLATERY E, et al. Spatial profiling of early primate gastrulation in utero [J]. Nature, 2022, 609(7925): 136-143. |
| 113 | ZHAI J L, GUO J, WAN H F, et al. Primate gastrulation and early organogenesis at single-cell resolution[J]. Nature, 2022, 612(7941): 732-738. |
| 114 | PAN J X, LI Y J, LIN Z L, et al., Spatiotemporal transcriptome atlas of human embryos after gastrulation[EB/OL]. bioRxiv, 2023: 2023.04.22.537909. (2023-04-22)[2025-02-01]. . |
| 115 | XU Y C, ZHANG T J, ZHOU Q, et al. A single-cell transcriptome atlas profiles early organogenesis in human embryos[J]. Nature Cell Biology, 2023, 25(4): 604-615. |
| 116 | ZENG B, LIU Z Y, LU Y F, et al. The single-cell and spatial transcriptional landscape of human gastrulation and early brain development[J]. Cell Stem Cell, 2023, 30(6): 851-866.e7. |
| 117 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 118 | SIMONS K T, BONNEAU R, RUCZINSKI I, et al. Ab initio protein structure prediction of CASP Ⅲ targets using ROSETTA[J]. Proteins, 1999, : 171-176. |
| 119 | EDMAN N I, PHAL A, REDLER R L, et al. Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies[J]. Cell, 2024, 187(14): 3726-3740.e43. |
| 120 | PIRANER D I, ABEDI M H, DURAN GONZALEZ M J, et al. Engineered receptors for soluble cellular communication and disease sensing[J]. Nature, 2025, 638(8051): 805-813. |
| [1] | HU Bowen, TAN Jiaping, LIU Xiaodong. Advances in the development of human embryo models [J]. Synthetic Biology Journal, 2024, 5(4): 719-733. |
| [2] | AI Zongyong, ZHANG Chengting, NIU Baohua, YIN Yu, YANG Jie, LI Tianqing. Early human embryo development and stem cells [J]. Synthetic Biology Journal, 2024, 5(4): 700-718. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||