Synthetic Biology Journal ›› 2024, Vol. 5 ›› Issue (6): 1300-1318.DOI: 10.12211/2096-8280.2024-013
• Invited Review • Previous Articles Next Articles
Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds
ZHAO Liang1,2, LI Zhenshuai1,3, FU Liping1,2, LYU Ming1,2, WANG Shi’an1,4, ZHANG Quan5, LIU Licheng3, LI Fuli1, LIU Ziyong1,2
- 1.CAS Key Laboratory of Biofuel,Qingdao C1 refinery Research Engineering Center,Qingdao Institute of Bioenergy and Bioprocess Technology,Chinese Academy of Sciences,Qingdao 266101,Shandong,China
2.Shandong Energy Institute,Qingdao 266101,Shandong,China
3.Ocean University of China,Qingdao 266100,Shandong,China
4.Qingdao New Energy Shandong Laboratory,Qingdao 266101,Shandong,China
5.Sinopec Dalian Research Institute of Petroleum and Petrochemicals,Dalian 116045,Liaoning,China
-
Received:2024-02-04Revised:2024-05-08Online:2025-01-10Published:2024-12-31 -
Contact:LIU Ziyong
生物转化一碳化合物原料产油脂与单细胞蛋白研究进展
赵亮1,2, 李振帅1,3, 付丽平1,2, 吕明1,2, 王士安1,4, 张全5, 刘立成3, 李福利1, 刘自勇1,2
- 1.中国科学院青岛生物能源与过程研究所,青岛市碳一炼制工程研究中心,中国科学院生物燃料重点实验室,山东 青岛 266101
2.山东能源研究院,山东 青岛 266101
3.中国海洋大学,山东 青岛 266100
4.青岛新能源山东省实验室,山东 青岛 266101
5.中国石油化工股份有限公司大连研究院,辽宁 大连 116045
-
通讯作者:刘自勇 -
作者简介:赵亮 (1996—),男,硕士,助理工程师。研究方向为解脂耶氏酵母油脂与蛋白发酵工艺优化。E-mail:zhaoliang@qibebt.ac.cn刘自勇 (1983—),男,副研究员,硕士生导师。研究方向为厌氧梭菌高效转化木质纤维素和合成气生产生物乙醇、丁醇和长链油脂等。E-mail:liuzy@qibebt.ac.cn -
基金资助:国家重点研发计划(2023YFA0914400);国家自然科学基金面上项目(32370039);中国科学院战略性先导科技专项(C类)(XDC0110302)
CLC Number:
Cite this article
ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds[J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318.
赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2024-013
| 微生物类型 | 菌种 | 底物 | 代谢途径 | 代谢改造 | 产量/产率 | 产物 | 参考文献 | |
|---|---|---|---|---|---|---|---|---|
| 天然甲醇酵母 | P. pastoris | 甲醇 | XuMP途径 | — | 蛋白80.6 g/L | 脂肪酸23.4 g/L | 脂肪酸,单细胞蛋白 | [ |
| O. polymorpha | 甲醇 | — | 生物量26.6 g/L | 脂肪酸15.9 g/L | [ | |||
| C. boidinii | 甲醇 | — | 3.4 g/L | 单细胞蛋白 | [ | |||
| 天然甲醇细菌 | M. extorquens | 甲醇 | RuMP途径,丝氨酸循环 | — | 干重561 g/L | 单细胞蛋白 | [ | |
| B. methanolicus | 甲醇 | — | 干重30~144 g/L | 单细胞蛋白 | [ | |||
| 非天然甲醇酵母 | S. cerevisiae | 甲醇 | XuMP途径 | Mdh与XuMP途径共表达 | — | 单细胞蛋白 | [ | |
| Y. lipolytica | 甲醇 | RuMP、XuMP途径 | 表达杂合RuMP和XuMP途径基因,敲除内源甲醛脱氢酶 | — | 单细胞蛋白 | [ | ||
| 大肠杆菌 | E. coli | 甲醇、甲酸、CO2 | 还原性甘氨酸途径、CBB循环 | 利用还原性甘氨酸途径重新设计了中心碳代谢 | (2.8 ± 0.8) g干重/mol 甲酸 | 单细胞蛋白 | [ | |
| 天然甲酸利用微生物 | P. communis | 甲酸 | 丝氨酸循环、四氢叶酸循环、糖酵解途径、TCA循环 | — | 1.7 g/L | 单细胞蛋白 | [ | |
| 需钠弧菌 | V. natriegens | 甲酸 | 丝氨酸循环、TCA循环 | 重新连接丝氨酸循环和TCA循环 | — | 单细胞蛋白 | [ | |
| 产乙酸菌 | Clostridium autoethanogenum | CO、CO2 | Wood-Ljungdahl途径 | — | 1 × 108 t/a | 单细胞蛋白 | [ | |
| Clostridium ljungdahlii | CO、CO2 | Wood-Ljungdahl途径 | — | 2 g/L | 单细胞蛋白 | [ | ||
| 氢氧化细菌 | Hydrogenophaga | CO、CO2 | 反向三羧酸循环、CBB循环、Wood-Ljungdahl途径 | — | 0.9~1.7 g/L | 单细胞蛋白 | [ | |
| Xanthobacter | CO、CO2 | — | 单细胞蛋白 | |||||
| Aquamicrobium | CO、CO2 | — | 单细胞蛋白 | |||||
| Defluviimonas | CO、CO2 | — | 单细胞蛋白 | |||||
| 甲烷氧化菌 | Proteobacteria-γ亚型 | CH4 | RuMP途径 | — | 4 kg/(m3·h) | 单细胞蛋白 | [ | |
| Proteobacteria-α亚型 | CH4 | 丝氨酸循环 | — | — | 单细胞蛋白 | |||
| Verrucomicrobia | CH4 | CBB循环 | — | — | 单细胞蛋白 | [ | ||
Table 1 Microorganisms capable of utilizing one-carbon compounds to synthesize fatty acids and single-cell proteins
| 微生物类型 | 菌种 | 底物 | 代谢途径 | 代谢改造 | 产量/产率 | 产物 | 参考文献 | |
|---|---|---|---|---|---|---|---|---|
| 天然甲醇酵母 | P. pastoris | 甲醇 | XuMP途径 | — | 蛋白80.6 g/L | 脂肪酸23.4 g/L | 脂肪酸,单细胞蛋白 | [ |
| O. polymorpha | 甲醇 | — | 生物量26.6 g/L | 脂肪酸15.9 g/L | [ | |||
| C. boidinii | 甲醇 | — | 3.4 g/L | 单细胞蛋白 | [ | |||
| 天然甲醇细菌 | M. extorquens | 甲醇 | RuMP途径,丝氨酸循环 | — | 干重561 g/L | 单细胞蛋白 | [ | |
| B. methanolicus | 甲醇 | — | 干重30~144 g/L | 单细胞蛋白 | [ | |||
| 非天然甲醇酵母 | S. cerevisiae | 甲醇 | XuMP途径 | Mdh与XuMP途径共表达 | — | 单细胞蛋白 | [ | |
| Y. lipolytica | 甲醇 | RuMP、XuMP途径 | 表达杂合RuMP和XuMP途径基因,敲除内源甲醛脱氢酶 | — | 单细胞蛋白 | [ | ||
| 大肠杆菌 | E. coli | 甲醇、甲酸、CO2 | 还原性甘氨酸途径、CBB循环 | 利用还原性甘氨酸途径重新设计了中心碳代谢 | (2.8 ± 0.8) g干重/mol 甲酸 | 单细胞蛋白 | [ | |
| 天然甲酸利用微生物 | P. communis | 甲酸 | 丝氨酸循环、四氢叶酸循环、糖酵解途径、TCA循环 | — | 1.7 g/L | 单细胞蛋白 | [ | |
| 需钠弧菌 | V. natriegens | 甲酸 | 丝氨酸循环、TCA循环 | 重新连接丝氨酸循环和TCA循环 | — | 单细胞蛋白 | [ | |
| 产乙酸菌 | Clostridium autoethanogenum | CO、CO2 | Wood-Ljungdahl途径 | — | 1 × 108 t/a | 单细胞蛋白 | [ | |
| Clostridium ljungdahlii | CO、CO2 | Wood-Ljungdahl途径 | — | 2 g/L | 单细胞蛋白 | [ | ||
| 氢氧化细菌 | Hydrogenophaga | CO、CO2 | 反向三羧酸循环、CBB循环、Wood-Ljungdahl途径 | — | 0.9~1.7 g/L | 单细胞蛋白 | [ | |
| Xanthobacter | CO、CO2 | — | 单细胞蛋白 | |||||
| Aquamicrobium | CO、CO2 | — | 单细胞蛋白 | |||||
| Defluviimonas | CO、CO2 | — | 单细胞蛋白 | |||||
| 甲烷氧化菌 | Proteobacteria-γ亚型 | CH4 | RuMP途径 | — | 4 kg/(m3·h) | 单细胞蛋白 | [ | |
| Proteobacteria-α亚型 | CH4 | 丝氨酸循环 | — | — | 单细胞蛋白 | |||
| Verrucomicrobia | CH4 | CBB循环 | — | — | 单细胞蛋白 | [ | ||
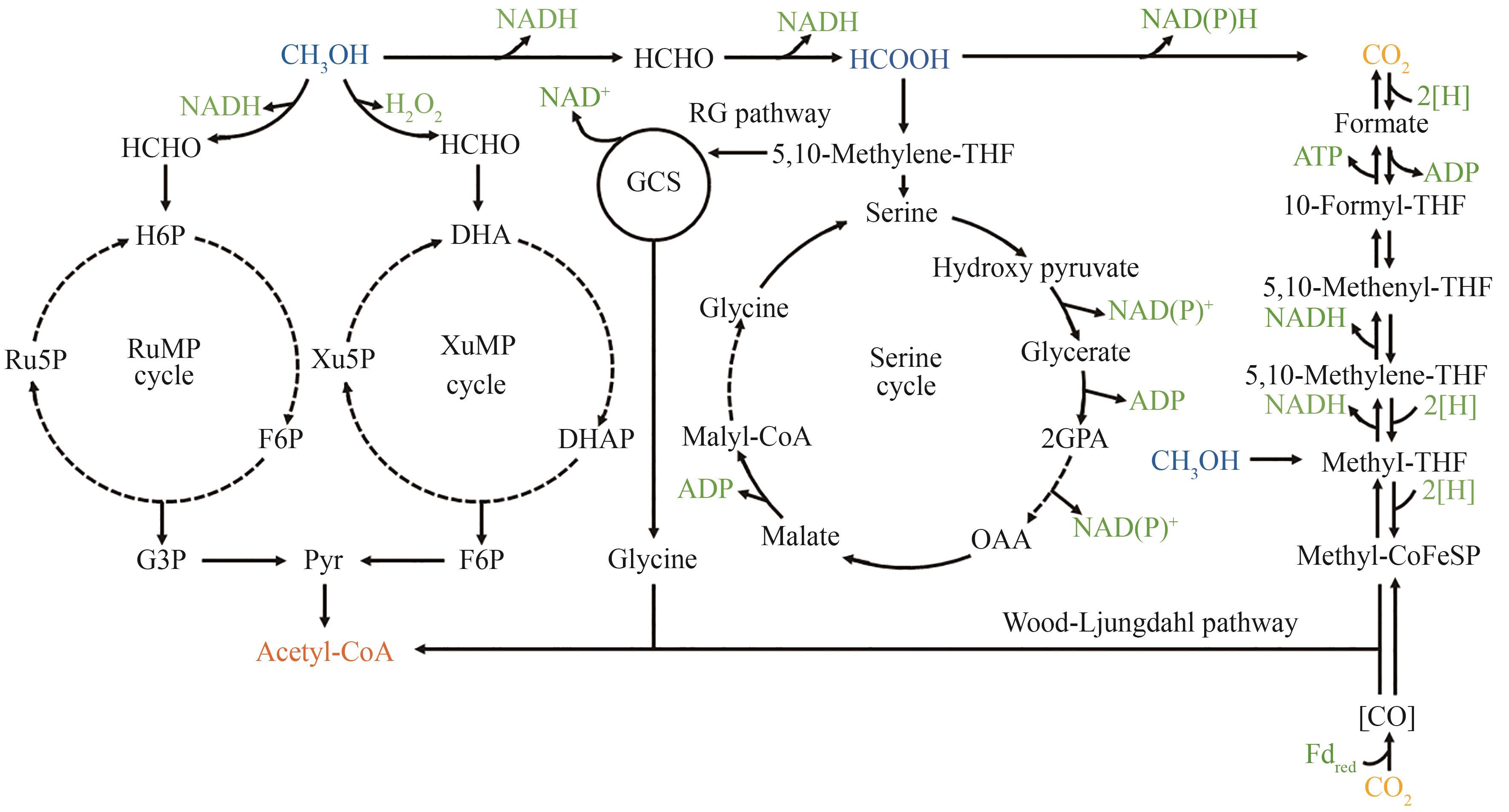
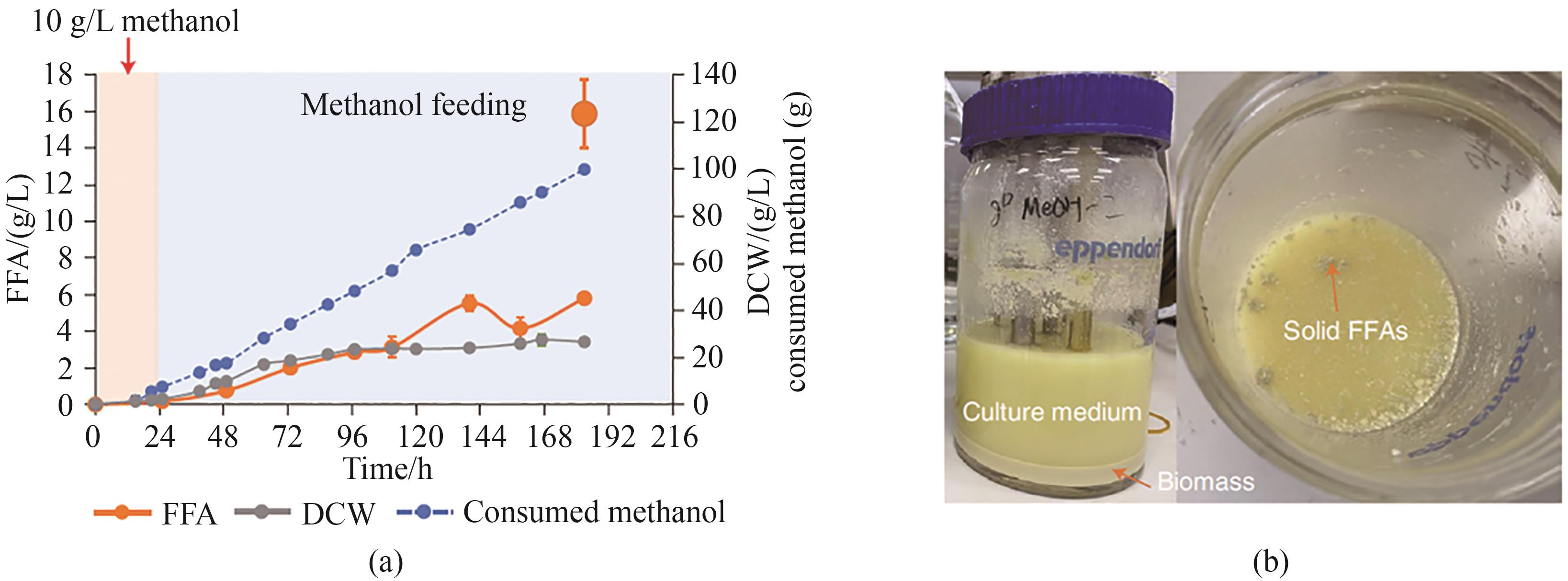
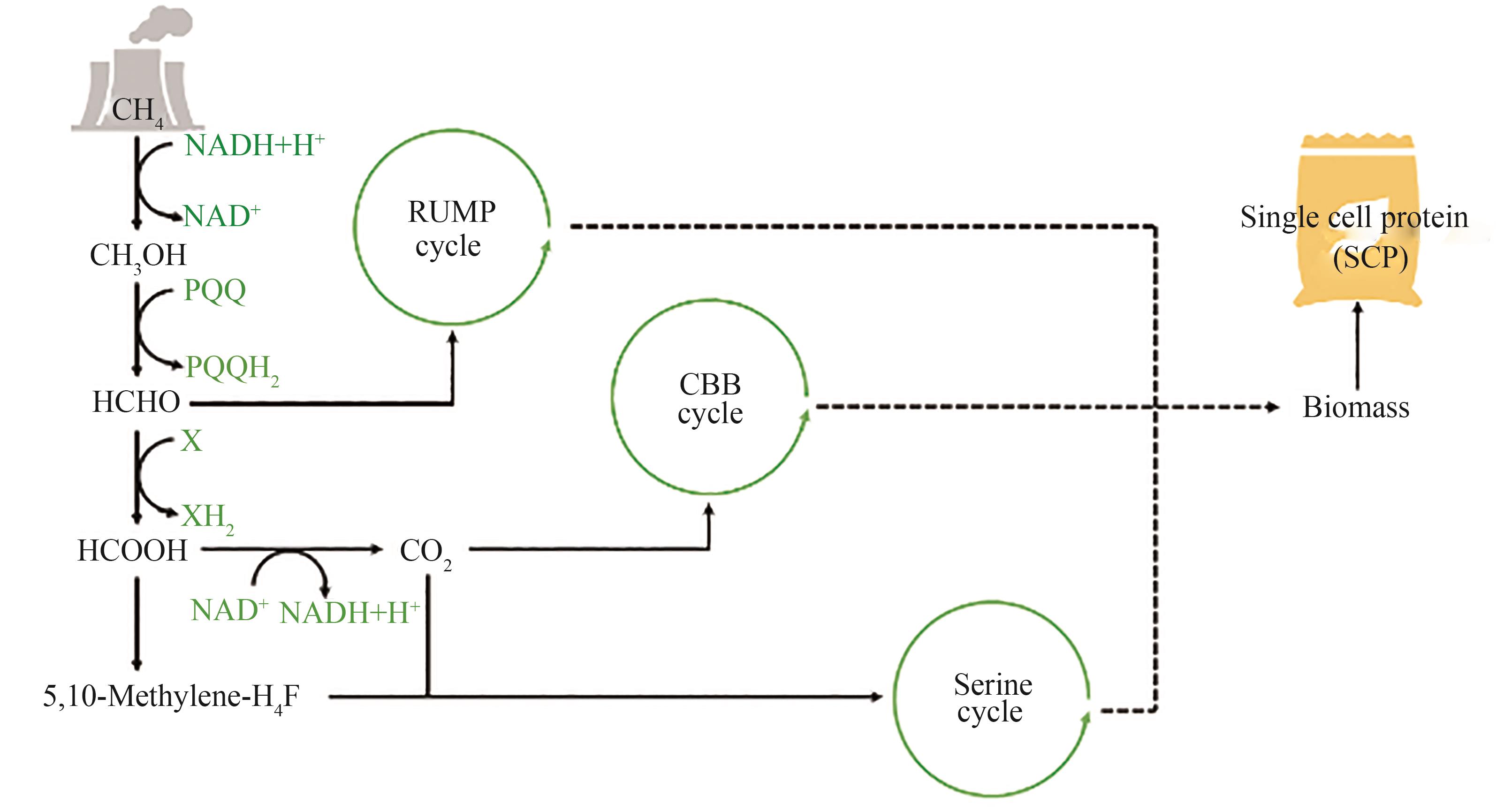
Fig. 1 Metabolic pathways of microorganisms utilizing liquid carbon material substrates(The dashed line represents a multi-step reaction) H6P—Hexulose-6-phosphate; Ru5P—Ribulose-5-phosphate; F6P—Fructose-6-phosphate; DHA—Dihydroxyacetone; Xu5P—Ribulose-5-phosphate; DHAP—Dihydroxyacetone phosphate; G3P—Glyceraldehyde-3-phosphate; Pyr—Pyruvate; GCS—Glycine cleavage system; 2GPA—Glycerate 2P; OAA—Oxaloacetic acid
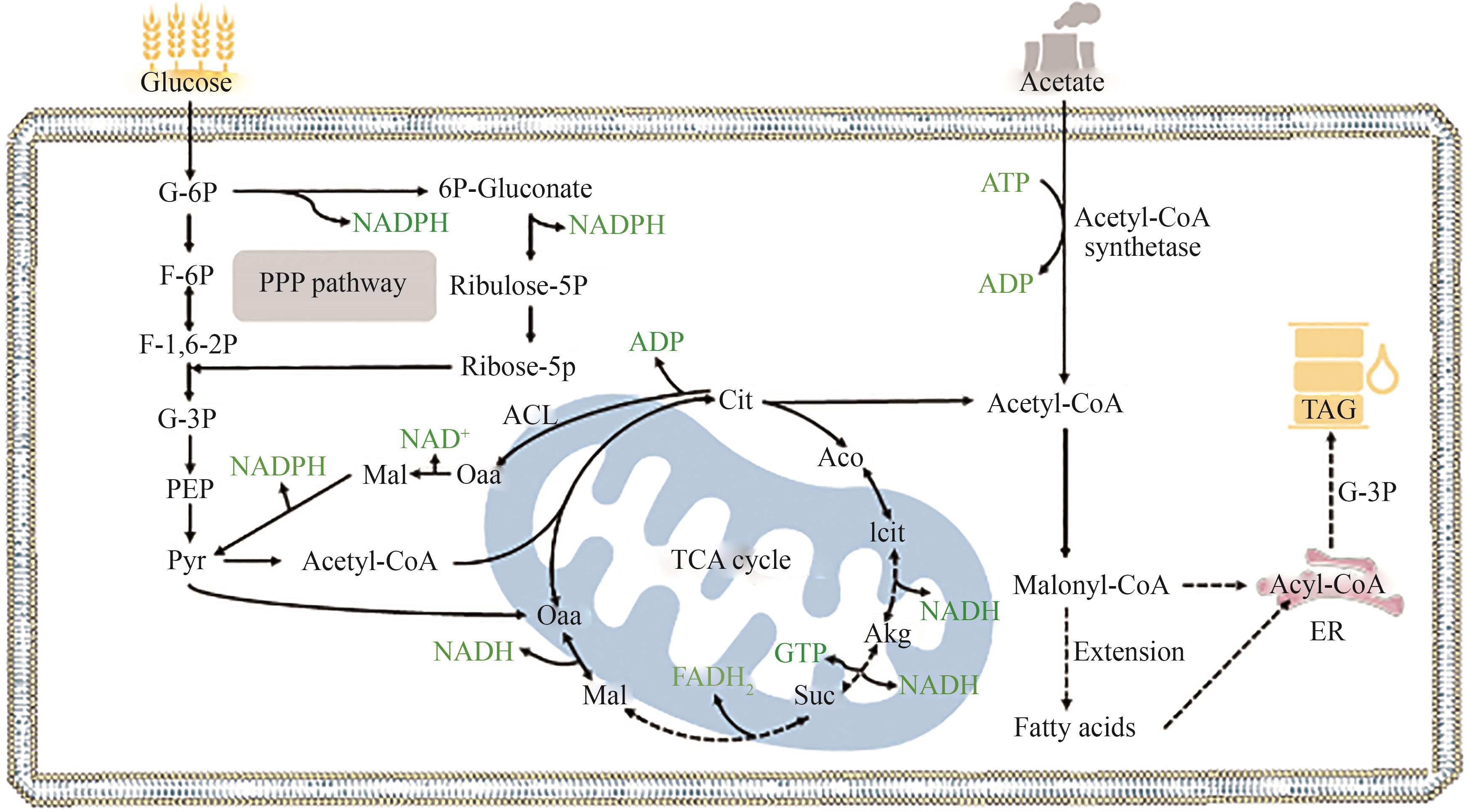
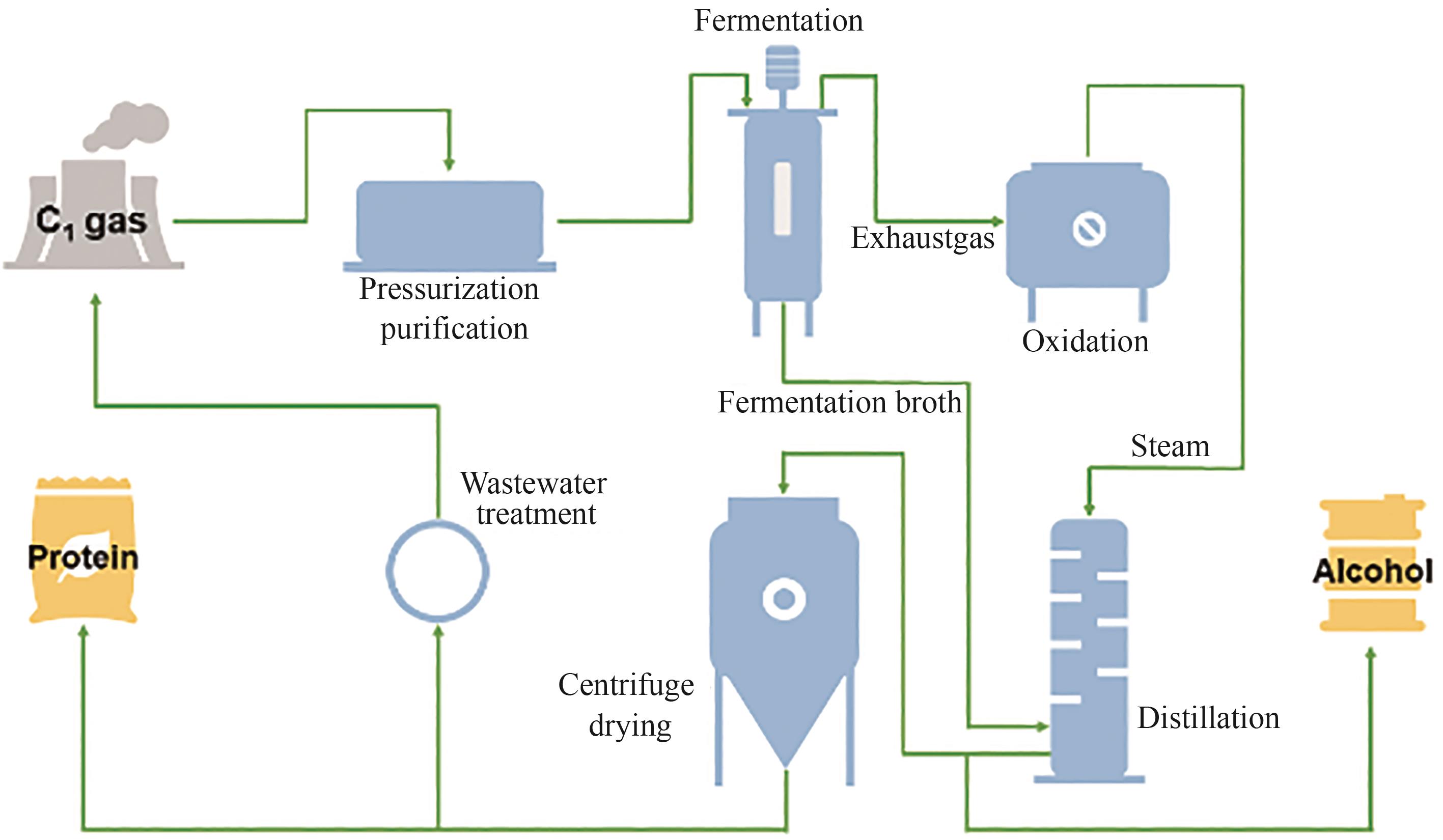
Fig. 3 Pathways of lipid synthesis using glucose or acetate as substrate(The dashed line represents a multi-step reaction) G-6P—Glucose-6-phosphate; 6P-Gluconate—6-Phosphogluconate; Ribulose-5P—Ribulose-5-phosphate; Ribose-5P—Ribose-5-phosphate; F-6P—Fructose-6-phosphate; F-1,6-2P—Fructose-1,6-phosphate; G-3P—3-Phosphoglyceraldehyde; PEP—Phosphoenolpyruvate; Pyr—Pyruvate; Cit—Citrate; Aco—cis: aconitate; Icit—Isocitrate; Akg—2-Oxo-glutarate; Suc—Succinate; Mal—Malate; Oaa—Oxaloacetate; ACL—ATP-citrate lyase; ER—Endoplasmic reticulum; TAG—Triglyceride
| 1 | 国家统计局. 中华人民共和国2023年国民经济和社会发展统计公报[R/OL]. (2024-02-29)[2024-03-01]. . |
| National Bureau of Statistics. Statistical Communiqué of the People’s Republic of China on the 2023 National Economic and Social Development[R/OL]. (2024-02-29)[2024-03-01]. . | |
| 2 | 中华人民共和国海关总署. 海关统计:2023年1至12月部分进口商品主要贸易方式量值表(人民币值)[EB/OL]. [2024-03-01]. . |
| General Administration of Customs of the People’s Republic of China. Customs Statistics: Table of Quantities and Values of Main Trade Modes of Selected Imported Commodities from January to December 2023 (in RMB)[EB/OL]. [2024-03-01]. . | |
| 3 | 中华人民共和国自然资源部. 全国石油天然气资源勘查开采通报(2020年度)[R/OL]. (2021-09-17)[2024-01-01]. . |
| Ministry of Natural Resources of the People’s Republic of China. National Bulletin on Oil and Gas Resources Exploration and Exploitation (2020) [R/OL]. (2021-09-17)[2024-01-01]. . | |
| 4 | Agency International Energy. CO2 Emissions in 2022[R/OL]. IEA. [2024-01-01]. . |
| 5 | 肖俊夫, 陈德敏, 高艳红. “双碳”目标下再生资源利用的碳减排效应及作用机制研究[J]. 重庆大学学报(社会科学版), 2023, 46(10): 2. |
| XIAO J F, CHEN D M, GAO Y H. Research on the effect and mechanism of renewable resources utilization on carbon emission reduction under “double-carbon” target[J]. Journal of Chongqing University(Social Science Edition), 2023, 46(10): 2. | |
| 6 | JIANG W, LI C, LI Y J, et al. Metabolic engineering strategies for improved lipid production and cellular physiological responses in yeast Saccharomyces cerevisiae [J]. Journal of Fungi, 2022, 8(5): 427. |
| 7 | YAN D, YE S Y, HE Y, et al. Fatty acids and lipid mediators in inflammatory bowel disease: from mechanism to treatment[J]. Frontiers in Immunology, 2023, 14: 1286667. |
| 8 | CHEUNG P C K. Chemical evaluation of some lesser known edible mushroom mycelia produced in submerged culture from soy milk waste[J]. Food Chemistry, 1997, 60(1): 61-65. |
| 9 | ATHANASIADIS I, BOSKOU D, KANELLAKI M, et al. Effect of carbohydrate substrate on fermentation by kefir yeast supported on delignified cellulosic materials[J]. Journal of Agricultural and Food Chemistry, 2001, 49(2): 658-663. |
| 10 | CHOI M H, PARK Y H. Growth of Pichia guilliermondii A9, an osmotolerant yeast, in waste brine generated from kimchi production[J]. Bioresource Technology, 1999, 70(3): 231-236. |
| 11 | ANUPAMA, RAVINDRA P. Value-added food: single cell protein[J]. Biotechnology Advances, 2000, 18(6): 459-479. |
| 12 | CAI P, WU X Y, DENG J, et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris [J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(29): e2201711119. |
| 13 | GAO L, MENG J, DAI W L, et al. Deciphering cell wall sensors enabling the construction of robust P. pastoris for single-cell protein production[J]. Biotechnology for Biofuels and Bioproducts, 2023, 16(1): 178. |
| 14 | 高琳惠, 蔡鹏, 周雍进. 甲醇酵母代谢工程研究进展[J]. 生物工程学报, 2021, 37(3): 966-979. |
| GAO L H, CAI P, ZHOU Y J. Advances in metabolic engineering of methylotrophic yeasts[J]. Chinese Journal of Biotechnology, 2021, 37(3): 966-979. | |
| 15 | GAO J Q, LI Y X, YU W, et al. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol[J]. Nature Metabolism, 2022, 4(7): 932-943. |
| 16 | ZHOU H L, WANG F, MAO H L, et al. Efficient expression of heterologous protein by engineered Komagataella phaffii by harnessing a bioelectrical CO2 reduction system[J]. Biochemical Engineering Journal, 2023, 191: 108762. |
| 17 | VOGL T, FISCHER J E, HYDEN P, et al. Orthologous promoters from related methylotrophic yeasts surpass expression of endogenous promoters of Pichia pastoris [J]. AMB Express, 2020, 10(1): 38. |
| 18 | SAKAI Y, AKIYAMA M, KONDOH H, et al. High-level secretion of fungal glucoamylase using the Candida boidinii gene expression system[J]. Biochimica et Biophysica Acta, 1996, 1308(1): 81-87. |
| 19 | SCHNEIDER K, PEYRAUD R, KIEFER P, et al. The ethylmalonyl-CoA pathway is used in place of the glyoxylate cycle by Methylobacterium extorquens AM1 during growth on acetate[J]. Journal of Biological Chemistry, 2012, 287(1): 757-766. |
| 20 | MO X H, ZHANG H, WANG T M, et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis[J]. Applied Microbiology and Biotechnology, 2020, 104(10): 4515-4532. |
| 21 | BÉLANGER L, FIGUEIRA M M, BOURQUE D, et al. Production of heterologous protein by Methylobacterium extorquens in high cell density fermentation[J]. FEMS Microbiology Letters, 2004, 231(2): 197-204. |
| 22 | KLEIN V J, BRITO L F, PEREZ-GARCIA F, et al. Metabolic engineering of thermophilic Bacillus methanolicus for riboflavin overproduction from methanol[J]. Microbial Biotechnology, 2023, 16(5): 1011-1026. |
| 23 | WENDISCH V F, KOSEC G, HEUX S, et al. Aerobic utilization of methanol for microbial growth and production[J]. Advances in Biochemical Engineering/Biotechnology, 2022, 180: 169-212. |
| 24 | SCHRADER J, SCHILLING M, HOLTMANN D, et al. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria[J]. Trends in Biotechnology, 2009, 27(2): 107-115. |
| 25 | ZHAN C J, LI X W, LAN G X, et al. Reprogramming methanol utilization pathways to convert Saccharomyces cerevisiae to a synthetic methylotroph[J]. Nature Catalysis, 2023, 6(5): 435-450. |
| 26 | WANG G K, OLOFSSON-DOLK M, HANSSON F G, et al. Engineering yeast Yarrowia lipolytica for methanol assimilation[J]. ACS Synthetic Biology, 2021, 10(12): 3537-3550. |
| 27 | GLEIZER S, BEN-NISSAN R, BAR-ON Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 [J]. Cell, 2019, 179(6): 1255-1263.e12. |
| 28 | KIM S H, LINDNER S N, ASLAN S, et al. Growth of E. coli on formate and methanol via the reductive glycine pathway[J]. Nature Chemical Biology, 2020, 16(5): 538-545. |
| 29 | TONG S, ZHAO L Z, ZHU D L, et al. From formic acid to single-cell protein: genome-scale revealing the metabolic network of Paracoccus communis MA5[J]. Bioresources and Bioprocessing, 2022, 9(1): 55. |
| 30 | 赵立枝, 朱大玲, 李德茂. 利用甲酸产单细胞蛋白菌株的筛选及发酵工艺[J]. 饲料研究, 2021, 44(9): 88-93. |
| ZHAO L Z, ZHU D L, LI D M. Screening and fermentation process of single cell protein-producing strains by formic acid[J]. Feed Research, 2021, 44(9): 88-93. | |
| 31 | TIAN J Z, DENG W S Y, ZHANG Z W, et al. Discovery and remodeling of Vibrio natriegens as a microbial platform for efficient formic acid biorefinery[J]. Nature Communications, 2023, 14(1): 7758. |
| 32 | WAN S, LAI M C, GAO X Y, et al. Recent progress in engineering Clostridium autoethanogenum to synthesize the biochemicals and biocommodities[J]. Synthetic and Systems Biotechnology, 2024, 9(1): 19-25. |
| 33 | HEFFERNAN J K, MAHAMKALI V, VALGEPEA K, et al. Analytical tools for unravelling the metabolism of gas-fermenting Clostridia[J]. Current Opinion in Biotechnology, 2022, 75: 102700. |
| 34 | 王永伟, 施晶晶, 段涛, 等. 生物技术在粮油饲料资源增值转化中的应用研究进展[J]. 粮油食品科技, 2023, 31(5): 152-159. |
| WANG Y W, SHI J J, DUAN T, et al. Research progress on the application of biotechnology in value-added conversion of grain, oil and feed resources[J]. Science and Technology of Cereals, Oils and Foods, 2023, 31(5): 152-159. | |
| 35 | YI J H, HUANG H Y, LIANG J Y, et al. A heterodimeric reduced-ferredoxin-dependent methylenetetrahydrofolate reduc-tase from syngas-fermenting Clostridium ljungdahlii [J]. Microbiology Spectrum, 2021, 9(2): e0095821. |
| 36 | ZHU H F, LIU Z Y, ZHOU X, et al. Energy conservation and carbon flux distribution during fermentation of CO or H2/CO2 by Clostridium ljungdahlii [J]. Frontiers in Microbiology, 2020, 11: 416. |
| 37 | 路伟源. 甲烷氧化菌与氢氧化细菌共培养生产微生物蛋白工艺优化及机理研究[D]. 北京:北京化工大学, 2023. |
| LU W Y. Process optimization and mechanism of microbial protein production by co-culture of methane oxidizing bacteria and hydrogen oxidizing bacteria[D]. Beijing: Beijing University of Chemical Technology, 2023. | |
| 38 | 严程, 梅娟, 赵由才. 好氧甲烷氧化菌及其工程应用进展[J]. 生物工程学报, 2022, 38(4): 1322-1338. |
| YAN C, MEI J, ZHAO Y C. Engineering application of aerobic methane oxidizing bacteria(methanotrophs): a review[J]. Chinese Journal of Biotechnology, 2022, 38(4): 1322-1338. | |
| 39 | RITALA A, HÄKKINEN S T, TOIVARI M, et al. Single cell protein-state-of-the-art, industrial landscape and patents 2001-2016[J]. Frontiers in Microbiology, 2017, 8: 2009. |
| 40 | 傅晓莹, 乔玮博, 史硕博. 微生物利用一碳底物生产单细胞蛋白研究进展[J]. 食品科学, 2023, 44(3): 1-11. |
| FU X Y, QIAO W B, SHI S B. Microbial production of single cell proteins from single carbon substrates: a review[J]. Food Science, 2023, 44(3): 1-11. | |
| 41 | 蔡朝阳, 何崭飞, 胡宝兰. 甲烷氧化菌分类及代谢途径研究进展[J]. 浙江大学学报(农业与生命科学版), 2016, 42(3): 273-281. |
| CAI C Y, HE Z F, HU B L. Progresses in the classification and mechanism of methane-oxidizing bacteria[J]. Journal of Zhejiang University (Agriculture and Life Sciences), 2016, 42(3): 273-281. | |
| 42 | WANG X, QIN J L, MA C, et al. Methanol assimilation with CO2 reduction in Butyribacterium methylotrophicum and development of genetic toolkits for its engineering[J]. ACS Sustainable Chemistry & Engineering, 2021, 9(36): 12079-12090. |
| 43 | 冯叨, 高教琪, 龚志伟, 等. 多形汉逊酵母代谢改造生产脂肪酸及发酵条件优化[J]. 生物工程学报, 2022, 38(2): 760-771. |
| FENG D, GAO J Q, GONG Z W, et al. Production of fatty acids by engineered Ogataea polymorpha [J]. Chinese Journal of Biotechnology, 2022, 38(2): 760-771. | |
| 44 | WU X Y, CAI P, GAO L H, et al. Efficient bioproduction of 3-hydroxypropionic acid from methanol by a synthetic yeast cell factory[J]. ACS Sustainable Chemistry & Engineering, 2023, 11(16): 6445-6453. |
| 45 | MENG J, LIU S F, GAO L, et al. Economical production of Pichia pastoris single cell protein from methanol at industrial pilot scale[J]. Microbial Cell Factories, 2023, 22(1): 198. |
| 46 | NATTERMANN M, WENK S, PFISTER P, et al. Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo [J]. Nature Communications, 2023, 14(1): 2682. |
| 47 | SINGH H B, KANG M K, KWON M, et al. Developing methylotrophic microbial platforms for a methanol-based bioindustry[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 1050740. |
| 48 | BANG J H, HWANG C H, AHN J H, et al. Escherichia coli is engineered to grow on CO2 and formic acid[J]. Nature Microbiology, 2020, 5(12): 1459-1463. |
| 49 | ZHANG S Q, ZHANG L J, XU G, et al. A review on biodiesel production from microalgae: influencing parameters and recent advanced technologies[J]. Frontiers in Microbiology, 2022, 13: 970028. |
| 50 | WEUSTER-BOTZ D. Process engineering aspects for the microbial conversion of C1 gases[M/OL]//One-carbon feedstocks for sustainable bioproduction. Cham: Springer International Publishing, 2021, 180: 33-56. (2021-07-22)[2024-01-01]. . |
| 51 | 万赛, 王皓明, 马小清, 等. 碳一气体生物转化中的产乙酸菌改造与发酵工艺优化[J]. 生物工程学报, 2023, 39(6): 2410-2429. |
| WAN S, WANG H M, MA X Q, et al. Genetic modification of acetogens and optimization of fermentation process in C1-gas bioconversion[J]. Chinese Journal of Biotechnology, 2023, 39(6): 2410-2429. | |
| 52 | JIN T Z, WANG Y J, YAO S, et al. Bioconversion of carbon dioxide to succinate by Citrobacter [J]. Chemical Engineering Journal, 2023, 452: 139668. |
| 53 | ALBERTO R, WELTER C, KUBISCH C, et al. Co-fermenting pyrolysis aqueous condensate and pyrolysis syngas with anaerobic microbial communities enables L-malate production in a secondary fermentative stage[J]. Fermentation, 2022, 8(10): 512. |
| 54 | OH H J, GONG G T, AHN J H, et al. Effective hexanol production from carbon monoxide using extractive fermentation with Clostridium carboxidivorans P7[J]. Bioresource Technology, 2023, 367: 128201. |
| 55 | BAE J Y, SONG Y S, LEE H S, et al. Valorization of C1 gases to value-added chemicals using acetogenic biocatalysts[J]. Chemical Engineering Journal, 2022, 428: 131325. |
| 56 | DAI Z X, GU H L, ZHANG S J, et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae [J]. Bioresource Technology, 2017, 245(Pt B): 1407-1412. |
| 57 | 姚伦, 周雍进. 一碳化合物生物利用和转化研究进展[J]. 化工进展, 2023, 42(1): 16-29. |
| YAO L, ZHOU Y J. Progress in microbial utilization of one-carbon feedstocks for biomanufacturing[J]. Chemical Industry and Engineering Progress, 2023, 42(1): 16-29. | |
| 58 | CAI T, SUN H B, QIAO J, et al. Cell-free chemoenzymatic starch synthesis from carbon dioxide[J]. Science, 2021, 373(6562): 1523-1527. |
| 59 | YANG J G, SONG W, CAI T, et al. De novo artificial synthesis of hexoses from carbon dioxide[J]. Science Bulletin, 2023, 68(20): 2370-2381. |
| 60 | GAO F, LIU G Y, CHEN A B, et al. Artificial photosynthetic cells with biotic-abiotic hybrid energy modules for customized CO2 conversion[J]. Nature Communications, 2023, 14(1): 6783. |
| 61 | MOON J, WASCHINGER L M, MÜLLER V. Lactate formation from fructose or C1 compounds in the acetogen Acetobacterium woodii by metabolic engineering[J]. Applied Microbiology and Biotechnology, 2023, 107(17): 5491-5502. |
| 62 | BENITO-VAQUERIZO S, NOUSE N, SCHAAP P J, et al. Model-driven approach for the production of butyrate from CO2/H2 by a novel co-culture of C. autoethanogenum and C. beijerinckii [J]. Frontiers in Microbiology, 2022, 13: 1064013. |
| 63 | POULALIER-DELAVELLE M, BAKER J P, MILLARD J, et al. Endogenous CRISPR/Cas systems for genome engineering in the acetogens Acetobacterium woodii and Clostridium autoethanogenum [J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1213236. |
| 64 | SAMRANRIT T, TEEKA J, NGERNSOMBAT K, et al. Modulation of yeast oil production by Pseudozyma parantarctica CHC28 using xylose and organic acids and its conversion feasibility to bio-polyurethane foam[J]. Biochemical Engineering Journal, 2023, 198: 109025. |
| 65 | HUANG C C, CHEN Y R, CHENG S, et al. Enhanced acetate utilization for value-added chemicals production in Yarrowia lipolytica by integration of metabolic engineering and microbial electrosynthesis[J]. Biotechnology and Bioengineering, 2023, 120(10): 3013-3024. |
| 66 | ZHENG T T, ZHANG M L, WU L H, et al. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering[J]. Nature Catalysis, 2022, 5: 388-396. |
| 67 | GAO Q, YANG J L, ZHAO X R, et al. Yarrowia lipolytica as a metabolic engineering platform for the production of very-long-chain wax esters[J]. Journal of Agricultural and Food Chemistry, 2020, 68(39): 10730-10740. |
| 68 | ZHAO X R, CHEN X L, YANG J L, et al. De novo synthesis of nervonic acid and optimization of metabolic regulation by Yarrowia lipolytica [J]. Bioresources and Bioprocessing, 2023, 10(1): 70. |
| 69 | LIU F X, LU Z W, LU T T, et al. Metabolic engineering of oleaginous yeast in the lipogenic phase enhances production of nervonic acid[J]. Metabolic Engineering, 2023, 80: 193-206. |
| 70 | WANG K F, LIN L, WEI P, et al. Combining orthogonal plant and non-plant fatty acid biosynthesis pathways for efficient production of microbial oil enriched in nervonic acid in Yarrowia lipolytica [J]. Bioresource Technology, 2023, 378: 129012. |
| 71 | SU H, SHI P H, SHEN Z S, et al. High-level production of nervonic acid in the oleaginous yeast Yarrowia lipolytica by systematic metabolic engineering[J]. Communications Biology, 2023, 6(1): 1125. |
| 72 | RAVI R K, NEERAJ A, YADAV R H. Assessment of microbial biomass for production of ecofriendly single-cell protein, bioenergy, and other useful products[M/OL]//Microbes in land use change management. Amsterdam: Elsevier, 2021: 267-284. (2021-08-27)[2024-01-01]. . |
| 73 | JONES S W, KARPOL A, FRIEDMAN S, et al. Recent advances in single cell protein use as a feed ingredient in aquaculture[J]. Current Opinion in Biotechnology, 2020, 61: 189-197. |
| 74 | MOLITOR B, MISHRA A, ANGENENT L T. Power-to-protein: converting renewable electric power and carbon dioxide into single cell protein with a two-stage bioprocess[J]. Energy & Environmental Science, 2019, 12(12): 3515-3521. |
| 75 | FU Y F, LI Y, LIDSTROM M. The oxidative TCA cycle operates during methanotrophic growth of the TypeⅠmethanotroph Methylomicrobium buryatense 5GB1[J]. Metabolic Engineering, 2017, 42: 43-51. |
| 76 | KHOSHNEVISAN B, TSAPEKOS P, ZHANG Y F, et al. Urban biowaste valorization by coupling anaerobic digestion and single cell protein production[J]. Bioresource Technology, 2019, 290: 121743. |
| 77 | NWAOKORIE U J, REINMETS K, DE LIMA L A, et al. Deletion of genes linked to the C1-fixing gene cluster affects growth, by-products, and proteome of Clostridium autoethanogenum [J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1167892. |
| 78 | 舒芹, 李凯凯, 全拓, 等. 微生物蛋白作为优质替代蛋白资源的应用研究[J]. 未来食品科学, 2022(2): 96-106. |
| SHU Q, LI K K, QUAN T, et al. Application and development of microbial proteins as high-quality alternative protein resources - a view[J]. Future Food Science, 2022(2): 96-106. | |
| 79 | LIU Y Q, ZHANG Z W, JIANG W H, et al. Protein acetylation-mediated cross regulation of acetic acid and ethanol synthesis in the gas-fermenting Clostridium ljungdahlii [J]. Journal of Biological Chemistry, 2022, 298(2): 101538. |
| 80 | VALGEPEA K, TALBO G, TAKEMORI N, et al. Absolute proteome quantification in the gas-fermenting acetogen Clostridium autoethanogenum [J]. mSystems, 2022, 7(2): e00026-22. |
| 81 | DETSIOS N, MARAGOUDAKI L, ATSONIOS K, et al. Design considerations of an integrated thermochemical/biochemical route for aviation and maritime biofuel production[J]. Biomass Conversion and Biorefinery,2024,14,27537-27555. |
| 82 | ROBLES-IGLESIAS R, NICAUD J M, VEIGA M C, et al. Integrated fermentative process for lipid and β-carotene production from acetogenic syngas fermentation using an engineered oleaginous Yarrowia lipolytica yeast[J]. Bioresource Technology, 2023, 389: 129815. |
| 83 | ROBLES-IGLESIAS R, VEIGA M C, KENNES C. Sequential bioconversion of C1-gases (CO, CO2, syngas) into lipids, through the carboxylic acid platform, with Clostridium aceticum and Rhodosporidium toruloides [J]. Journal of Environmental Management, 2023, 347: 119097. |
| 84 | ODUOR W W, WANDERA S M, MURUNGA S I, et al. Enhancement of anaerobic digestion by co-digesting food waste and water hyacinth in improving treatment of organic waste and bio-methane recovery[J]. Heliyon, 2022, 8(9): e10580. |
| 85 | KÜÇÜKAĞA Y, FACCHIN A, KARA S, et al. Conversion of pyrolysis products into volatile fatty acids with a biochar-packed anaerobic bioreactor[J]. Industrial & Engineering Chemistry Research, 2022, 61(45): 16624-16634. |
| 86 | REGIS F, MONTEVERDE A H A, FINO D. A techno-economic assessment of bioethanol production from switchgrass through biomass gasification and syngas fermentation[J]. Energy, 2023, 274: 127318. |
| 87 | MA S, DONG C Q, HU X Y, et al. Techno-economic evaluation of a combined biomass gasification-solid oxide fuel cell system for ethanol production via syngas fermentation[J]. Fuel, 2022, 324: 124395. |
| 88 | PATI S, DE S, CHOWDHURY R. Exploring the hybrid route of bio-ethanol production via biomass co-gasification and syngas fermentation from wheat straw and sugarcane bagasse: model development and multi-objective optimization[J]. Journal of Cleaner Production, 2023, 395: 136441. |
| 89 | HEFFERNAN J K, LAI C Y, GONZALEZ-GARCIA R A, et al. Biogas upgrading using Clostridium autoethanogenum for value-added products[J]. Chemical Engineering Journal, 2023, 452: 138950. |
| 90 | PUIMAN L, ELISIÁRIO M P, CRASBORN L M L, et al. Gas mass transfer in syngas fermentation broths is enhanced by ethanol[J]. Biochemical Engineering Journal, 2022, 185: 108505. |
| 91 | PUIMAN L, ABRAHAMSON B, VAN DER LANS R G J M, et al. Alleviating mass transfer limitations in industrial external-loop syngas-to-ethanol fermentation[J]. Chemical Engineering Science, 2022, 259: 117770. |
| 92 | KÜÇÜKAĞA Y, FACCHIN A, STEFANELLI V, et al. Innovative char-sparger for improving volatile fatty acids (VFA) production in homoacetogenic fermentation of H2/CO2 with microbial mixed cultures (MMC)[J]. Chemical Engineering Journal, 2023, 471: 144165. |
| 93 | PERRET L, BOUKIS N, SAUER J. Influence of increased cell densities on product ratio and productivity in syngas fermentation[J]. Industrial & Engineering Chemistry Research, 2023, 62(35): 13799-13810. |
| 94 | VELVIZHI G, SARKAR O, ROVIRA-ALSINA L, et al. Conversion of carbon dioxide to value added products through anaerobic fermentation and electro fermentation: a comparative approach[J]. International Journal of Hydrogen Energy, 2022, 47(34): 15442-15455. |
| 95 | KIM J H, LEE M, JEONG H, et al. Recycling of minerals with acetate separation in biological syngas fermentation with an electrodialysis system[J]. Chemical Engineering Journal, 2023, 459: 141555. |
| 96 | MARIËN Q, REGUEIRA A, GANIGUÉ R. Steerable isobutyric and butyric acid production from CO2 and H2 by Clostridium luticellarii [J]. Microbial Biotechnology, 2024, 17(1): e14321. |
| 97 | KATAKOJWALA R, THARAK A, SARKAR O, et al. Design and evaluation of gas fermentation systems for CO2 reduction to C2 and C4 fatty acids: non-genetic metabolic regulation with pressure, pH and reaction time[J]. Bioresource Technology, 2022, 351: 126937. |
| 98 | LIU J, ZHOU W T, HE Q N, et al. Microbial lipid production from high concentration of volatile fatty acids via Trichosporon cutaneum for biodiesel preparation[J]. Applied Biochemistry and Biotechnology, 2022, 194(7): 2968-2979. |
| 99 | ZHOU W, WANG Y N, ZHANG J L, et al. A metabolic model of Lipomyces starkeyi for predicting lipogenesis potential from diverse low-cost substrates[J]. Biotechnology for Biofuels, 2021, 14(1): 148. |
| 100 | SUN S M, DING Y M, LIU M, et al. Comparison of glucose, acetate and ethanol as carbon resource for production of poly(3-hydroxybutyrate) and other acetyl-CoA derivatives[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 833. |
| 101 | PEREIRA A S, LOPES M, MIRANDA S M, et al. Bio-oil production for biodiesel industry by Yarrowia lipolytica from volatile fatty acids in two-stage batch culture[J]. Applied Microbiology and Biotechnology, 2022, 106(8): 2869-2881. |
| 102 | REROP Z S, STELLNER N I, GRABAN P, et al. Bioconversion of a lignocellulosic hydrolysate to single cell oil for biofuel production in a cost-efficient fermentation process[J]. Fermentation, 2023, 9(2): 189. |
| 103 | BURGSTALLER L, OLIVER L, DIETRICH T, et al. Pilot scale production of single cell oil by Apiotrichum brassicae and Pichia kudriavzevii from acetic acid and propionic acid[J]. Applied Sciences, 2023, 13(8) : 4674. |
| 104 | MORENO J F, OULEGO P, COLLADO S, et al. Production of biolipids from volatile fatty acids of sewage sludge by Yarrowia lipolytica [J]. Fuel, 2023, 348: 128488. |
| 105 | NAVEIRA-PAZOS C, VEIGA M C, KENNES C. Accumulation of lipids by the oleaginous yeast Yarrowia lipolytica grown on carboxylic acids simulating syngas and carbon dioxide fermentation[J]. Bioresource Technology, 2022, 360: 127649. |
| 106 | MORALES-PALOMO S, GONZÁLEZ-FERNÁNDEZ C, TOMÁS-PEJÓ E. Prevailing acid determines the efficiency of oleaginous fermentation from volatile fatty acids[J]. Journal of Environmental Chemical Engineering, 2022, 10(2): 107354. |
| 107 | Kiverdi, Inc. [EB/OL]. [2023-12-29]. . |
| 108 | Air Protein Inc. [EB/OL]. [2024-01-02]. |
| 109 | Newswire Contact PR. FGen AG, Ginkgo Bioworks Subsidiarya, Awarded Funding Through European Innovation Council’s Pathfinder Programme For Sustainable Milk Protein Production Project[N/OL]. News from ginkgo bioworks. (2023-12-19)[2024-01-01]. . |
| 110 | 中华人民共和国农业农村部. 中华人民共和国农业农村部公告第465号[EB/OL]. (2021-12-07)[2024-03-06]. . |
| Ministy of Agnculure and Rural Afairs of the People’s Republic of China. Announcement No. 465 of the Ministry of Agriculture and Rural Affairs of the People’s Republic of China[EB/OL]. (2021-12-07)[2024-03-06]. . | |
| 111 | GORLA G, FERRER A, GIUSSANI B. Process understanding and monitoring: a glimpse into data strategies for miniaturized NIR spectrometers[J]. Analytica Chimica Acta, 2023, 1281: 341902. |
| 112 | SCHORN-GARCÍA D, EZENARRO J, ACEÑA L, et al. Spatially offset Raman spectroscopic (SORS) analysis of wine alcoholic fermentation: a preliminary study[J]. Fermentation, 2023, 9(2): 115. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | YING Hanjie, LIU Dong, WANG Zhenyu, SHEN Tao, ZHUANG Wei, ZHU Chenjie. Exploring industrial biomanufacturing and the goal of “carbon neutrality” [J]. Synthetic Biology Journal, 2025, 6(1): 1-7. |
| [3] | ZHANG Yi-Heng P. Job, CHEN Xuemei, SHI Ting. Price to Cost-of-raw-materials Ratio (PC) of biomanufacturing: definition and application [J]. Synthetic Biology Journal, 2025, 6(1): 8-17. |
| [4] | ZHANG Yi-Heng P. Job. The enlightenment of the Chinese philosophy “Tao-Fa-Shu-Qi” to industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1231-1241. |
| [5] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [6] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [7] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [8] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [9] | LIU Jianming, ZHANG Chijian, ZHANG Bing, ZENG Anping. Clostridium pasteurianum as an industrial chassis for efficient production of 1,3-propanediol: from metabolic engineering to fermentation and product separation [J]. Synthetic Biology Journal, 2024, 5(6): 1386-1403. |
| [10] | CHENG Feng, ZOU Shuping, XU Jianmiao, TANG Heng, XUE Yaping, ZHENG Yuguo. BioHPP®: a benchmark of biomanufacturing for high optically pure L-phosphinothricin [J]. Synthetic Biology Journal, 2024, 5(6): 1404-1418. |
| [11] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [12] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [15] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||