Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (6): 1262-1276.DOI: 10.12211/2096-8280.2022-016
• Invited Review • Previous Articles Next Articles
Identification of RiPPs precursor peptides and cleavage sites based on deep learning
LYU Jingwei1, DENG Zixin1, ZHANG Qi2, DING Wei1
- 1.State Key Laboratory of Microbial Metabolism,School of Life Sciences and Biotechnology,Shanghai Jiao Tong University,Shanghai,200240,China
2.Department of Chemistry,Fudan University,shanghai 200243,China
-
Received:2022-03-07Revised:2022-04-23Online:2023-01-17Published:2022-12-31 -
Contact:DING Wei
基于深度学习识别RiPPs前体肽及裂解位点
吕靖伟1, 邓子新1, 张琪2, 丁伟1
- 1.上海交通大学生命科学技术学院,微生物代谢国家重点实验室,上海 200030
2.复旦大学化学系,上海 200243
-
通讯作者:丁伟 -
作者简介:吕靖伟 (1996—),硕士研究生。研究方向为天然产物合成基因挖掘。E-mail:jingwei_lv@sjtu.edu.cn丁伟 (1981—),男,博士,副教授,博士生导师。研究方向为微生物代谢及合成生物学。E-mail:weiding@sjtu.edu.cn -
基金资助:国家重点研发计划(2018Y F A0900402)
CLC Number:
Cite this article
LYU Jingwei, DENG Zixin, ZHANG Qi, DING Wei. Identification of RiPPs precursor peptides and cleavage sites based on deep learning[J]. Synthetic Biology Journal, 2022, 3(6): 1262-1276.
吕靖伟, 邓子新, 张琪, 丁伟. 基于深度学习识别RiPPs前体肽及裂解位点[J]. 合成生物学, 2022, 3(6): 1262-1276.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-016

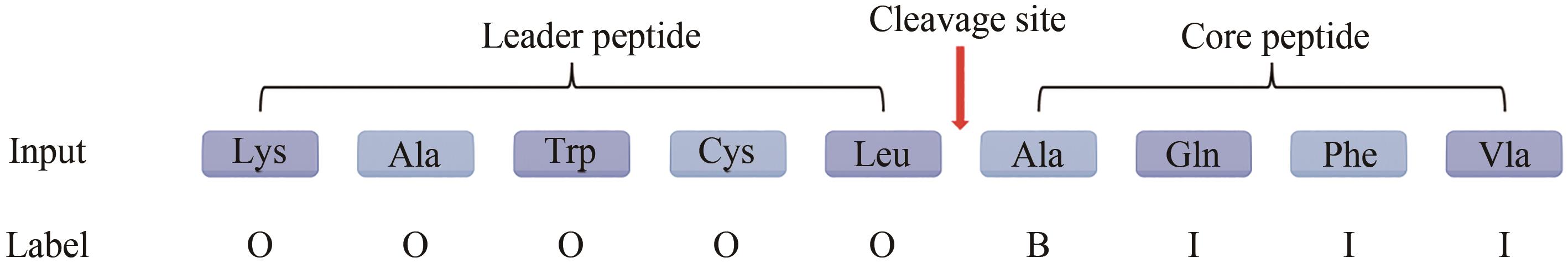
Fig. 1 Biosynthetic pathways of RiPPs(The proportion of each part of the sequence in the figure is for illustration only and does not represent its actual length)
| Model | Precision | Recall | F1 |
|---|---|---|---|
BERT (top 1 layers initialized) | 0.9056 | 0.8962 | 0.9009 |
BERT (top 2 layers initialized) | 0.8938 | 0.9031 | 0.8985 |
BERT (fully initialized) | 0.8710 | 0.8408 | 0.8556 |
BERT (pre-trained) | 0.9031 | 0.9031 | 0.9031 |
| BERT-CNN | 0.9123 | 0.8997 | 0.9059 |
| BERT-DPCNN | 0.9126 | 0.9031 | 0.9078 |
| BERT-RCNN | 0.9127 | 0.8685 | 0.8901 |
BERiPPs (BERT-BiLSTM) | 0.9331 | 0.9170 | 0.9250 |
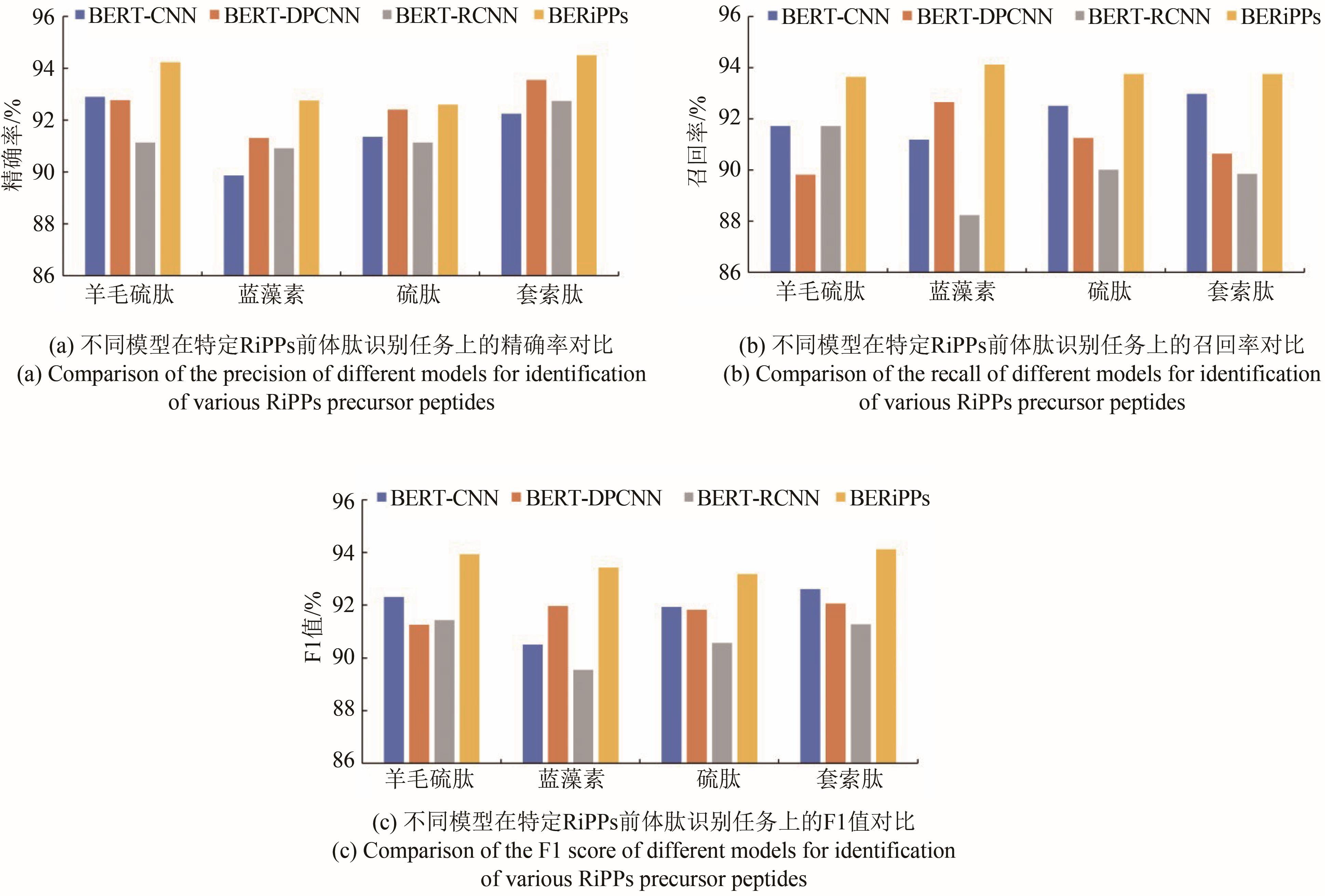
Tab. 1 Comparison of different algorithms in identification of RiPPs precursor peptides
| Model | Precision | Recall | F1 |
|---|---|---|---|
BERT (top 1 layers initialized) | 0.9056 | 0.8962 | 0.9009 |
BERT (top 2 layers initialized) | 0.8938 | 0.9031 | 0.8985 |
BERT (fully initialized) | 0.8710 | 0.8408 | 0.8556 |
BERT (pre-trained) | 0.9031 | 0.9031 | 0.9031 |
| BERT-CNN | 0.9123 | 0.8997 | 0.9059 |
| BERT-DPCNN | 0.9126 | 0.9031 | 0.9078 |
| BERT-RCNN | 0.9127 | 0.8685 | 0.8901 |
BERiPPs (BERT-BiLSTM) | 0.9331 | 0.9170 | 0.9250 |

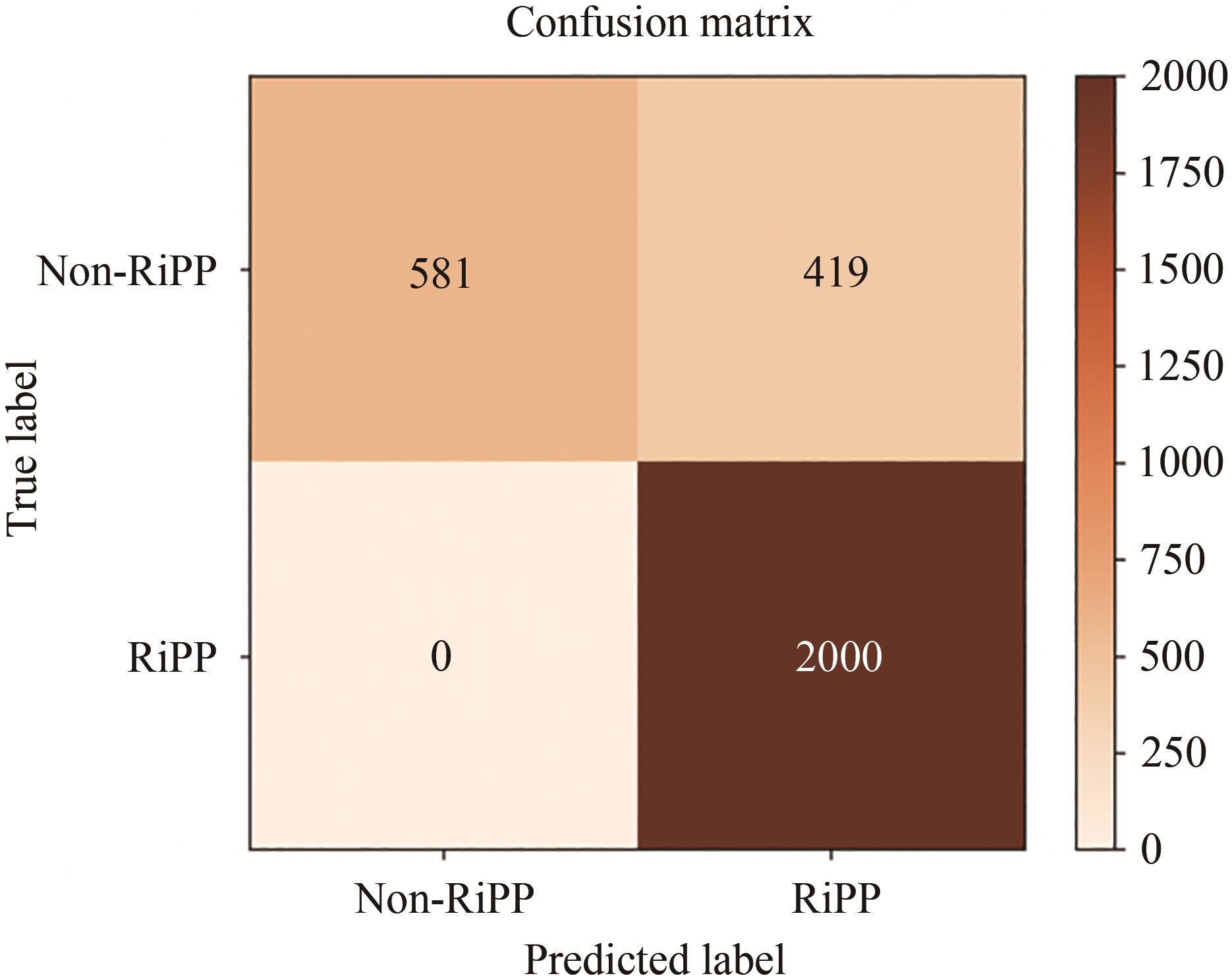
Fig. 7 Comparison of prediction results of RiPPs classes between BERiPPs under different training methods and DeepRiPP(Due to the different number of various RiPPs samples in the test set, the values in the confusion matrix are normalized and then accurate to one decimal point according to the rounding principle.)
| Class | Loss Function | Precision | Recall | F1 |
|---|---|---|---|---|
| Autoinducing peptides | Focal Loss | 1.0000 | 0.9583 | 0.9787 |
| Cross Entropy Loss | 0.9231 | 1.0000 | 0.9600 | |
| Thiopeptides | Focal Loss | 0.8947 | 0.8947 | 0.8947 |
| Cross Entropy Loss | 0.8500 | 0.8947 | 0.8718 | |
| Lasso peptides | Focal Loss | 0.8873 | 0.9130 | 0.9000 |
| Cross Entropy Loss | 0.8889 | 0.9275 | 0.9078 | |
| Class Ⅲ_Ⅳ Lanthipeptides | Focal Loss | 0.9483 | 0.9649 | 0.9565 |
| Cross Entropy Loss | 0.9655 | 0.9825 | 0.9739 |
Tab. 2 Comparison of prediction results of partial RiPPs classes under different loss functions
| Class | Loss Function | Precision | Recall | F1 |
|---|---|---|---|---|
| Autoinducing peptides | Focal Loss | 1.0000 | 0.9583 | 0.9787 |
| Cross Entropy Loss | 0.9231 | 1.0000 | 0.9600 | |
| Thiopeptides | Focal Loss | 0.8947 | 0.8947 | 0.8947 |
| Cross Entropy Loss | 0.8500 | 0.8947 | 0.8718 | |
| Lasso peptides | Focal Loss | 0.8873 | 0.9130 | 0.9000 |
| Cross Entropy Loss | 0.8889 | 0.9275 | 0.9078 | |
| Class Ⅲ_Ⅳ Lanthipeptides | Focal Loss | 0.9483 | 0.9649 | 0.9565 |
| Cross Entropy Loss | 0.9655 | 0.9825 | 0.9739 |
| 1 | MARTENS E, DEMAIN A L. The antibiotic resistance crisis, with a focus on the United States[J]. The Journal of Antibiotics, 2017, 70(5): 520-526. |
| 2 | HUTCHINGS M I, TRUMAN A W, WILKINSON B. Antibiotics: past, present and future[J]. Current Opinion in Microbiology, 2019, 51: 72-80. |
| 3 | HUDSON G A, MITCHELL D A. RiPP antibiotics: Biosynthesis and engineering potential[J]. Current Opinion in Microbiology, 2018, 45: 61-69. |
| 4 | WANG F T, WEI W Q, ZHAO J F, et al. Genome mining and biosynthesis study of a type B linaridin reveals a highly versatile α-N-methyltransferase[J]. CCS Chemistry, 2021, 3(3): 1049-1057. |
| 5 | SKINNIDER M A, JOHNSTON C W, EDGAR R E, et al. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(42): E6343-E6351. |
| 6 | ARNISON P G, BIBB M J, BIERBAUM G, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature[J]. Natural Product Reports, 2013, 30(1): 108-160. |
| 7 | YU Y, ZHANG Q, VAN DER DONK W A. Insights into the evolution of lanthipeptide biosynthesis[J]. Protein Science, 2013, 22(11): 1478-1489. |
| 8 | ZHONG Z, HE B B, LI J, et al. Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs)[J]. Synthetic and Systems Biotechnology, 2020, 5(3): 155-172. |
| 9 | BLIN K, SHAW S, STEINKE K, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline[J]. Nucleic Acids Research, 2019, 47(W1): W81-W87. |
| 10 | HETRICK K J, VAN DER DONK W A. Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era[J]. Current Opinion in Chemical Biology, 2017, 38: 36-44. |
| 11 | HYATT D, CHEN G L, LOCASCIO P F, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification[J]. BMC Bioinformatics, 2010, 11: 119. |
| 12 | DELCHER A L, BRATKE K A, POWERS E C, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer[J]. Bioinformatics, 2007, 23(6): 673-679. |
| 13 | VAN HEEL A J, DE JONG A, MONTALBÁN-LÓPEZ M, et al. BAGEL3: automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides[J]. Nucleic Acids Research, 2013, 41(W1): W448-W453. |
| 14 | TIETZ J I, SCHWALEN C J, PATEL P S, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape[J]. Nature Chemical Biology, 2017, 13(5): 470-478. |
| 15 | MERWIN N J, MOUSA W K, DEJONG C A, et al. DeepRiPP integrates multiomics data to automate discovery of novel ribosomally synthesized natural products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(1): 371-380. |
| 16 | AGRAWAL P, KHATER S, GUPTA M, et al. RiPPMiner: a bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links[J]. Nucleic Acids Research, 2017, 45(W1): W80-W88. |
| 17 | DE LOS SANTOS E L C. NeuRiPP: Neural network identification of RiPP precursor peptides[J]. Scientific Reports, 2019, 9: 13406. |
| 18 | SHIN H C, ROTH H R, GAO M C, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning[J]. IEEE Transactions on Medical Imaging, 2016, 35(5): 1285-1298. |
| 19 | SUNDERMEYER M, SCHLÜTER R, NEY H. LSTM neural networks for language modeling[C]// 13th Annual conference of the International Speech Communication Association 2012 (INTERSPEECH 2012). Portland, OR, USA: International Speech Communications Association, 2012:194-197. |
| 20 | DEVLIN J, CHANG M W, LEE K, et al. BERT: pre-training of Deep Bidirectional Transformers for Language Understanding[C]//Proceedings of NAACL-HLT. 2019: 4171-4186. |
| 21 | TENNEY I, DAS D, PAVLICK E. BERT rediscovers the classical NLP pipeline[C]//Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Florence, Italy. Stroudsburg, PA, USA: Association for Computational Linguistics, 2019: 4593-4601. |
| 22 | CHO K, VAN MERRIENBOER B, GULCEHRE C, et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation[C]//Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Doha, Qatar. Stroudsburg, PA, USA: Association for Computational Linguistics, 2014: 1724-1734. |
| 23 | Bahdanau D, Cho K H, Bengio Y. Neural machine translation by jointly learning to align and translate[C]//3rd International Conference on Learning Representations, ICLR 2015. 2015. |
| 24 | Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need[C]//Advances in neural information processing systems. 2017: 5998-6008. |
| 25 | SHERSTINSKY A. Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network[J]. Physica D: Nonlinear Phenomena, 2020, 404: 132306. |
| 26 | WANG Q, LI B, XIAO T, et al. Learning deep transformer models for machine translation[C]//Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Florence, Italy. Stroudsburg, PA, USA: Association for Computational Linguistics, 2019: 1810-1822. |
| 27 | SÖDING J. Protein homology detection by HMM-HMM comparison[J]. Bioinformatics, 2005, 21(7): 951-960. |
| 28 | LIU L Y, REN X, SHANG J B, et al. Efficient contextualized representation: Language model pruning for sequence labeling[C]//Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. Brussels, Belgium. Stroudsburg, PA, USA: Association for Computational Linguistics, 2018: 1215-1225. |
| 29 | Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging[EB/OL]. arXiv preprint: 2015, arXiv:1508.01991. |
| 30 | ZHAO H S, JIA J Y, KOLTUN V. Exploring self-attention for image recognition[C]//2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Seattle: IEEE, 2020, 10073-10082. |
| 31 | Dodge J, Ilharco G, Schwartz R, et al. Fine-tuning pretrained language models: Weight initializations, data orders, and early stopping[EB/OL]. arXiv preprint: 2020, arXiv:2002.06305. |
| 32 | YADAV S, SHUKLA S. Analysis of k-fold cross-validation over hold-out validation on colossal datasets for quality classification[C]//2016 IEEE 6th International Conference on Advanced Computing. Bhimavaram, India: IEEE, 2016: 78-83. |
| 33 | ZHANG Z L, SABUNCU M. Generalized cross entropy loss for training deep neural networks with noisy labels[J]. Montréal: NeurIPS, 2018, 31. |
| 34 | LIN T Y, GOYAL P, GIRSHICK R, et al. Focal loss for dense object detection[C]//2017 IEEE International Conference on Computer Vision. Venice: IEEE, 2017: 2999-3007. |
| 35 | LI B, SHER D, KELLY L, et al. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(23): 10430-10435. |
| 36 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 37 | XIE Q Z, LUONG M T, HOVY E, et al. Self-training with noisy student improves ImageNet classification[C]//2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Seattle: IEEE,2020 : 10684-10695. |
| [1] | XIE Xiangqian, GUO Wen, WANG Huan, LI Jin. Biosynthesis and chemical synthesis of ribosomally synthesized and post-translationally modified peptides containing aminovinyl cysteine [J]. Synthetic Biology Journal, 2024, 5(5): 981-996. |
| [2] | ZHU Jingyong, LI Junxiang, LI Xuhui, ZHANG Jin, WU Wenjing. Advances in applications of deep learning for predicting sequence-based protein interactions [J]. Synthetic Biology Journal, 2024, 5(1): 88-106. |
| [3] | CHEN Zhihang, JI Menglin, QI Yifei. Research progress of artificial intelligence in desiging protein structures [J]. Synthetic Biology Journal, 2023, 4(3): 464-487. |
| [4] | SONG Yidong, YUAN Qianmu, YANG Yuedong. Application of deep learning in protein function prediction [J]. Synthetic Biology Journal, 2023, 4(3): 488-506. |
| [5] | Yuanjun HAN, Tianlu MO, Zixin DENG, Qi ZHANG, Wei DING. Study on the post-translational modification of RiPPs Xye catalyzed by CyFE PacB [J]. Synthetic Biology Journal, 2021, 2(5): 826-836. |
| [6] | Ye WANG, Haochen WANG, Minghao YAN, Guanhua HU, Xiaowo WANG. Design of biomolecular sequences by artificial intelligence [J]. Synthetic Biology Journal, 2021, 2(1): 1-14. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||