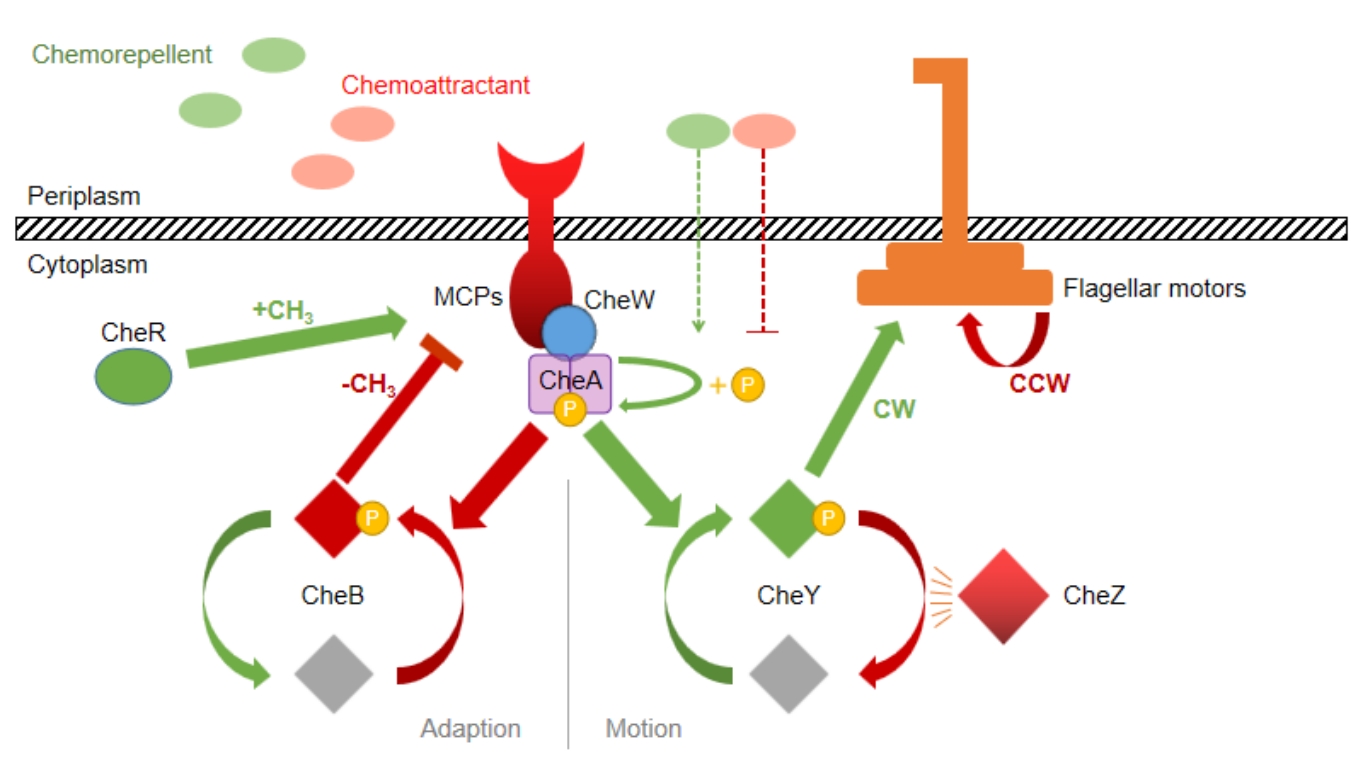
A preliminary study on the regulation and analysis of indole mediated bidirectional chemotaxis behavior of Escherichia coli
WANG Kunkun1, CHEN Hongyu2, ZHANG Andong1, LU Xiaoyun1, TAN Dan1
- 1.Key Laboratory of Biomedical Information Engineering of Ministry of Education,School of Life Science and Technology,Xi'an Jiaotong University,Xi'an 710049,China
2.School of Life Sciences,Tsinghua University,Beijing 100084,China
-
Received:2025-04-29Revised:2025-07-31Published:2025-08-01 -
Contact:TAN Dan
大肠杆菌中吲哚介导的双向趋化行为的调控与分析
汪昆昆1, 陈泓宇2, 张安栋1, 卢晓云1, 谭丹1
- 1.西安交通大学生命科学与技术学院,生物医学信息工程教育部重点实验室,陕西 西安 710049
2.清华大学生命科学学院,北京 100084
-
通讯作者:谭丹 -
作者简介:汪昆昆 (1999—),男,硕士研究生。研究方向为基于嗜盐微生物的合成生物学。E-mail:3122113016@stu.xjtu.edu.cn谭丹 (1986—),女,副教授,博士,硕士生导师。研究方向为基于嗜盐微生物的合成生物学与定量生物学、新活性天然产物的生物合成及关键酶挖掘与研究等。E-mail:tandan@mail.xjtu.edu.cn
第一联系人:汪昆昆、陈泓宇为同等贡献作者
CLC Number:
Cite this article
WANG Kunkun, CHEN Hongyu, ZHANG Andong, LU Xiaoyun, TAN Dan. A preliminary study on the regulation and analysis of indole mediated bidirectional chemotaxis behavior of Escherichia coli[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-036.
汪昆昆, 陈泓宇, 张安栋, 卢晓云, 谭丹. 大肠杆菌中吲哚介导的双向趋化行为的调控与分析[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-036.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-036
| Name | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F- lacZΔ lacΔ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK+,mk+) phoA supE44 Y-thi-1 gyrA96 relA1 | TransGen Biotech |
| E. coli RP437 | F- thr-1 araC14 leuB6(Am) fhuA31 lacY1 tsx-78λ- eda-50 hisG4(Oc) rfbC1 rpsL136(strR) xylA5 metF159(Am) mtl-1 thiE1 | [ |
| E. coli RP437ΔTar | E. coli RP437 mutant with tar gene knockdown | This study |
| Plasmids | ||
| pUC19 (Bsa I free) | AmpR, pBR322 origin, no Bsa I restriction enzyme site | Tsingke |
| pUC-eGFP | pUC19-derived plasmid carrying eGFP, for cell visualization in Transwell and microfluidics experiment | This study |
| pdCas9 | CmR, backbone plasmid for CRISPRi, carrying dCas9 gene (tetracycline-induced) with sgRNA scaffold (arabinose-induced), and mRFP expression cassette | [ |
| pdCas9-Spacer 1~6 | pdCas9-derived plasmid inserted with Spacer 1~6 for tar gene knockdown | This study |
| Oligomers | Tsingke | |
| Spacer-F1 | TTCAGCCATACTTTTCATACTCCC | |
| Spacer-R1 | GAAGCGGAATATATCCCTAGGTAT | |
| Spacer-extend-F1 | GCAAGGCGATTAAGTTGGGTAA | |
| Spacer-extend-R1 | TGAGTTAGCTCACTCATTAGGCAC | |
| Spacer-extend-F2 | CGTAAGGAGAAAATACCGCATCAG | |
| Spacer-extend-R2 | ACAGGAAACAGCTATGACCAGAATT | |
| GFP-F-EcoR I | CCCAATCCGGAATTCCGCAATTAATGTGAGTTAGCTCAC | |
| GFP-R-EcoR I | CCCGGACACGAATTCGATCCGGATATAGTTCCTCCTTTC | |
| GFP-R-Spe I | CCCGGACACACTAGTGATCCGGATATAGTTCCTCCT | |
| Spacer-1 | CCAGGGAATGCAAAATGCAA | |
| Spacer-2 | CCAGCACGGCGGCAAAGTGG | |
| Spacer-3 | CGCACGCGCGGCTTCAACCG | |
| Spacer-4 | TGCTCACTGGCAGGACGGGA | |
| Spacer-5 | CGATAGCGCCAGGAAAACAT | |
| Spacer-6 | TTCAGTACGGGAGGAAAGAT |
Table 1 Strains, Plasmids and Oligomers used in this study
| Name | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F- lacZΔ lacΔ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK+,mk+) phoA supE44 Y-thi-1 gyrA96 relA1 | TransGen Biotech |
| E. coli RP437 | F- thr-1 araC14 leuB6(Am) fhuA31 lacY1 tsx-78λ- eda-50 hisG4(Oc) rfbC1 rpsL136(strR) xylA5 metF159(Am) mtl-1 thiE1 | [ |
| E. coli RP437ΔTar | E. coli RP437 mutant with tar gene knockdown | This study |
| Plasmids | ||
| pUC19 (Bsa I free) | AmpR, pBR322 origin, no Bsa I restriction enzyme site | Tsingke |
| pUC-eGFP | pUC19-derived plasmid carrying eGFP, for cell visualization in Transwell and microfluidics experiment | This study |
| pdCas9 | CmR, backbone plasmid for CRISPRi, carrying dCas9 gene (tetracycline-induced) with sgRNA scaffold (arabinose-induced), and mRFP expression cassette | [ |
| pdCas9-Spacer 1~6 | pdCas9-derived plasmid inserted with Spacer 1~6 for tar gene knockdown | This study |
| Oligomers | Tsingke | |
| Spacer-F1 | TTCAGCCATACTTTTCATACTCCC | |
| Spacer-R1 | GAAGCGGAATATATCCCTAGGTAT | |
| Spacer-extend-F1 | GCAAGGCGATTAAGTTGGGTAA | |
| Spacer-extend-R1 | TGAGTTAGCTCACTCATTAGGCAC | |
| Spacer-extend-F2 | CGTAAGGAGAAAATACCGCATCAG | |
| Spacer-extend-R2 | ACAGGAAACAGCTATGACCAGAATT | |
| GFP-F-EcoR I | CCCAATCCGGAATTCCGCAATTAATGTGAGTTAGCTCAC | |
| GFP-R-EcoR I | CCCGGACACGAATTCGATCCGGATATAGTTCCTCCTTTC | |
| GFP-R-Spe I | CCCGGACACACTAGTGATCCGGATATAGTTCCTCCT | |
| Spacer-1 | CCAGGGAATGCAAAATGCAA | |
| Spacer-2 | CCAGCACGGCGGCAAAGTGG | |
| Spacer-3 | CGCACGCGCGGCTTCAACCG | |
| Spacer-4 | TGCTCACTGGCAGGACGGGA | |
| Spacer-5 | CGATAGCGCCAGGAAAACAT | |
| Spacer-6 | TTCAGTACGGGAGGAAAGAT |
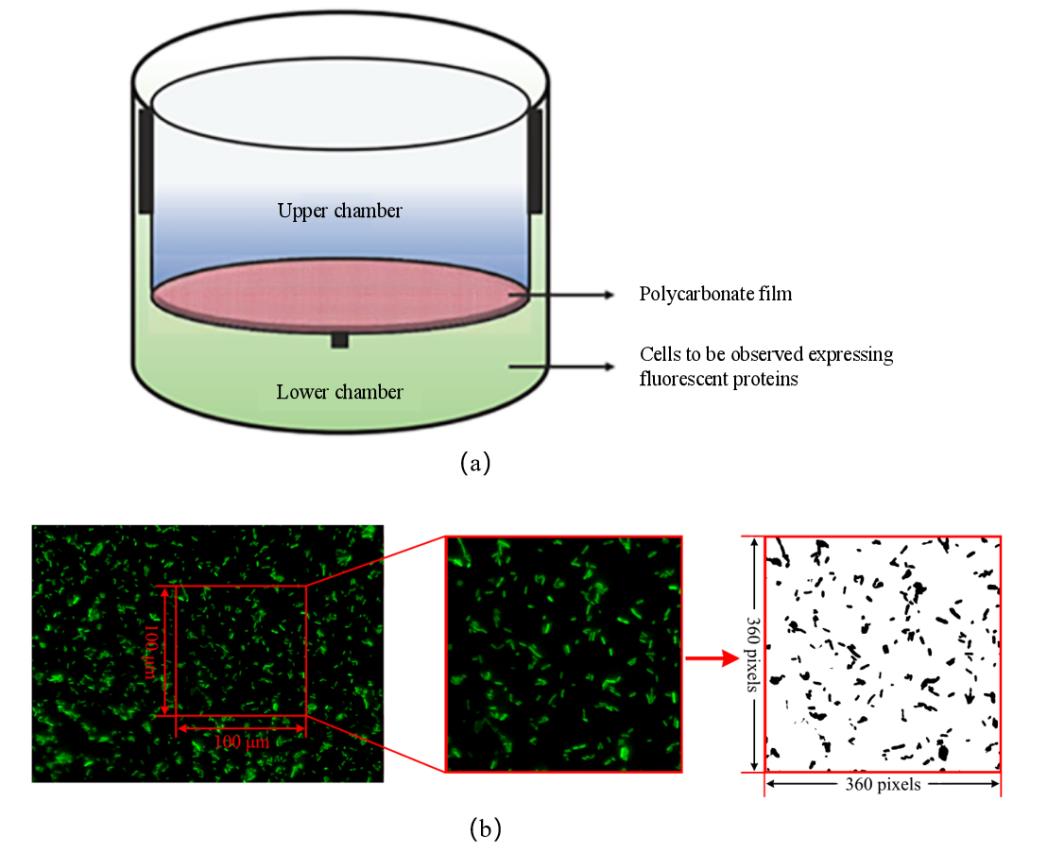
Fig. 2 Transwell migration experiment and image processing procedure(a)The Transwell bacterial migration device [24];(b)Transwell fluorescence image processing procedure
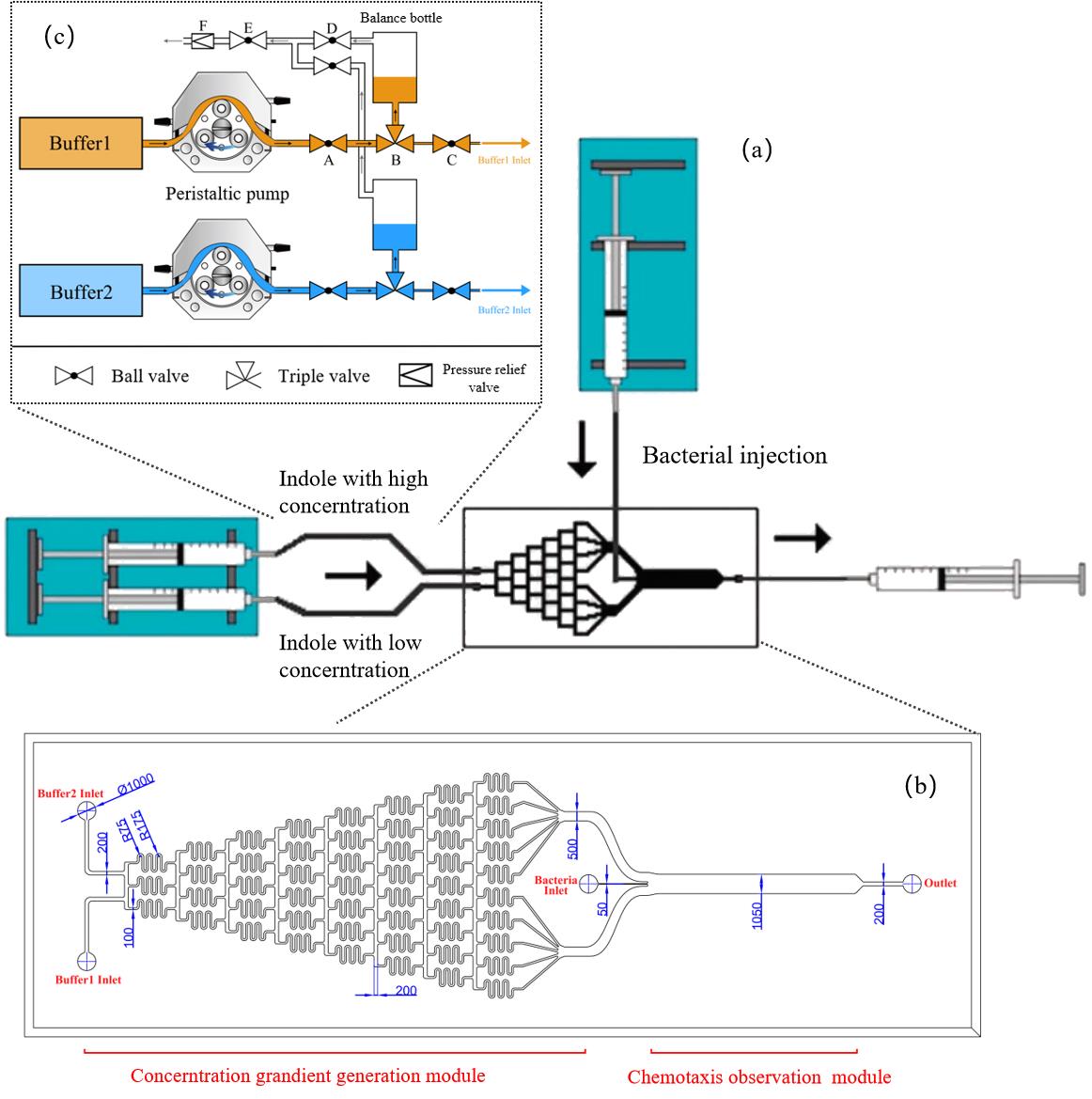
Fig. 3 The design and optimization of microfluidics(a)μFlow design of microfluidics [27];(b)The detail of microfluidics;(c)The injection system of microfluidics
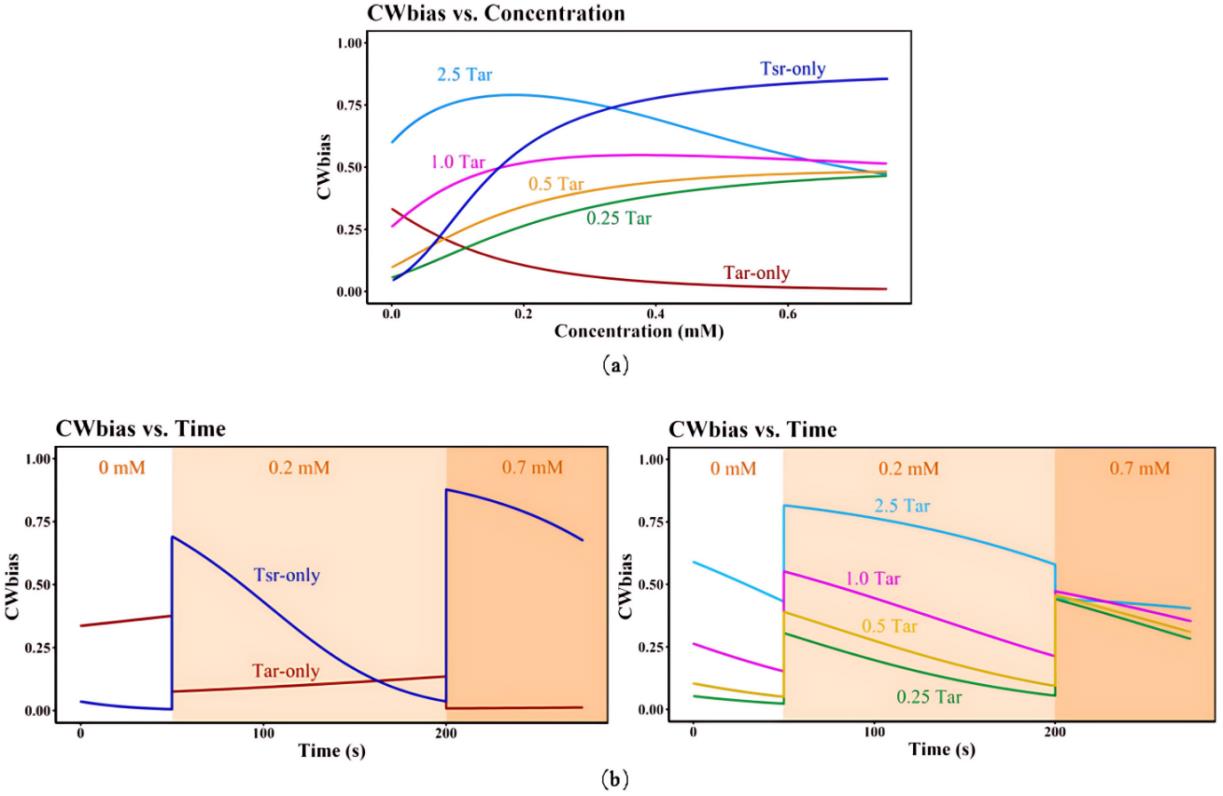
Fig. 4 Simulation results of signal transduction kinetic model(a)CWbias-c relationship at different Tsr/Tar receptor ratios and different indole concentrations;(b)CWbias-t relationship at different Tsr/Tar receptor ratios and different indole concentrations;Tar-only and Tsr-only mean physiological concentration of Tar receptors and Tsr receptors. The number before Tar represent the corresponding increase/decrease fold when the physiological concentration of Tar served as 1Tar.
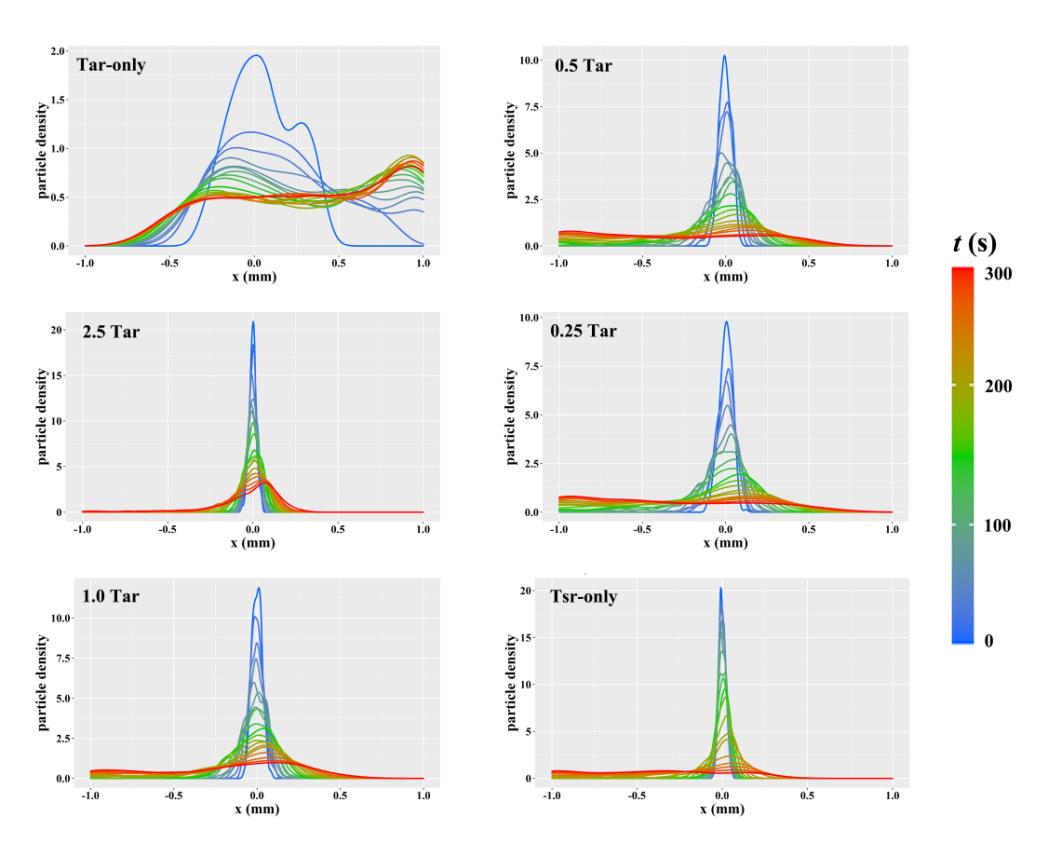
Fig. 5 Density distribution curves of particles in simulated motion of random walk modelTar-only and Tsr-only mean physiological concentration of Tar receptors and Tsr receptors. The number before Tar represent the corresponding increase/decrease fold when the physiological concentration of Tar served as 1Tar.
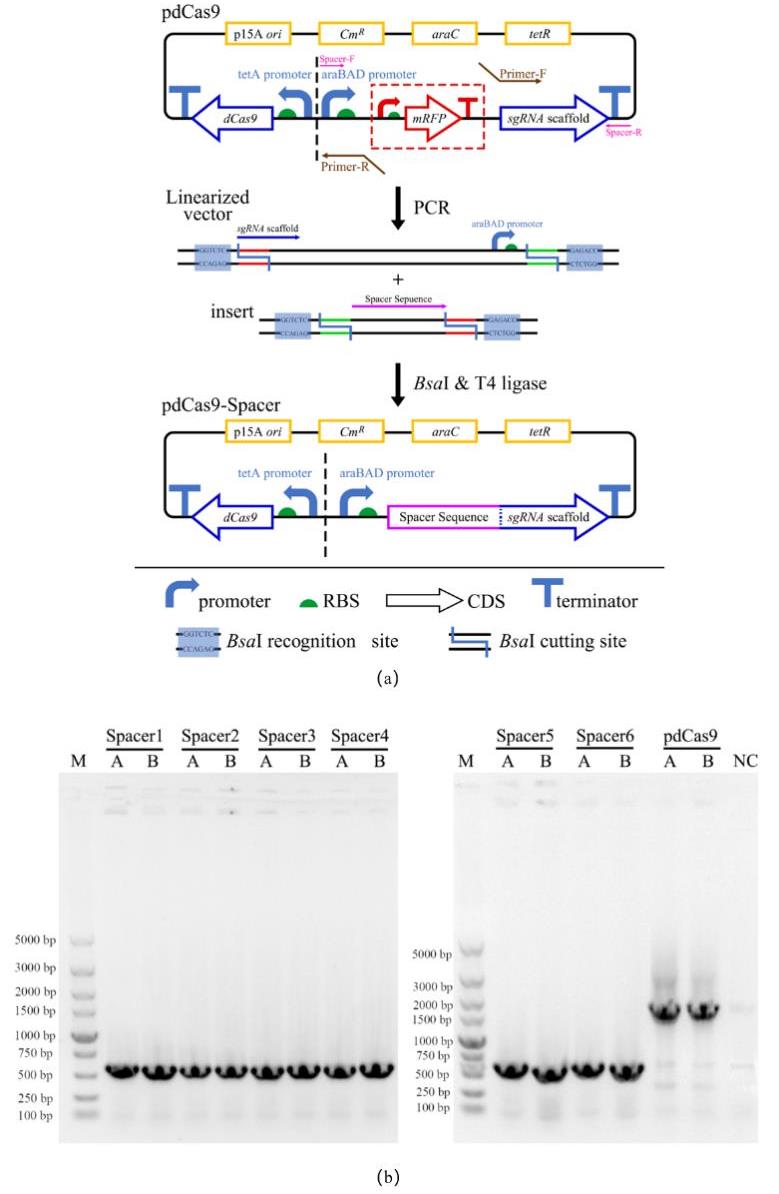
Fig. 6 Design, construction and verification of CRISPRi plasmids(a) Design of CRISPRi plasmid and its construction via Golden Gate of pdCas9 linear vector and spacer sequence;(b)Results of colony PCR after transformation;M:DL5000 DNA Marker;Spacer 1~6:Colony PCR results of recombinant strains with pdCas9-Spacer 1~6 plasmids;A、B:Two parallel single colonies on each screening plate;NC:negative control, that is, E. coli DH5α wild type without any plasmid
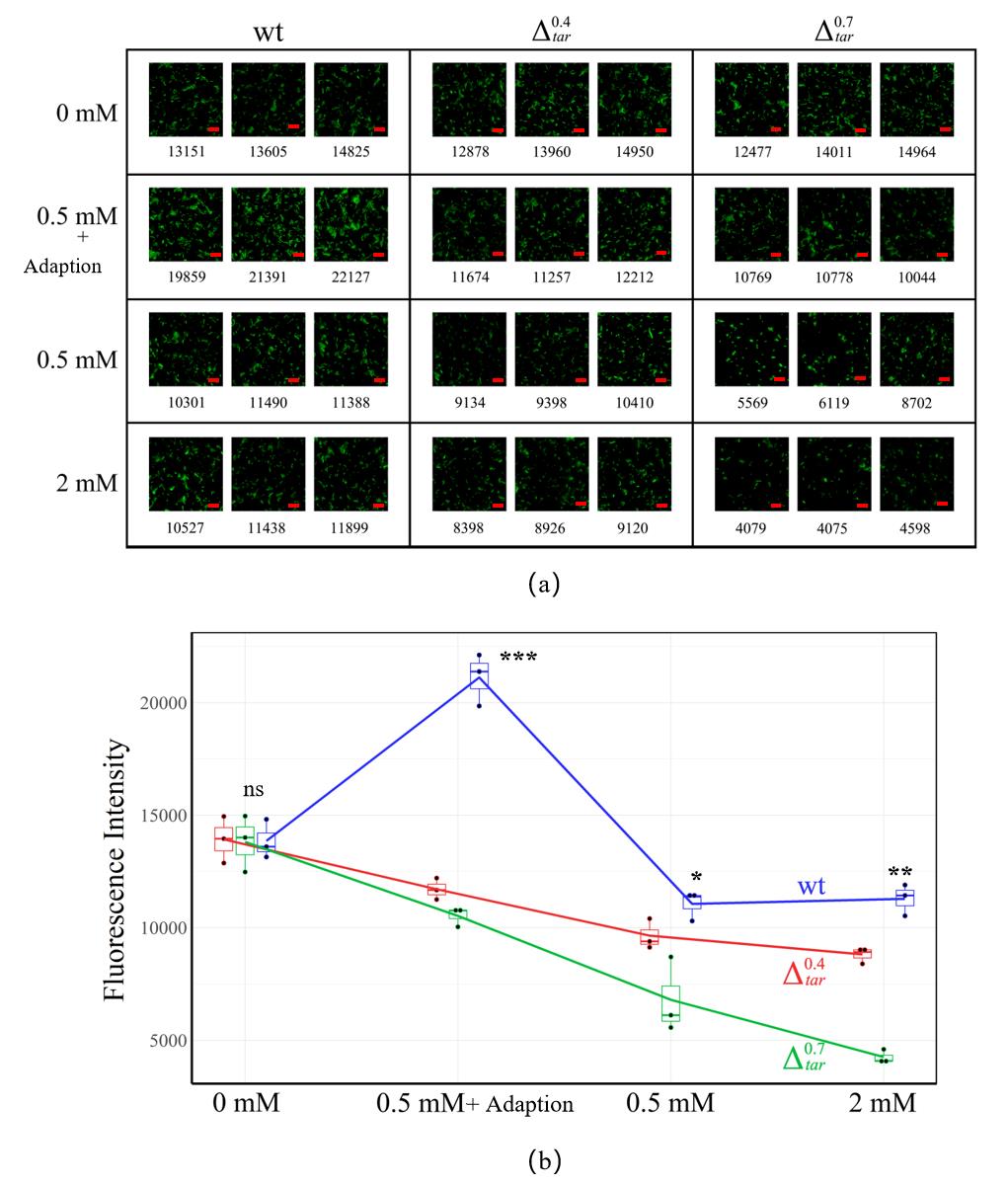
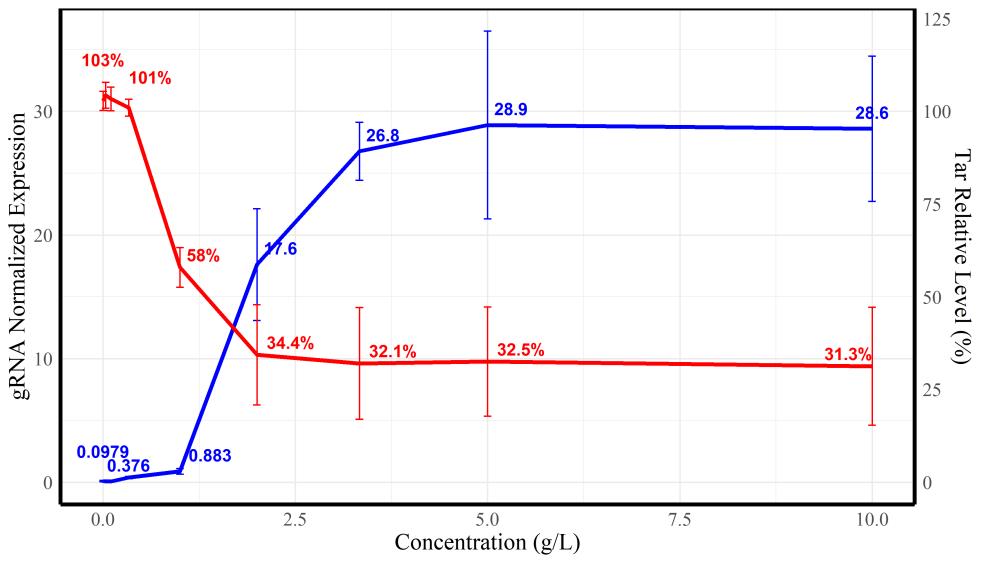
Fig. 8 Results of Transwell migration experiments(a)Transwell imaging results of tar knockdown mutants under different indole concentrations, which is 0 mM、0.5mM、0.5mM+adoption and 2 mM, respectively. Each group contains three parallel view fields with dimension of 100 μm × 100 μm. Number below each image means the fluorescence intensity in the view field after statistical processing. Bar is 2 μm;(b)Quantitative trend of the response of different tar knockdown mutants to indole concentration in Transwell experiments
| 菌株类型 | tar相对转录水平 | tsr相对转录水平 | tsr/tar |
|---|---|---|---|
| E. coli RP437野生型 | 7.80±0.10 | 14.66±0.03 | 1.88±0.06 |
| Tar 受体敲低菌株 | 4.68±0.08 | 14.81±0.02 | 3.16±0.05 |
| Tar 受体敲低菌株 | 2.53±0.02 | 14.31±0.11 | 5.66±0.03 |
Table 2 The relative transcriptional level of tar and tsr in E. coli used in Transwell experiment tested by RT-qPCR
| 菌株类型 | tar相对转录水平 | tsr相对转录水平 | tsr/tar |
|---|---|---|---|
| E. coli RP437野生型 | 7.80±0.10 | 14.66±0.03 | 1.88±0.06 |
| Tar 受体敲低菌株 | 4.68±0.08 | 14.81±0.02 | 3.16±0.05 |
| Tar 受体敲低菌株 | 2.53±0.02 | 14.31±0.11 | 5.66±0.03 |
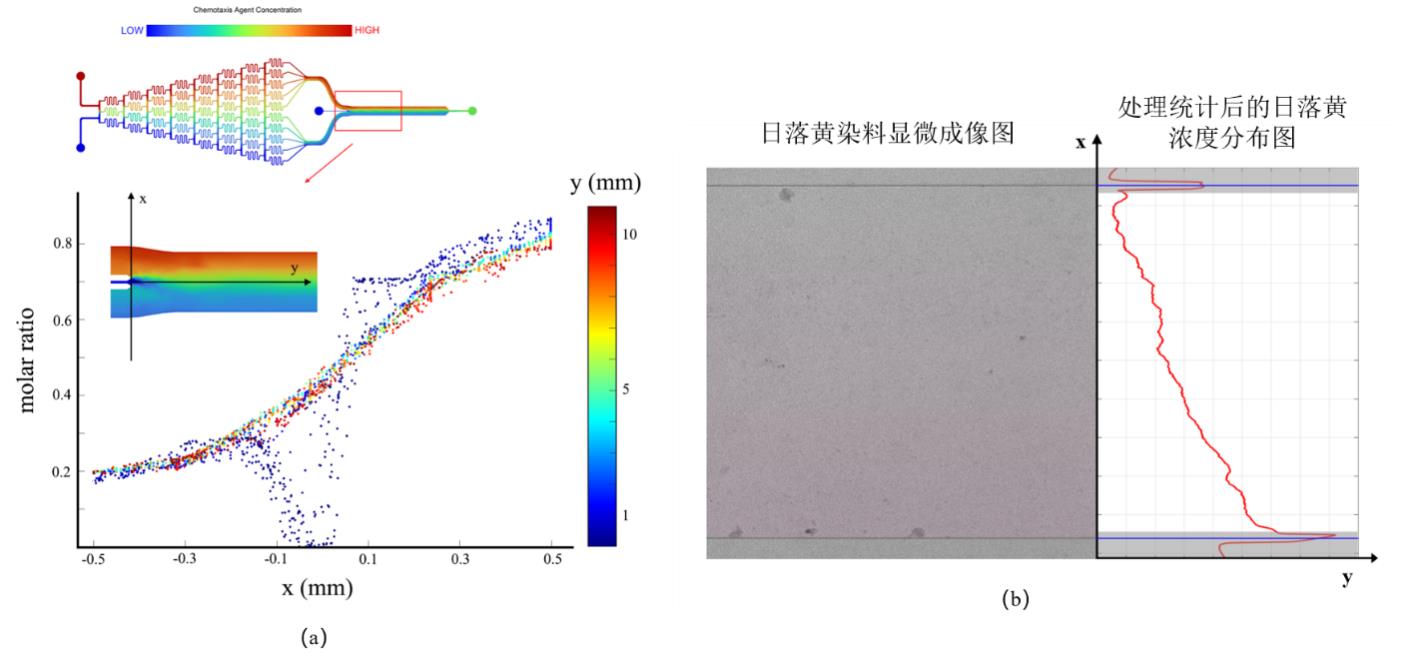
Fig. 9 Simulation and experimental verification results of concentration field in microfluidic devices(a)Simulation results of the distribution of concentration field in all directions; (b) Experimental verification of concentration field distribution in microfluidic device using sunset yellow as a dye
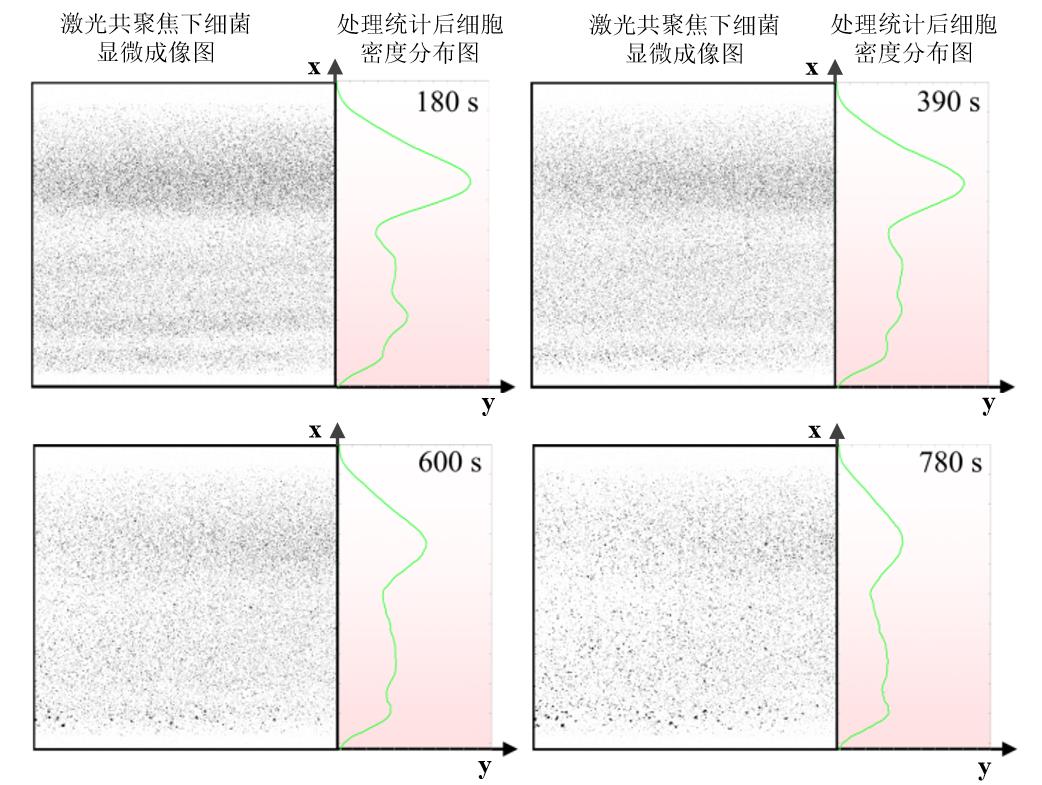
Fig. 10 Density distribution over time of Escherichia coli wild-type in microfluidic channel(The left part of each image represents the bacterial imaging results under confocal microscopy, and the right part represents the bacterial density after statistical analysis. The color of the background in right part indicates the concentration of indole, and the darker color means the higher indole concentration.)
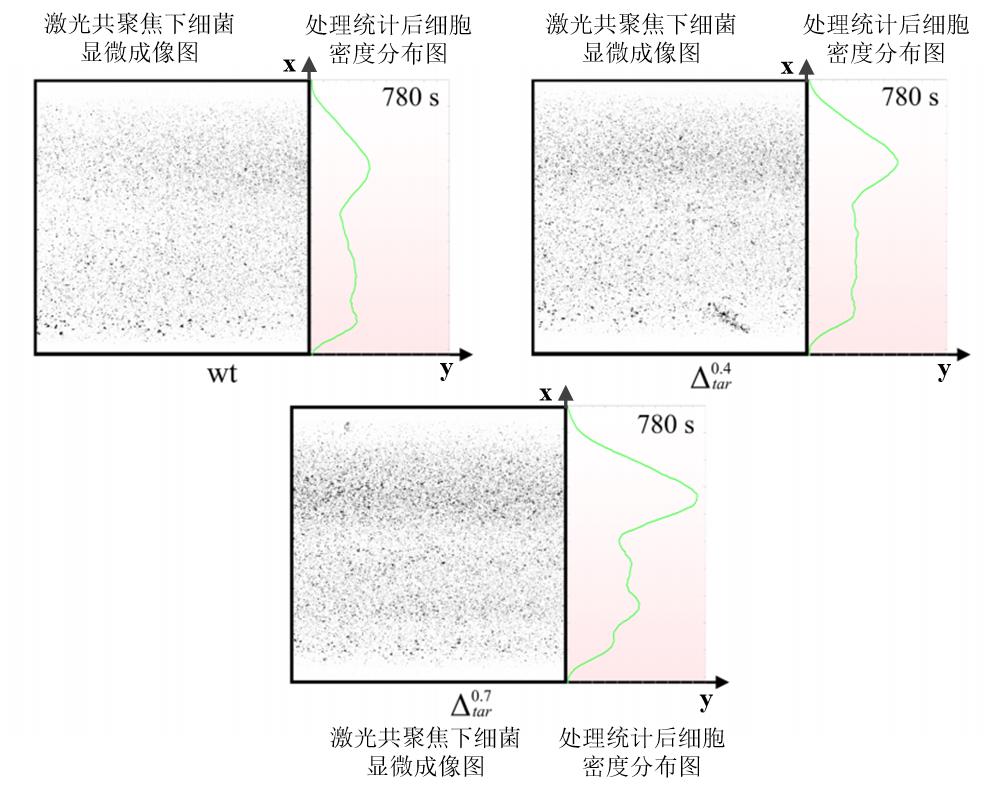
Fig. 11 Density distribution of strains with different Tar receptor expression levels in microfluidic channels at the same timeThe left part of each image represents the bacterial imaging results under confocal microscopy, and the right part represents the bacterial density after statistical analysis. The color of the background in right part indicates the concentration of indole, and the darker color means the higher indole concentration.
| [1] | ADLER J. Chemotaxis in bacteria [J]. Science, 1966, 153(3737): 708-16. |
| [2] | IMAE Y, OOSAWA K, MIZUNO T, et al. Phenol: A complex chemoeffector in bacterial chemotaxis [J]. J Bacteriol, 1987, 169(1): 371-9. |
| [3] | AIHARA E, CLOSSON C, MATTHIS A L, et al. Motility and chemotaxis mediate the preferential colonization of gastric injury sites by helicobacter pylori [J]. PLoS Pathog, 2014, 10(7): e1004275. |
| [4] | RAMOS H C, RUMBO M, SIRARD J C. Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa [J]. Trends Microbiol, 2004, 12(11): 509-17. |
| [5] | WADHAMS G H, ARMITAGE J P. Making sense of it all: Bacterial chemotaxis [J]. Nat Rev Mol Cell Biol, 2004, 5(12): 1024-37. |
| [6] | HOCH J A. Two-component and phosphorelay signal transduction [J]. Curr Opin Microbiol, 2000, 3(2): 165-70. |
| [7] | ADLER J. Chemotaxis in bacteria [J]. Annu Rev Biochem, 1975, 44: 341-56. |
| [8] | MACNAB R M. Examination of bacterial flagellation by dark-field microscopy [J]. J Clin Microbiol, 1976, 4(3): 258-65. |
| [9] | GREBE T W, STOCK J. Bacterial chemotaxis: The five sensors of a bacterium [J]. Curr Biol, 1998, 8(5): R154-7. |
| [10] | ZHULIN I B. The superfamily of chemotaxis transducers: From physiology to genomics and back [J]. Adv Microb Physiol, 2001, 45: 157-98. |
| [11] | NINFA E G, STOCK A, MOWBRAY S, et al. Reconstitution of the bacterial chemotaxis signal transduction system from purified components [J]. J Biol Chem, 1991, 266(15): 9764-70. |
| [12] | BOURRET R B, BORKOVICH K A, SIMON M I. Signal transduction pathways involving protein phosphorylation in prokaryotes [J]. Annu Rev Biochem, 1991, 60: 401-41. |
| [13] | MCEVOY M M, BREN A, EISENBACH M, et al. Identification of the binding interfaces on chey for two of its targets, the phosphatase CheZ and the flagellar switch protein flim [J]. J Mol Biol, 1999, 289(5): 1423-33. |
| [14] | DJORDJEVIC S, STOCK A M. Crystal structure of the chemotaxis receptor methyltransferase cher suggests a conserved structural motif for binding S-adenosylmethionine [J]. Structure, 1997, 5(4): 545-58. |
| [15] | YANG J, CHAWLA R, RHEE K Y, et al. Biphasic chemotaxis of Escherichia coli to the microbiota metabolite indole [J]. Proc Natl Acad Sci U S A, 2020, 117(11): 6114-20. |
| [16] | NORRIS N, ALCOLOMBRI U, KEEGSTRA J M, et al. Bacterial chemotaxis to saccharides is governed by a trade-off between sensing and uptake [J]. Biophys J, 2022, 121(11): 2046-59. |
| [17] | JIANG L, OUYANG Q, TU Y. Quantitative modeling of Escherichia coli chemotactic motion in environments varying in space and time [J]. PLoS Comput Biol, 2010, 6(4): e1000735. |
| [18] | KELLER E F, SEGEL L A. Traveling bands of chemotactic bacteria: A theoretical analysis [J]. J Theor Biol, 1971, 30(2): 235-48. |
| [19] | ZHU B, LI H, ZHANG L, et al. A markov random field model-based approach for differentially expressed gene detection from single-cell RNA-seq data [J]. Brief Bioinform, 2022, 23(5). |
| [20] | PARKINSON J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis [J]. J Bacteriol, 1978, 135(1): 45-53. |
| [21] | BAI Y, HE C, CHU P, et al. Spatial modulation of individual behaviors enables an ordered structure of diverse phenotypes during bacterial group migration [J]. Elife, 2021, 10. |
| [22] | BRADLEY R W. An easy-to-use crispri plasmid tool for inducible knockdown in E. coli [J]. Biotechnol Rep (Amst), 2021, 32: e00680. |
| [23] | ROCHA D, CASTRO T L P, AGUIAR E, et al. Gene expression analysis in bacteria by RT-qPCR [J]. Methods Mol Biol, 2020, 2065: 119-37. |
| [24] | JANI S. Visualizing chemoattraction of planktonic cells to a biofilm [J]. Methods Mol Biol, 2018, 1729: 61-9. |
| [25] | JUSTUS C R, MARIE M A, SANDERLIN E J, et al. Transwell in vitro cell migration and invasion assays [J]. Methods Mol Biol, 2023, 2644: 349-59. |
| [26] | ZANG X Q, LI Z Y, ZHANG X Y, et al. Advance in bacteria chemotaxis on microfluidic devices [J]. Chinese Journal of Analytical Chemistry, 2017, 45(11): 1734-44. |
| [27] | ENGLERT D L, MANSON M D, JAYARAMAN A. Investigation of bacterial chemotaxis in flow-based microfluidic devices [J]. Nat Protoc, 2010, 5(5): 864-72. |
| [28] | HINKAMP P E. A table of fikentscher K values versus relative viscosities for a concentration of 1·0 [J]. Polymer, 1967, 8: 381-4. |
| [29] | ANDERS C, NIEWOEHNER O, DUERST A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease [J]. Nature, 2014, 513(7519): 569-73. |
| [30] | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an rna-guided platform for sequence-specific control of gene expression [J]. Cell, 2021, 184(3): 844. |
| [31] | NISHIMASU H, RAN F A, HSU P D, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA [J]. Cell, 2014, 156(5): 935-49. |
| [32] | DOMINGUEZ A A, LIM W A, QI L S. Beyond editing: Repurposing CRISPR-Cas9 for precision genome regulation and interrogation [J]. Nat Rev Mol Cell Biol, 2016, 17(1): 5-15. |
| [33] | BYUNGJIN LEE H-H J, KANG KYOUNG-KU, LEE CHANG-SOO, LEE SANG-HO. Improvement of a diffusion-based microfluidic chemotaxis assay through stable formation of a chemical gradient [J]. Chemical Engineering Science, 2019, 202: 130-7. |
| [34] | LI JEON N, BASKARAN H, DERTINGER S K, et al. Neutrophil chemotaxis in linear and complex gradients of interleukin-8 formed in a microfabricated device [J]. Nat Biotechnol, 2002, 20(8): 826-30. |
| [35] | DARNTON N C, TURNER L, ROJEVSKY S, et al. On torque and tumbling in swimming escherichia coli [J]. J Bacteriol, 2007, 189(5): 1756-64. |
| [36] | PING L. Cell orientation of swimming bacteria: From theoretical simulation to experimental evaluation [J]. Sci China Life Sci, 2012, 55(3): 202-9. |
| [37] | YANG Y, SOURJIK V. Opposite responses by different chemoreceptors set a tunable preference point in escherichia coli ph taxis [J]. Mol Microbiol, 2012, 86(6): 1482-9. |
| [38] | HU B, TU Y. Precision sensing by two opposing gradient sensors: How does Escherichia coli find its preferred ph level? [J]. Biophys J, 2013, 105(1): 276-85. |
| [39] | KHAN S, SPUDICH J L, MCCRAY J A, et al. Chemotactic signal integration in bacteria [J]. Proc Natl Acad Sci U S A, 1995, 92(21): 9757-61. |
| [40] | SALMAN H, LIBCHABER A. A concentration-dependent switch in the bacterial response to temperature [J]. Nat Cell Biol, 2007, 9(9): 1098-100. |
| [41] | PAULICK A, JAKOVLJEVIC V, ZHANG S, et al. Mechanism of bidirectional thermotaxis in Escherichia coli [J]. Elife, 2017, 6. |
| [42] | LAGANENKA L, LEE J W, MALFERTHEINER L, et al. Chemotaxis and autoinducer-2 signalling mediate colonization and contribute to co-existence of Escherichia coli strains in the murine gut [J]. Nat Microbiol, 2023, 8(2): 204-17. |
| [43] | SONG J, ZHANG Y, ZHANG C, et al. A microfluidic device for studying chemotaxis mechanism of bacterial cancer targeting [J]. Sci Rep, 2018, 8(1): 6394. |
| [1] | WANG Cuizhen, CHEN Tiao, WANG Jianbo. Enzyme-catalyzed Hetero-Diels-Alder reactions [J]. Synthetic Biology Journal, 2024, 5(1): 107-125. |
| [2] | GUO Xiaojie, JIAN Xingjin, WANG Liyan, ZHANG Chong, XING Xinhui. Progress in bioreactors and instruments for phenotype testing with synthetic biology research [J]. Synthetic Biology Journal, 2024, 5(1): 16-37. |
| [3] | QIN Weitong, YANG Guangyu. Research and application progress of microdroplets high throughput screening methods [J]. Synthetic Biology Journal, 2023, 4(5): 966-979. |
| [4] | TU Ran, LI Shixin, LI Haoni, WANG Meng. Advances and applications of droplet-based microfluidics in evolution and screening of engineered microbial strains [J]. Synthetic Biology Journal, 2023, 4(1): 165-184. |
| [5] | GUO Wei, FU Yuhao, FAN Yingying, ZHOU Jialing, LI Xin, WEI Ping. Artificial control of mammalian cell chemotaxis and motility [J]. Synthetic Biology Journal, 2022, 3(6): 1109-1125. |
| [6] | ZHAO Xiaoyu, ZHANG Hao, LI Xuefei, HU Zheng. An evolutionary perspective on quantitative biological principles and synthetic life design [J]. Synthetic Biology Journal, 2022, 3(1): 6-21. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||